Liquid biopsy in clinical outcomes and detection of T790M mutation in metastatic non-small cell lung cancer after progression to EGFR-TKI
Abstract
BACKGROUND:
Liquid biopsy (LB) is used to detect epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) and has been demonstrated to have prognostic and predictive value.
OBJECTIVE:
To associate the rates of EGFR and T790M mutations detected by LB during disease progression after first- or second-generation EGFR-TKIs with clinical characteristics and survival outcomes.
METHODS:
From January 2018 to December 2021, 295 patients with advanced EGFR mutant (EGFRm) NSCLC treated with first- or second-generation EGFR-TKIs were retrospectively analyzed. LB was collected at the time of progression. The frequency of EGFR
RESULTS:
The prevalence of EGFR
CONCLUSIONS:
Our findings showed that LB positivity was associated with worse survival outcomes and specific clinical characteristics. This study also confirmed the feasibility and detection rate of T790M mutation in a Latin American population.
1.Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer death worldwide [1, 2]. The majority of NSCLC patients are diagnosed with advanced-stage disease [3, 4]. Patients with NSCLC are treated based on a personalized approach, according to genomic profile, to ensure better oncological outcomes [5]. The detection of epidermal growth factor receptor (EGFR) mutations produces a paradigm shift in the treatment of NSCLC, which varies according to the geographic region in Latin America, and. oscillates between 32–54%. In Mexico, EGFR mutations represent around 36% of druggable alterations, which is higher compared with North America and Europe, ranking 10–15% [6, 7].
NSCLC patients harboring EGFR mutations have improved oncological outcomes with EGFR-tyrosine kinase inhibitor (TKI) therapy [8, 9, 10, 11]. According to the results of the FLAURA trial, osimertinib is currently the preferred upfront treatment [8]. However, in many other countries, the upfront therapy for patients with EGFRm NSCLC still depends on first- and second-generation TKIs followed by osimertinib in patients who developed a positive T790M mutation at disease progression. Among the common mechanism of acquired resistance after this treatment is the acquisition of EGFR T790M mutation in approximately 50–60% of cases [12, 13, 14].
EGFR T790M resistance mutation can be detected in tumor tissue (surgical biopsy or cytology specimens) or cell-free circulating tumor DNA (ctDNA) extracted from peripheral blood through liquid biopsy (LB) [15]. New tumor tissue biopsy is the most reliable procedure for detecting the T790M mutation at progression. However, this is not always feasible and might be associated with complications due to the invasiveness of tumor biopsy. Other factors, such as tumor heterogeneity and unwillingness, make this method challenging for many patients [16, 17, 18].
LB refers to any tumor-derived material circulating through the blood or other body fluid. In lung cancer, circulating tumor cells and ctDNA are the most widely studied substrates [19]. LB has been shown to represent an innovative alternative for patients unable to undergo re-biopsy as a non-invasive method that allows the detection of EGFR T790M [20]. Other advantages over tissue samples include the reduced cost, rapid turnaround time, and potential for longitudinal monitoring by serial biopsies, showing a high concordance with standard tissue genotyping [20, 21]. In addition, some studies have demonstrated that detection of EGFRm ctDNA is associated with worse outcomes and disease burden, which could be predictive of response, increasing ctDNA levels which may anticipate progression to standard imaging progression by RECIST [22, 23].
Scarce information is available on the utility of LB to detect T790M in the LATAM population in a real-world context and its association with clinical outcomes. Thus, this study aimed to determine the rate of T790M mutation as a mechanism of resistance to first or second-generation EGFR-TKI according to plasma ctDNA detected on LB, associate positiveness to clinical characteristics, and evaluate outcomes in those who received osimertinib.
2.Materials and methods
2.1Ethics approval
This study was approved by the ethics committee of participating medical institution and was authorized under the number Rev/032/20.
2.2Experimental subjects
In this retrospective cohort, we evaluated 295 patients with Stage IV NSCLC treated in one institution between January 2018–December 2021. Patients harboring a sensitive EGFR mutation (delexon19 or L858R) and disease progression in the first-line setting with one first or second-generation EGFR-TKI were included. Patients included had 1) confirmed diagnosis of NSCLC, 2) EGFR mutation, 3) disease progression according to RECIST 1.1 criteria, 4) Treatment with first or second-generation EGFR-TKI. 5) All patients required standard laboratory workups before starting therapy and during treatment according to local policies to monitor adverse events.
All patients performed an LB at radiological progression with the Biocept Target Selector TM ctDNA platform [24]. Each blood samples consist of 20 ml of peripheral blood collected in the CEE-Sure TM tubes (Biocept, San Diego, California, USA). EGFR mutation assay is a quantitative Real-Time PCR-based mutant enrichment assay with selective blocking of EGFR wild-type amplification. The amplification products are purified and subject to sequencing to confirm the presence of the EGFR mutation.
Two expert medical oncologists extracted all clinical and pathological data from electronic medical records. Collected data included the patient’s clinical characteristics at baseline and disease progression, and date, results, and number of LBs before a positive result.
Table 1
Baseline characteristics
| Characteristics |
| % |
| Sex, female | 196 | 66.4 |
| Age, years | ||
| Mean | 62.5 | |
| Smoking status | ||
| Ever smokers | 61 | 20.6 |
| Nonsmokers | 243 | 79.4 |
| Tobacco index, pack/yrs. | ||
| Mean | 10.42 | |
| Body Mass Index (BMI) | ||
| Mean | 24.24 | |
| Comorbidities | ||
| Hypertension | 85 | 28.0 |
| Diabetes Mellitus Type 2 | 44 | 15.0 |
| Obesity | 19 | 15.5 |
| Chronic Obstructive pulmonary disease | 6 | 2.0 |
| Heart failure | 4 | 1.3 |
| ECOG PS scale | ||
| 0–1 | 271 | 92.1 |
| | 24 | 7.9 |
| Adenocarcinoma | 285 | 96.1 |
| Squamous or mixed histology | 10 | 3.9 |
| LADC subtype | ||
| Lepidic | 25 | 8.5 |
| Acinar | 84 | 28.5 |
| Papillary | 24 | 8.1 |
| Solid | 76 | 25.8 |
| Micropapillary | 6 | 2.0 |
| Not otherwise specified | 80 | 27.1 |
| Histological grade | ||
| Well-differentiated | 16 | 5.4 |
| Moderately differentiated | 116 | 39.3 |
| Poor differentiated | 113 | 38.3 |
| Not otherwise specified | 50 | 16.9 |
| Clinical stage IV | 295 | 100 |
| First line treatment | ||
| 1st generation TKI (Erlotinib, Gefitinib) | 164 | 55.6 |
| 2d generation TKI (Afatinib) | 131 | 44.4 |
| EGFR mutations at diagnosis | ||
| Exon 19 Del | 197 | 66.0 |
| L858R Exon 21 | 89 | 30.1 |
| Other mutations | 9 | 3.9 |
Notes. ECOG: Eastern Cooperative Oncology Group performance status scale. LADC: lung adenocarcinoma. EGFR: epidermal growth factor receptor.
2.3Statistical analysis
Categorical variables were summarized as frequencies and percentages, while continuous variables were reported as mean or median with their corresponding dispersion measures. Progression-free survival (PFS), intracranial progression free-survival (icPFS), and Overall survival (OS) were estimated by the Kaplan-Meier method, and statistical differences among groups of interest were calculated with the Log-Rank test method. PFS was defined from the beginning of the second line treatment until disease progression according to RECIST Criteria or death; icPFS was defined from diagnosis to the date of appearance of new brain metastasis. OS was defined as the period from diagnosis to death or loss of follow-up. A Cox proportional hazard model was performed for the multivariate analysis. A significant
3.Results
3.1Clinical characteristics
Two hundred ninety-five patients were analyzed. Of the total, 66.4% were women, 79.4% had never smoked, 92.1% had an ECOG performance status (PS) of 0 to 1, and 96.61% were adenocarcinomas. Regarding mutations at the time of diagnosis, 66% had exon 19 deletions (exon19del), and 30.1% had a point mutation of exon 21 L858R. The most frequent metastatic sites at diagnosis were pleura (50%), followed by the central nervous system (47.6%) and bones (38.1%). All patient demographics and clinical characteristics are summarized in Table 1.
3.2Clinical characteristics at disease progression
At the time of progression, 69.8% had extra-thoracic progression, whereas 30.2% had exclusively an intrathoracic affection (Table 2). All patients had an initial LB at the first disease progression, 23 underwent a second LB at the next progression, and one underwent a third LB after a third progression.
Table 2
Clinical characteristics at progression
| Characteristics |
| % |
|---|---|---|
| Metastatic sites at progression | ||
| CNS | 30 | 47.6 |
| Bone | 28 | 38.1 |
| Pleura | 32 | 50.8 |
| Adrenal glands | 12 | 19 |
| Liver | 13 | 20.6 |
| Type of progression | ||
| Intrathoracic | 89 | 30.2 |
| Extra-thoracic | 206 | 69.8 |
| Second-line therapy | ||
| Carboplatin Pemetrexed | 139 | 47.1 |
| Other systemic treatment | 30 | 10.2 |
| Other TKI | 51 | 17.3 |
| Osimertinib | 22 | 7.5 |
| TKI | 19 | 6.4 |
| None | 26 | 8.8 |
Notes. CNS: central nervous system. TKI: tyrosine kinase inhibitor.
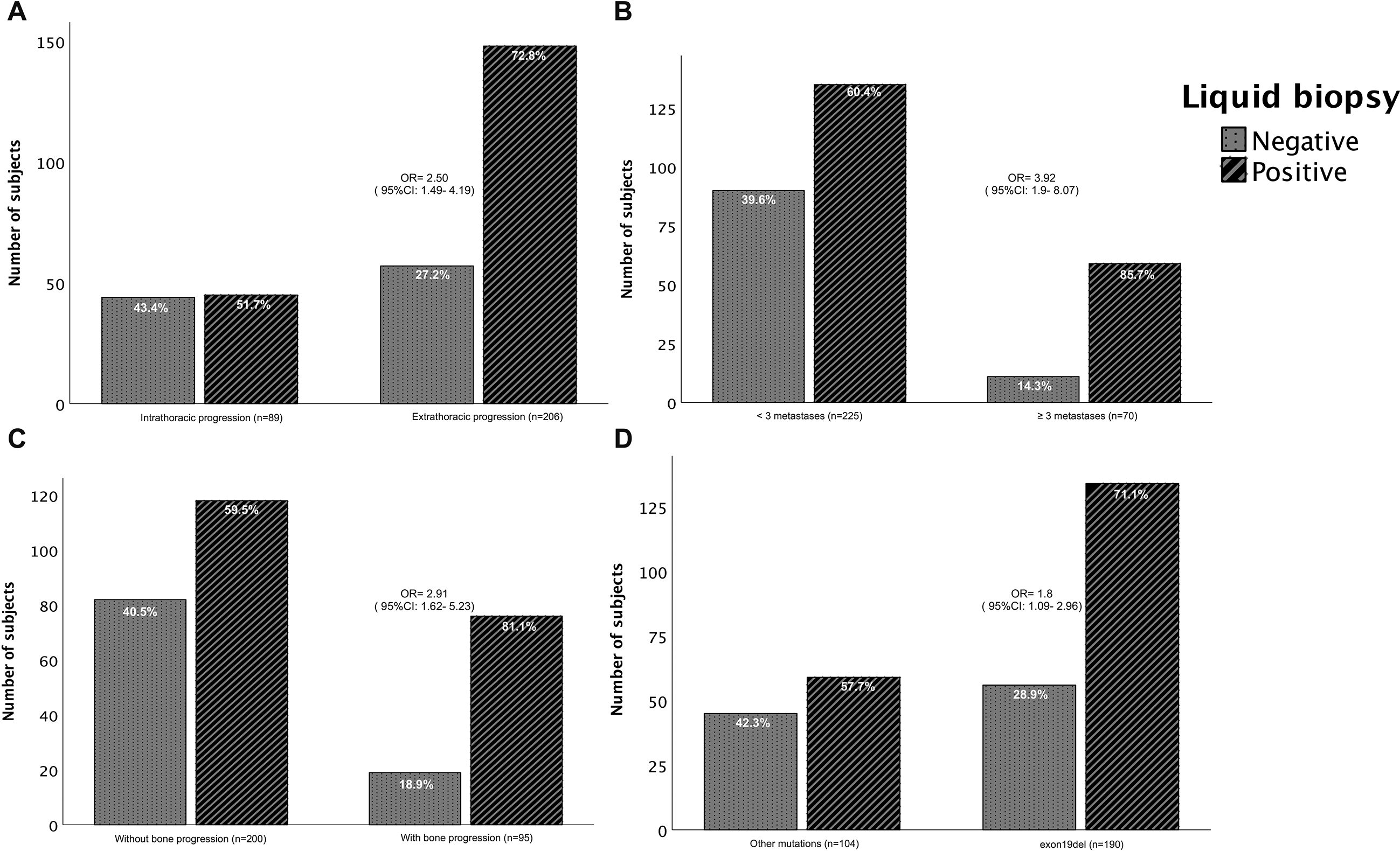
Figure 1.
Detection of ctDNA positivity or negativity by liquid biopsy. Based on clinicopathological characteristics: a) type of thoracic progression (
3.3Detection of T790M in liquid biopsy
Of the 295 analyzed patients, 122 (41.4%) had positive T790M at the first disease progression. In most of the cases, T790M was not the only detected alteration; 69 (56.4%) combined with exon19del, 27 (22.3%) with L858R, and 2 (0.7%) with exon19del and L858R. T790M alone was present in 24 (19.6%) patients. In additional nine patients, subsequent LBs were performed, and the T790M was detected for a total of 131 patients (44%) (Table 3).
Figure 2.
Overall survival (OS) according to liquid biopsy (LB) results. CI: confidence interval. HR: hazard ratio. Statistical significance was set at
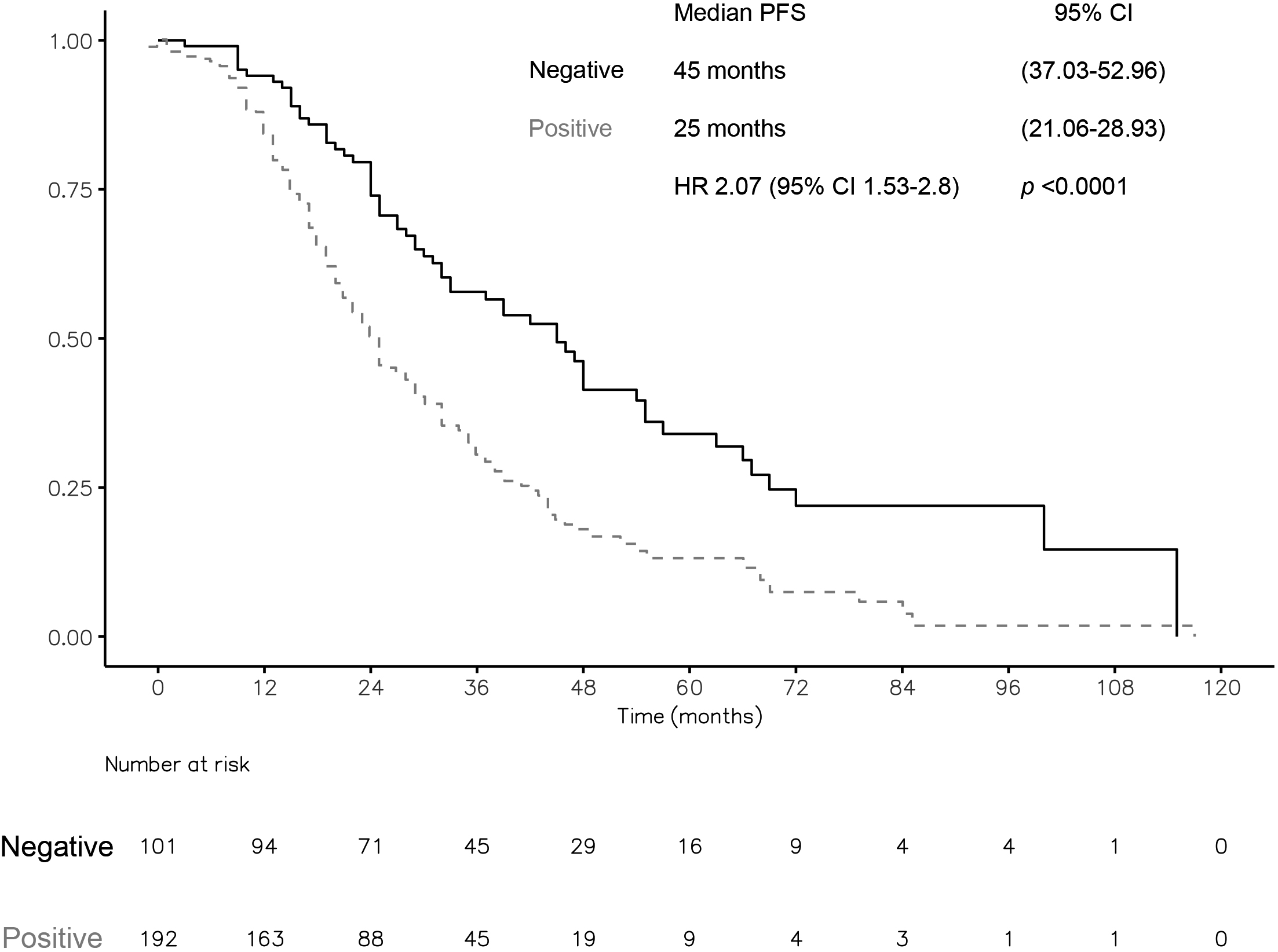
Table 3
Results of liquid biopsy
|
| % | |
| T790M exon19del | 69 | 56.4 |
| T790M L858R | 27 | 22.3 |
| T790M exon19del | 2 | 0.7 |
| T790M | 24 | 19.6 |
Notes.
3.4Characteristics associated with positivity in liquid biopsy
The patients with exon19del at extra-thoracic progression, more than three sites of metastases involved at baseline, and the presence of bone metastases at progression had a higher LB detection rate. In patients with initial exon19del, LB positivity was 71%. Meanwhile, the positive rate in patients with other mutations was 57.7% (Fig. 1). Similarly, patients with extra-thoracic progression had a higher LB detection rate (72.8%) compared with patients with intrathoracic progression (51.7%) (
Figure 3.
Survival of patients with T790M mutation at disease progression. A) PFS of patients treated with osimertinib versus those treated with other drugs. B) Overall survival (OS) in patients treated with osimertinib vs. other treatments. PFS: progression-free survival. CI: confidence interval. NR: not rated. HR: hazard ratio. Statistical significance was set at a
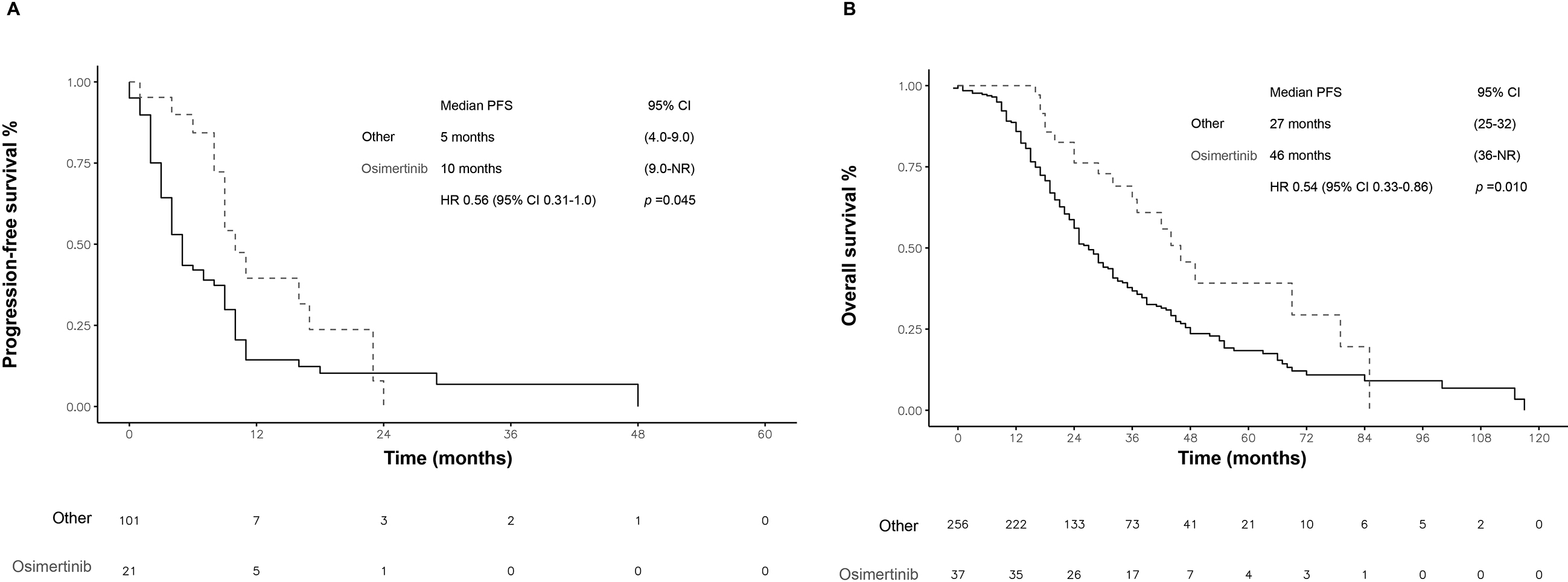
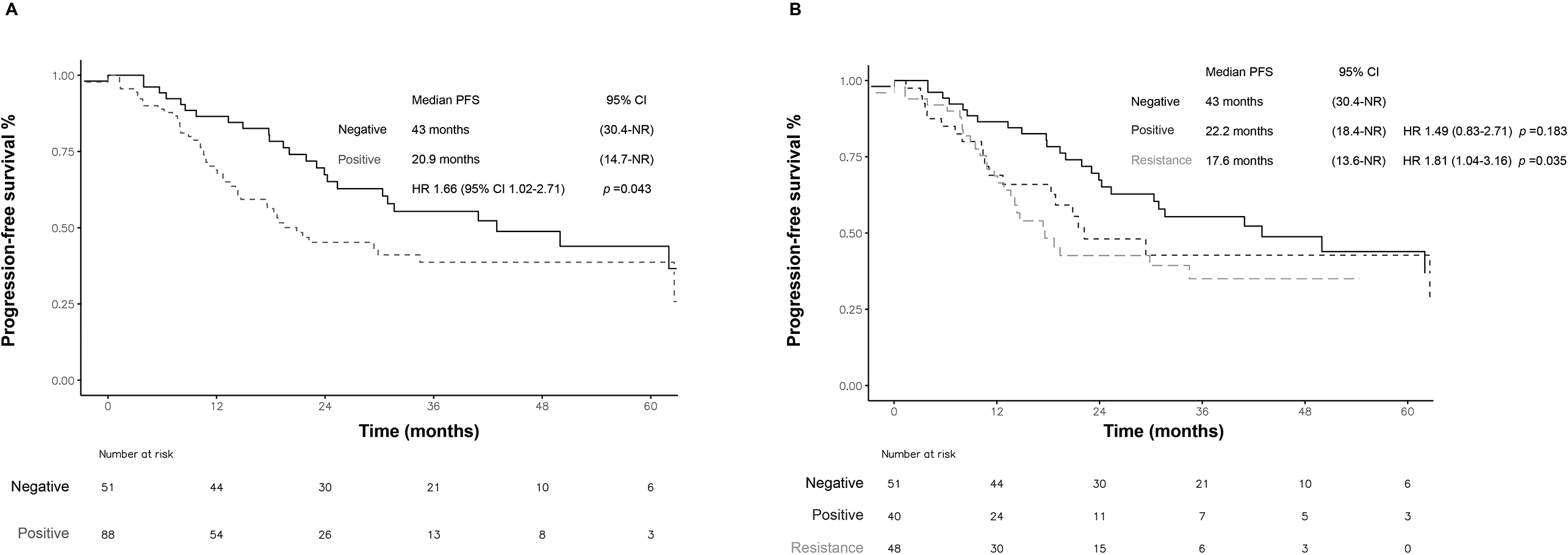
Figure 4.
icPFS according to liquid biopsy results. A) positive and negative. B) Positivity, sensitizing, or resistance mutations. icPFS: intracranial progression-free survival. CI: confidence interval. NR: not rated. HR: hazard ratio. Statistical significance was set at a
3.5Survival outcomes
The median OS for all patients was 30 months (95% CI: 26.06–33.93). After TKI progression, the median PFS was eight months (95% CI: 6.49–9.50). The median OS was 45.0 months (95% CI: 37.03–52.96) vs. 25.0 months [95% CI: 21.06–28.93; HR 2.00 (1.48, 2.71);
The median PFS in patients with a T790M mutation detected at first progression (
In patients treated with osimertinib at any time (
In the case of intracranial disease progression in patients without baseline brain metastases (
According to the type of mutation detected at progression in the LB, patients with a positive T790M mutation had significantly lower icPFS (17.6 months; HR 1.81 95% CI 1.04–3.16;
4.Discussion
In our study, we demonstrated that the positivity of liquid biopsy at disease progression to a first or second-generation TKI was associated with worse survival outcomes including a shorter icPFS, and the detection rate of T790M mutation as the main mechanism of resistance in metastatic EGFRm NSCLC was 44% in Hispanic population. Moreover, we could identify that those patients with more than three metastatic extra-thoracic diseases, and bone metastasis were associated with a greater probability of a positive result on LB.
A T790M as main resistance mechanism after first-line EGFR-TKI treatment has been reported to be around 47.1% in Hispanic population in tumor specimens [14]. In largest series that assessed mechanisms of acquired resistance to EGFR-TKI therapy, 63% of the patients developed T790M mutations. Of note, biopsies were obtained by the least invasive procedure and typically consisted of either a fine-needle aspiration or image-guided core biopsy, but not employed LB [13].
More complex scenarios have been identified in pretreated patients, in which the presence of T790M and persistence of common EGFR-activating mutations (exon 19 del and L858R) has been associated with poor prognosis in patients with advanced EGFRm NSCLC [25]. Our study found that in patients with a positive LB, the detection of T790M mutation alone was observed in 19.6%. In contrast, most patients have persistence of EGFR sensitive mutations, T790M plus exon 19 del in 56.4%, T790M plus L858R in 22.3%, and more complex combinations (T790M plus exon 19 del plus L858R) were extremely rare (
One meta-analysis confirmed that the T790M mutation used to coexist more frequently with an L858R than with the exon19del [odds ratio 1.65; 95% CI (1.17 to 2.32)], which potentially might explain the more aggressive biology of L858R tumors [25]. However, this information has been contradictory with further evidence which suggested a stronger association between the emergence of T790M and the persistence of the primary activating alteration, exon19del, among patients with acquired resistance to TKI (53% vs. 36%; OR 1.87;
Although the feasibility and reliability of LB have been confirmed in clinical practice [28, 29, 30], the test’s sensitivity is related to the detection limits of the technique and the characteristics of ctDNA. Based on the different platforms, in some assays (BEAMing,) the detection rate of LB is around 70% [31], along with this, a relatively low sensitivity (60%) in comparison with high specificity (80–90%) has been reported in an independent meta-analysis [20, 32]. Due to this possible low sensitivity, the current recommendation is that patients with negative LB should undergo tissue testing for T790M [33, 34]. In our study, the LB detection rate was 41.4%, then the detection rate went up to 44% with subsequent testing with the same technique. This reinforces that even the LB could be a suitable option for some patients to detect additional resistance mechanisms if tissue is unavailable or the risk of undergoing an invasive procedure is too high. In line with our work, previous evidence has confirmed an additional 12.5% of detection rate in patients with an initial negative LB and then tested positive for T790M at a second LB [35]. In another study that used a median of 3 LB, the prevalence of T790M was 34.5% [36].
In addition to the procedure itself and technique employed in the ctDNA analysis, some clinical characteristics have gained relevance in the performance and sensitivity of the LB. In previous studies, the number of metastatic sites which reflects disease burden was associated with a higher positive rate [37]. We were able to identify the most common metastatic sites of metastases in the present cohort; pleura (47%), central nervous system (47%), and bone (38.1%). Remarkably, most patients had a moderate disease burden between 2 or 3 involved sites. We found that in patients with three or more sites of metastasis, especially when the affection was extra-thoracic and the bone was involved, the detection rate of the LB was significantly higher.
Patterns of progression and sites have gained relevance in clinical practice to consider subsequent strategies and overcome resistance in EGFR NSCLC extra-thoracic disease, as the main site of progression has been associated with a positive LB [38]. However, one-half of patients progressed within the lung as the initial progression after TKI treatment [38, 39]. Al Halabil et al., in a retrospective study of patients with NSCLC treated with first and second-generation TKI, found that the main sites of progression were those initially involved in 50% of cases, being the intra-thoracic the most common site [38]. This was confirmed by Patel and colleagues, in which 17.5% had extra-thoracic failure, and 22.2% had intra- and extra-thoracic progression [38]. In contrast with these previous reports, our study found that extra-thoracic progression was the main site of progression (69%), and this finding was associated with a higher detection rate by LB and bone lesions, in line with previous reports [28, 35].
ctDNA shedding is related to the disease burden [22, 40] and can help distinguish between residual disease or anticipated imaging changes [41]. This might explain why LB has been associated with worse survival, especially in patients with a persistent EGFR-sensitive mutation. This was replicated in our study in which patients with a positive LB had worse survival independent of the type of mutation [42]. Despite these findings, starting osimertinib at the moment of the T790M mutation detection is not justified, which precedes the radiological progression [43]. In this regard, shedding ctDNA with persistent EGFR-sensitive mutation could be the most essential factor associated with survival.
We found that CNS disease was not associated with LB positivity. However, in patients without baseline brain metastases, the icPFS was significantly shorter in patients with a positive LB at progression. This lower icPFS was statistically significant in patients with a positive T790M mutation. However, a trend to a worse icPFS was also observed for patients with a persistent EGFR sensitive mutation (delexon19 and L858R) regardless of the T790M status. The prognostic utility of liquid biopsy for intracranial PFS is particularly interesting, considering the increased risk of brain affection in patients with EGFR alterations. Very limited data have assessed this key point; based on our findings, it will be very attractive to assess prospectively the prognostic and predictive value of liquid biopsy regarding central nervous system efficacy in this subpopulation. Also, the potential utility of liquid biopsy is to guide clinicians to develop strategies with high brain penetration in the subgroup of patients with the highest risk.
Acquired T790M has been associated with indolent growth and a favorable prognosis compared to other acquired resistance mechanisms. In this sense, the absence of T790M after progression probably indicates the presence of alternative resistance mechanisms, which are associated with a higher disease burden and poorer performance status, contributing to the shorter survival of these patients [42]. As demonstrated in our study, patients with better outcomes were those with a positive T790M mutation who received osimertinib treatment, highlighting the importance of detecting T790M and receiving the appropriate treatment.
In clinical trials, such as the AURA3 [44] trial, a low proportion of patients (51.2%) had T790M mutation, as assessed using plasma ctDNA. However, the T790M mutation detected by ctDNA has been demonstrated to be a surrogate marker for T790M in tumor tissues and is predictive of treatment response [27]. In our study, the median PFS of patients with the T790M mutation treated with osimertinib was similar to that reported in the AURA-3 trial [44, 45]. In another trial [31, 46], no difference in outcomes was found with osimertinib using plasma or tumor tissue to detect T790M, supporting the preferred use of LB and leaving tissue biopsy only for patients with negative LB [31, 46].
Some limitations of the present study were the retrospective single-institution design that could not reflect daily practice in other Latin American institutions, and the presence of some bias in our results due to the nature of the research. Even though, our institution is a high-reference center with a complete representation of the entire country, reflecting important real-world evidence. In addition, due to the retrospective design, we could not evaluate the concordance between the tissue and LB because most patients did not have available tissue samples after progression. We recognize that the standard procedure for patients with negative LB results is to perform tissue re-biopsy; however, in real-life, tissue re-biopsy is performed in 26.1% of cases [47]. Tissue confirmation after the first negative LB test was extremely low in our cohort because of the high proportion of patients who had more than one LB, and refused invasive procedures. We consider comparing real-world scenarios paramount in optimizing clinical practice, considering that awareness and knowledge of liquid biopsy use are still limited. This study is of high value because in real life, most patients do not have access to a biopsy at progression as demonstrated previously. With the short turnaround of liquid biopsy, most patients receive adequate treatment with good outcomes, and the rest of the patients can be considered candidates for clinical trials.
5.Conclusions
This study supports a comparable rate of T790M detection by liquid biopsy in the Latin American population, and a higher detection rate was associated with clinical characteristics. Moreover, it emphasizes that the detection of ctDNA by LB is feasible after progression to first-line EGFR therapy and has a prognostic and potentially predictive value in EGFRm NSCLC. Patients with positive LB results also had shorter intracranial progression-free intervals, a finding that might be confirmed in a further prospective analysis.
Author contributions
Conception: D.H., L.L.-M., O.A.
Interpretation and analysis of data: D.H., A.V.-V., E.V.-S., D.C.-F., G.C.-R., M.O.
Preparation of manuscript: D.H., A.V.-V., L.L.-M.
Revision for important intellectual content: O.A., D.H., L.B-G.
Supervision: O.A., D.H., L.L.-M.
Supplementary data
The supplementary files are available to download from http://dx.doi.org/10.3233/CBM-230124.
Acknowledgments
The authors would like to recognize the work of the team of the Thoracic Oncology Department at our institution for the support and care of patients.
References
[1] | The Global Cancer Observatory, Globocan 2020, International Agency for Research on Cancer 419: ((2020) ), 1–2. |
[2] | A. Jemal, F. Bray, M.M. Center, J. Ferlay, E. Ward and D. Forman, Global cancer statistics, CA Cancer J Clin 61: ((2011) ), 69–90. |
[3] | M.D.P. S S Birring, Symptoms and the early diagnosis of lung cancer, Thorax 60: ((2005) ), 268–269. |
[4] | T.S. Mok, Y.-L. Wu, S. Thongprasert, C.-H. Yang, D.-T. Chu, N. Saijo, P. Sunpaweravong, B. Han, B. Margono, Y. Ichinose, Y. Nishiwaki, Y. Ohe, J.-J. Yang, B. Chewaskulyong, H. Jiang, E.L. Duffield, C.L. Watkins, A.A. Armour and M. Fukuoka, Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma, N Engl J Med 361: ((2009) ), 947–957. |
[5] | F. Passiglia, S. Rizzo, M. Di Maio, A. Galvano, G. Badalamenti, A. Listì, L. Gulotta, M. Castiglia, V. Bazan, A. Russo and F. Fulfaro, The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: A systematic review and meta-analysis, Sci Rep 8: ((2018) ), 1–10. |
[6] | O. Arrieta, L.A. Ramírez-Tirado, R. Báez-Saldaña, O. Peña-Curiel, G. Soca-Chafre and E.O. Macedo-Perez, Different mutation profiles and clinical characteristics among Hispanic patients with non-small cell lung cancer could explain the “Hispanic paradox”, Lung Cancer 90: (2) ((2015) Nov), 161–6. doi: 10.1016/j.lungcan.2015.08.010. Epub 2015 Aug 22. PMID: 26358312. |
[7] | O. Arrieta, F. Andrés, C. Martín, L. Más-lópez, L. Corrales-rodríguez, G. Bramuglia, O. Castillo-fernandez, M. Meyerson, E. Amieva-rivera, A.D. Campos-parra, H. Carranza, J. Carlos, G. De, Y. Powazniak, F. Aldaco-sarvide, C. Vargas, M. Trigo, M. Magallanes-maciel, J. Otero, R. Sánchez-reyes and M. Cuello, Updated Frequency of EGFR and KRAS mutations in NonSmall-Cell Lung Cancer in Latin America The Latin-American Consortium for the Investigation of Lung Cancer, J Thorac Oncol 90: ((2015) ), 1–7. |
[8] | J.-C. Soria, Y. Ohe, J. Vansteenkiste, T. Reungwetwattana, B. Chewaskulyong, K.H. Lee, A. Dechaphunkul, F. Imamura, N. Nogami, T. Kurata, I. Okamoto, C. Zhou, B.C. Cho, Y. Cheng, E.K. Cho, P.J. Voon, D. Planchard, W.-C. Su, J.E. Gray, S.-M. Lee, R. Hodge, M. Marotti, Y. Rukazenkov and S.S. Ramalingam, Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer, New England Journal of Medicine 378: ((2018) ), 113–125. |
[9] | Y.-L. Wu, T.S.K. Mok, J.-Y. Han, M.-J. Ahn, A. Delmonte, S.S. Ramalingam, S.-W. Kim, F.A. Shepherd, J. Laskin, Y. He, H. Akamatsu, W.S.M.E. Theelen, W.-C. Su, T. John, M. Sebastian, H. Mann, M. Miranda, G. Laus, Y. Rukazenkov and V. Papadimitrakopoulou, Overall survival (OS) from the AURA3 phase III study: Osimertinib vs platinum-pemetrexed (plt-pem) in patients (pts) with EGFR T790M advanced non-small cell lung cancer (NSCLC) and progression on a prior EGFR-tyrosine kinase inhibitor (TKI), Annals of Oncology 30: ((2019) ), ix158. |
[10] | T.S. Mok, Y. Cheng, X. Zhou, K.H. Lee, K. Nakagawa, S. Niho, M. Lee, R. Linke, R. Rosell, J. Corral, M.R. Migliorino, A. Pluzanski, E.I. Sbar, T. Wang, J.L. White and Y.L. Wu, Improvement in overall survival in a randomized study that compared dacomitinib with gefitinib in patients with advanced non-small-cell lung cancer and EGFR-activating mutations, Journal of Clinical Oncology 36: ((2018) ), 2244–2250. |
[11] | H. Yoshioka, M. Shimokawa, T. Seto, S. Morita, Y. Yatabe, I. Okamoto, J. Tsurutani, M. Satouchi, T. Hirashima, S. Atagi, K. Shibata, H. Saito, S. Toyooka, N. Yamamoto, K. Nakagawa and T. Mitsudomi, Final overall survival results of WJTOG3405, a randomized phase III trial comparing gefitinib versus cisplatin with docetaxel as the first-line treatment for patients with stage IIIB/IV or postoperative recurrent EGFR mutation-positive non-small-cell lung, Annals of Oncology 30: ((2019) ), 1978–1984. |
[12] | S. Kobayashi, T.J. Boggon, T. Dayaram, P.A. Jänne, O. Kocher, M. Meyerson, B.E. Johnson, M.J. Eck, D.G. Tenen and B. Halmos, EGFR mutation and resistance of non-small-cell lung cancer to gefitinib, New England Journal of Medicine 352: ((2005) ), 786–792. |
[13] | H.A. Yu, M.E. Arcila, N. Rekhtman, C.S. Sima, M.F. Zakowski, W. Pao, M.G. Kris, V.A. Miller, M. Ladanyi and G.J. Riely, Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers, Clinical Cancer Research 19: ((2013) ), 2240–2247. |
[14] | A.F. Cardona, O. Arrieta, M.I. Zapata, L. Rojas, B. Wills, N. Reguart, N. Karachaliou, H. Carranza, C. Vargas, J. Otero, P. Archila, C. Martín, L. Corrales, M. Cuello, C. Ortiz, L.E. Pino, R. Rosell and Z.L. Zatarain-Barrón, Acquired Resistance to Erlotinib in EGFR Mutation-Positive Lung Adenocarcinoma among Hispanics (CLICaP), Target Oncol 12: ((2017) ), 513–523. |
[15] | T. Spence, S. Perera, J. Weiss, S. Grenier, L. Ranich, F. Shepherd and T.L. Stockley, Clinical implementation of circulating tumour DNA testing for EGFR T790M for detection of treatment resistance in non-small cell lung cancer, J Clin Pathol 74: ((2021) ), 91–97. |
[16] | K. Koyama, S. Miura, S. Watanabe, S. Shoji, J. Koshio, Y. Hayashi, D. Ishikawa, K. Sato, T. Miyabayashi, M. Okajima, T. Ota, T. Tanaka, N. Matsumoto, H. Kuriyama, T. Abe, K. Nozaki, K. Ichikawa, R. Kondo, H. Tanaka and T. Kikuchi, Observatiore-biopsyy of re-biopsy in EGFR-TKI-resistant patients with EGFR mutation-positive advanced NSCLC, Sci Rep 12: ((2022) ), 8–15. |
[17] | S.O. Takahisa Kawamura, Hirotsugu Kenmotsu*, Clinical Factors Predicting Detection of T790MRe-biopsyn in Re-biopsy for EGFR-Mutant Non-Small Cell Lung Cancer, Clin Lung Cancer 11: ((2010) ), 211–215. |
[18] | Q.S.C. Chu, A. Agha, N. Devost, R.N. Walton, S. Ghosh and C. Ho, Biopsy on progression in patients with egfr mutation-positive advanced non-small-cell lung cancer – a canadian experience, Current Oncology 27: ((2020) ), 27–33. |
[19] | J.W. Goldman, Z.S. Noor, J. Remon, B. Besse and N. Rosenfeld, Are liquid biopsies a surrogate for tissue EGFR testing, Annals of Oncology 29: ((2018) ), i38–i46. |
[20] | J. Luo, L. Shen and D. Zheng, Diagnostic value of circulating free DNA for detecting EGFR mutation status in NSCLC: A systematic review and meta-analysis, Sci Rep 4: ((2014) ). |
[21] | R. Zhang, B. Chen, X. Tong, Y. Wang, C. Wang, J. Jin, P. Tian and W. Li, Diagnostic accuracy of droplet digital PCR for detection of EGFR T790M mutation in circulating tumor DNA, Cancer Manag Res 10: ((2018) ), 1209–1218. |
[22] | K.S. Thress, A. Markovets, J.C. Barrett, J. Chmielecki, S.B. Goldberg, F.A. Shepherd, S. Vowler and G.R. Oxnard, Complete clearance of plasma EGFR mutations as a predictor of outcome on osimertinib in the AURA trial, Https://DoiOrg/ 101200/JCO20173515_suppl9018 35: ((2017) ), 9018–9018. |
[23] | A.G. Sacher, C. Paweletz, S.E. Dahlberg, R.S. Alden, A. O’Connell, N. Feeney, S.L. Mach, P.A. Jänne and O. Geoffrey, Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer, JAMA Oncol 2: ((2016) ), 1014–1022. |
[24] | L. Arnold, V. Alexiadis, T. Watanaskul, V. Zarrabi, J. Poole and V. Singh, Clinical validation of qPCR Target SelectorTM assays using highly specific switch-blockers for rare mutation detection, J Clin Pathol 73: ((2020) ), 648–655. |
[25] | L.Y. Chen, M.A. Molina-Vila, S.Y. Ruan, K.Y. Su, W.Y. Liao, K.L. Yu, C.C. Ho, J.Y. Shih, C.J. Yu, J.C.H. Yang, R. Rosell and P.C. Yang, Coexistence of EGFR T790M mutation and common activating mutations in pretreatment non-small cell lung cancer: A systematic review and meta-analysis, Lung Cancer 94: ((2016) ), 46–53. |
[26] | H. Liang, Z. Pan, W. Wang, C. Guo, D. Chen, J. Zhang, Y. Zhang, S. Tang, J. He and W. Liang, The alteration of T790M between 19 del and L858R in NSCLC in the course of EGFR-TKIs therapy: A literature-based pooled analysis, J Thorac Dis 10: ((2018) ), 2311–2320. |
[27] | J. Remon, C. Caramella, C. Jovelet, L. Lacroix, A. Lawson, S. Smalley, K. Howarth, D. Gale, E. Green, V. Plagnol, N. Rosenfeld, D. Planchard, M.V. Bluthgen, A. Gazzah, C. Pannet, C. Nicotra, E. Auclin, J.C. Soria and B. Besse, Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA, Ann Oncol 28: ((2017) ), 784–790. |
[28] | S. Mondaca, M. Offin, L. Borsu, M. Myers, S. Josyula, A. Makhnin, R. Shen, G.J. Riely, C.M. Rudin, M. Ladanyi, H.A. Yu, B.T. Li and M.E. Arcila, Lessons learned from routine, targeted assessment of liquid biopsies for EGFR T790M resistance mutation in patients with EGFR mutant lung cancers, Acta Oncol 58: ((2019) ), 1634. |
[29] | M. Reck, K. Hagiwara, B. Han, S. Tjulandin, C. Grohé, T. Yokoi, A. Morabito, S. Novello, E. Arriola, O. Molinier, R. McCormack, M. Ratcliffe and N. Normanno, ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study, J Thorac Oncol 11: ((2016) ), 1682–1689. |
[30] | A. Chiang, A. Fernandes, M. Pavilack, J. Wu, F. Laliberté, M.S. Duh, N. Chehab and J. Subramanian, MA15.11 Real World Biomarker Testing and Treatment Patterns in Patients with Advanced NSCLC Receiving EGFR-TKIs, Journal of Thoracic Oncology 13: ((2018) ), S410–S411. |
[31] | G.R. Oxnard, K.S. Thress, R.S. Alden, R. Lawrance, C.P. Paweletz, M. Cantarini, J.C.H. Yang, J.C. Barrett and P.A. Jänne, Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer, J Clin Oncol 34: ((2016) ), 3375–3382. |
[32] | M. Qiu, J. Wang, Y. Xu, X. Ding, M. Li, F. Jiang, L. Xu and R. Yin, Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: A meta-analysis, Cancer Epidemiol Biomarkers Prev 24: ((2015) ), 206–212. |
[33] | A. Passaro, N. Leighl, F. Blackhall, S. Popat, K. Kerr, M.J. Ahn, M.E. Arcila, O. Arrieta, D. Planchard, F. de Marinis, A.M. Dingemans, R. Dziadziuszko, C. Faivre-Finn, J. Feldman, E. Felip, G. Curigliano, R. Herbst, P.A. Jänne, T. John, T. Mitsudomi, T. Mok, N. Normanno, L. Paz-Ares, S. Ramalingam, L. Sequist, J. Vansteenkiste, I.I. Wistuba, J. Wolf, Y.L. Wu, S.R. Yang, J.C.H. Yang, Y. Yatabe, G. Pentheroudakis and S. Peters, ESMO expert consensus statements on the management of EGFR mutant non-small-cell lung cancer, Ann Oncol 33: ((2022) ), 466–487. |
[34] | C. Rolfo, P. Mack, G.V. Scagliotti, C. Aggarwal, M.E. Arcila, F. Barlesi, T. Bivona, M. Diehn, C. Dive, R. Dziadziuszko, N. Leighl, U. Malapelle, T. Mok, N. Peled, L.E. Raez, L. Sequist, L. Sholl, C. Swanton, C. Abbosh, D. Tan, H. Wakelee, I. Wistuba, R. Bunn, J. Freeman-Daily, M. Wynes, C. Belani, T. Mitsudomi and D. Gandara, Liquid Biopsy for Advanced NSCLC: A consensus statement from the international association for the study of lung cancer, Journal of Thoracic Oncology 16: ((2021) ), 1647–1662. |
[35] | R. Minari, G. Mazzaschi, P. Bordi, L. Gnetti, G. Alberti, A. Altimari, E. Gruppioni, F. Sperandi, C. Parisi, G. Guaitoli, S. Bettelli, L. Longo, F. Bertolini, M. Pagano, C. Bonelli, E. Tagliavini, D. Nicoli, A. Ubiali, A. Zangrandi, S. Trubini, M. Proietto, M. Fiorentino and M. Tiseo, Detection of EGFR-Activating and T790M Mutations Using Liquid Biopsy in Patients With EGFR-Mutated Non-Small-Cell Lung Cancer Whose Disease Has Progressed During Treatment With First- and Second-Generation Tyrosine Kinase Inhibitors: A Multicenter Real-Life Retrospective Study, Clin Lung Cancer 21: ((2020) ), e464–e473. |
[36] | A. Dal Maso, P. Del Bianco, F. Cortiula, G. Nardo, E. Zulato, L. Bonanno, A. Follador, G. De Maglio, G. Pasello and S. Indraccolo, EGFR T790M testing through repeated liquid biopsy over time: A real-world multicentric retrospective experience, J Thorac Dis 14: ((2022) ), 3364–3375. |
[37] | F. Passiglia, S. Rizzo, C. Rolfo, A. Galvano, E. Bronte, L. Incorvaia, A. Listi, N. Barraco, M. Castiglia, V. Calo, V. Bazan and A. Russo, Metastatic Site Location Influences the Diagnostic Accuracy of ctDNA EGFR-Mutation Testing in NSCLC Patients: A Pooled Analysis, Curr Cancer Drug Targets 18: ((2018) ), 697–705. |
[38] | H. Al-Halabi, K. Sayegh, S.R. Digamurthy, A. Niemierko, Z. Piotrowska, H. Willers and V. Sequist, Pattern of Failure Analysis in Metastatic EGFR-Mutant Lung Cancer Treated with Tyrosine Kinase Inhibitors to Identify Candidates for Consolidation Stereotactic Body Radiation Therapy, J Thorac Oncol 10: ((2015) ), 1601–1607. |
[39] | S.H. Patel, A. Rimner, A. Foster, Z. Zhang, K.M. Woo, H.A. Yu, G.J. Riely and A.J. Wu, Lung cancer, 108: ((2017) ), 109–114. |
[40] | P. Bordi, M. Del Re, R. Minari, E. Rofi, S. Buti, G. Restante, A. Squadrilli, S. Crucitta, C. Casartelli, L. Gnetti, C. Azzoni, L. Bottarelli, I. Petrini, A. Cosenza, L. Ferri, E. Rapacchi, R. Danesi and M. Tiseo, From the beginning to resistance: Study of plasma monitoring and resistance mechanisms in a cohort of patients treated with osimertinib for advanced T790M-positive NSCLC, Lung Cancer 131: ((2019) ), 78–85. |
[41] | A.M. Newman, S.V. Bratman, J. To, J.F. Wynne, N.C.W. Eclov, L.A. Modlin, C.L. Liu, J.W. Neal, H.A. Wakelee, R.E. Merritt, J.B. Shrager, B.W. Loo, A.A. Alizadeh and M. Diehn, An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage, Nat Med 20: ((2014) ), 548–554. |
[42] | G.R. Oxnard, M.E. Arcila, C.S. Sima, G.J. Riely, J. Chmielecki, M.G. Kris, W. Pao, M. Ladanyi and V.A. Miller, Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation, Clin Cancer Res 17: ((2011) ), 1616–1622. |
[43] | J. Remon, B. Besse, S.P. Aix, A. Callejo, K. Al-Rabi, R. Bernabe, L. Greillier, M. Majem, N. Reguart, I. Monnet, S. Cousin, P. Garrido, G. Robinet, R. Garcia Campelo, A. Madroszyk, J. Mazières, H. Curcio, B. Wasag, Y. Pretzenbacher, B. Fournier, A.M.C. Dingemans and R. Dziadziuszko, Osiertinib treatment based on plasma T790M monitoring in patients with EGFR-mutant non-small-cell lung cancer (NSCLC): EORTC Lung Cancer Group 1613 APPLE phase II randomized clinical trial, Ann Oncol 34: ((2023) ), 468–476. |
[44] | V.A. Papadimitrakopoulou, J.Y. Han, M.J. Ahn, S.S. Ramalingam, A. Delmonte, T.C. Hsia, J. Laskin, S.W. Kim, Y. He, C.M. Tsai, T. Hida, M. Maemondo, T. Kato, S. Jenkins, S. Patel, X. Huang, G. Laus, A. Markovets, K.S. Thress, Y.L. Wu and T. Mok, Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer, Cancer 126: ((2020) ), 373–380. |
[45] | T.S. Mok, Y.-L. Wu, M.-J. Ahn, M.C. Garassino, H.R. Kim, S.S. Ramalingam, F.A. Shepherd, Y. He, H. Akamatsu, W.S.M.E. Theelen, C.K. Lee, M. Sebastian, A. Templeton, H. Mann, M. Marotti, S. Ghiorghiu and V.A. Papadimitrakopoulou, Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer, N Engl J Med 376: ((2017) ), 629–640. |
[46] | D. Planchard, S. Popat, K. Kerr, S. Novello, E.F. Smit, C. Faivre-Finn, T.S. Mok, M. Reck, P.E. Van Schil, M.D. Hellmann and S. Peters, Corrigendum: Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up (Annals of Oncology (2018) 29 (iv192–iv237) DOI: 10.1093/annonc/mdy275), Annals of Oncology 30: ((2019) ), 863–870. |
[47] | P. Krawczyk, L. Grzycka-Kowalczyk, J. Błach, K. Reszka,I. Chmielewska, R. Kieszko, M. ójcik-Superczyńska, M. Szczyek, T. Jankowski and J. Milanowski, The efficacy of T790M mutation testing in liquid biopsy – Real clinic data, PLoS One 17: ((2022) ). |




