The prognostic and antitumor roles of key genes of ferroptosis in liver hepatocellular cancer and stomach adenocarcinoma
Abstract
BACKGROUND:
Liver hepatocellular cancer (LIHC) and stomach adenocarcinoma (STAD) are common malignancies with high lethal ratios worldwide. Great progress has been achieved by using diverse therapeutic strategies; however, these diseases still have an unfavourable prognosis. Ferroptosis inducer drugs, unlike apoptosis-related drugs, can overcome the resistance to cancer therapy caused by traditional chemicals. However, the relationship between overall survival (OS) and ferroptosis-related genes, as well as the mechanisms involved, are largely unclear.
METHODS:
The expression levels of AIFM2, GPX4, ACSL4, FTH1, NOS1, and PTGS2 in LIHC and STAD were obtained from UALCAN. The correlations of OS with these gene expression levels were obtained using the Kaplan-Meier Plotter database. The OS associated with genetic mutations of those genes compared to that of unchanged genes was analysed using the TIMER website. GO and KEGG enrichment analyses of ferroptosis-related genes and their coexpressed genes in LIHC and STAD were conducted using the STRING and DAVID databases. The relationship of PTGS2 and ACSL4 to immune cell infiltration was analysed using the TIMER website. The viability and GPX5 expression levels in LIHC cells treated with RSL3 and As2O3 were detected by MTT methods and western blotting, respectively.
RESULTS:
Our results showed that GPX4, FTH1 and AIFM2 were overexpressed in LIHC and STAD. High levels of GPX4, FTH1 and AIFM2 were prominently correlated with better prognosis in LIHC. However, GPX and FTH1 in STAD did not show significant correlations with OS. AIFM2 in STAD had the opposite trend with OS compared with that in LIHC. Moreover, a high mutation rate of these genes (35.74%) was also observed in LIHC patients, and genetic mutation of these genes was correlated with shorter OS. In contrast, the genetic mutation of these genes did not change OS in STAD. Enrichment analysis showed that the respiratory electron transport chain, cell chemotaxis and T-cell migration were related to ferroptosis. ASCL4 and PTGS2 coexpressed with cytokines associated with immune cell infiltration. Compared to RSL3 or As2O3 alone, As2O3 plus RSL3 significantly inhibited the growth of Huh7 cells. GPX4 was downregulated to an undetectable level when in combination with RSL3.
CONCLUSIONS:
Our results indicated that ferroptosis-related genes might play an important role in LIHC and STAD and might be risk factors for overall survival in LIHC and STAD.
1.Introduction
Digestive system neoplasms such as LIHC and STAD have received increasing attention for their malignancy [1, 2, 3, 4, 5]. Traditional chemicals cannot effectively eliminate cancer cells, so new strategies are urgently needed. Ferroptosis is a newly discovered form of cell death different from apoptosis, necrosis, and pyroptosis that can be triggered by iron-dependent lipid peroxidation [6, 7, 8, 9]. Knockdown or overexpression of ferroptosis-related genes such as PTGS2 and ACSL4 can overcome resistance to traditional drug therapy. In addition, ferroptosis inducers such as erastin, RSL3, and iron oxide nanoparticles can also trigger ferroptosis. The use of ferroptosis inducers is emerging as a promising therapeutic strategy [10, 11]. However, the correlation of ferroptosis-related genes with overall survival in LIHC and STAD patients and the potential mechanisms involved are largely unclear.
GPX4 overexpression can resist ferroptosis; however, knockdown of GPX4 could lead to lipid peroxide accumulation and trigger ferroptosis. GPX4 is an important enzyme for regulating ferroptosis by removing lipid peroxides. Inhibition of GPX4 overcomes resistance to lapatinib by promoting ferroptosis in NSCLC cells [12]. Targeting GPX4 might be a potential strategy to enhance the antitumor effects of lapatinib. AIFM2 (FSP1) is a ferroptosis regulator independent of GSH and can block RSL3-induced ferroptosis [13]. Knockdown of AIFM2 could enhance the sensitivity of sorafenib to antitumor therapy. ACSL4 is a member of the long chain family of acyl-CoA synthetase proteins, which play a key role in ferroptosis. Overexpression of ACSL4 inhibited the proliferation of glioma cells by regulating ferroptosis [14]. FTH1 conferred ferroptosis resistance by modifying iron metabolism and lipid peroxidation [15]. Knockdown of ferritin heavy chain-1 by RNA interference in HCC cells promoted ferroptosis in response to erastin and sorafenib. NOX1 encodes a member of the NADPH oxidase family of enzymes responsible for the catalytic one-electron transfer of oxygen to generate superoxide or hydrogen peroxide. NOX1 inhibition attenuates the development of a protumorigenic environment in experimental hepatocellular carcinoma [16]. PTGS2 is also a known regulator of ferroptosis [17]. PTGS2 acts both as a dioxygenase and as a peroxidase. The upregulation of ferroptosis marker PTGS2 mRNA was markedly prevented by the ferroptosis-specific inhibitor ferrostatin-1 [18].
In the present study, we comprehensively analysed the mRNA levels of AIFM2, GPX4, ACSL4, FTH1, NOS1 and PTGS2 using UALCAN. The correlations between the mRNA levels of these genes and prognosis in LIHC and STAD were determined using databases including UALCAN and gene expression profiling interactive analysis (GEPIA). We further explored the relationship between the expression levels of ACSL4 and PTGS2 and immune cell infiltration levels using the TIMER database. PTGS2 and ACSL4 in tumour tissues may be used as an indicator of sensitivity to immune checkpoint inhibitors.
2.Materials and methods
2.1GPX4 inhibitor in combination with As2O3
H-Huh7 cells were inoculated into 96-well plates or 6-well plates at a fusion rate of approximately 50%. Huh7 cells were treated with RSL3 or As2O3 for 48 hours with or without Vit. E and DFOM. Microscope images of Huh7 cells were observed. Cell viability was detected using MTT assays. Huh7 cells were treated with increasing concentrations of As2O3 or RSL3 plus As2O3. The expression levels of GPX4 and
2.2UALCAN
UALCAN is a comprehensive web resource for analysing cancer (TCGA, MET500 and CPTAC). It is built on PERL-CGI with high-quality graphics based on JavaScript and CSS. In our study, UALCAN was used to evaluate the diverse expression levels of genes in tumours and normal tissue. In addition, UALCAN was used to analyse the prognostic value of the mRNA expression of ferroptosis-related genes (GPX4, AIFM2, ACSL4, NOX1, FTH1 and PTGS2) in LIHC and STAD patients. Student’s
2.3GEPIA
GEPIA is a newly developed interactive web server for analysing the RNA sequencing expression data of 8587 normal samples and 9736 tumours. The GEPIA database was used to analyse the correlation of GPX4, AIFM2, ACSL4, NOX1, FTH1 and PTGS2 levels with tumour stage in LIHC and STAD. Student’s
2.4TCGA data and cBioPortal
cBioPortal is a comprehensive web resource for exploring, visualizing, and analysing multidimensional cancer genomics data. We analysed the genomic profiles of GPX4, AIFM2, ACSL4, NOX1, FTH1 and PTGS2, including putative copy number alterations, mutations from GISTIC, and mRNA expression z scores. Genetic mutations in GPX4, AIFM2, ACSL4, NOX1, FTH1 and PTGS2 and their correlation with overall survival are shown as Kaplan-Meier plots. The log-rank test was used to generate a
2.5DAVID
Sixty genes were significantly related to GPX4, AIFM2, ACSL4, NOX1, FTH1 and PTGS2, as analysed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Gene Ontology (GO) in the DAVID database. Gene ontology analyses have three main parts: molecular functions (MF), cellular components (CC), and biological processes (BP). The functional roles of the ferroptosis-related genes and the top coexpressed genes were analysed by this tool. KEGG analysis can define the pathways related to ferroptosis genes.
2.6TIMER database analysis
The TIMER database was used to analyse the correlation between the expression levels of ACSL4 and PTGS2 in LIHC and STAD and immune cell infiltration, including CD8
2.7Statistical analysis
Survival curves were obtained from the UALCAN database. The results of GEPIA analysis are presented with Cox P, P or HR values from a log-rank test. The Spearman’s correlation coefficient and the statistical significance are typical standards used to evaluate correlations between expressed genes.
3.Results
3.1Dysregulated expression of ferroptosis-related genes in patients with LIHC and STAD
Figure 1.
The mRNA expression of ferroptosis-related genes in LIHC or STAD tissues and adjacent normal tissues (UALCAN). A–C. The mRNA expression levels of GPX4, AIFM2, and ACSL4 were overexpressed in LIHC and STAD tissues compared to normal samples. D–F. The expression of NOX1, FTH1 and PTGS2 in tumour and normal tissues in LIHC or STAD. ***p < 0.001, **p < 0.01.
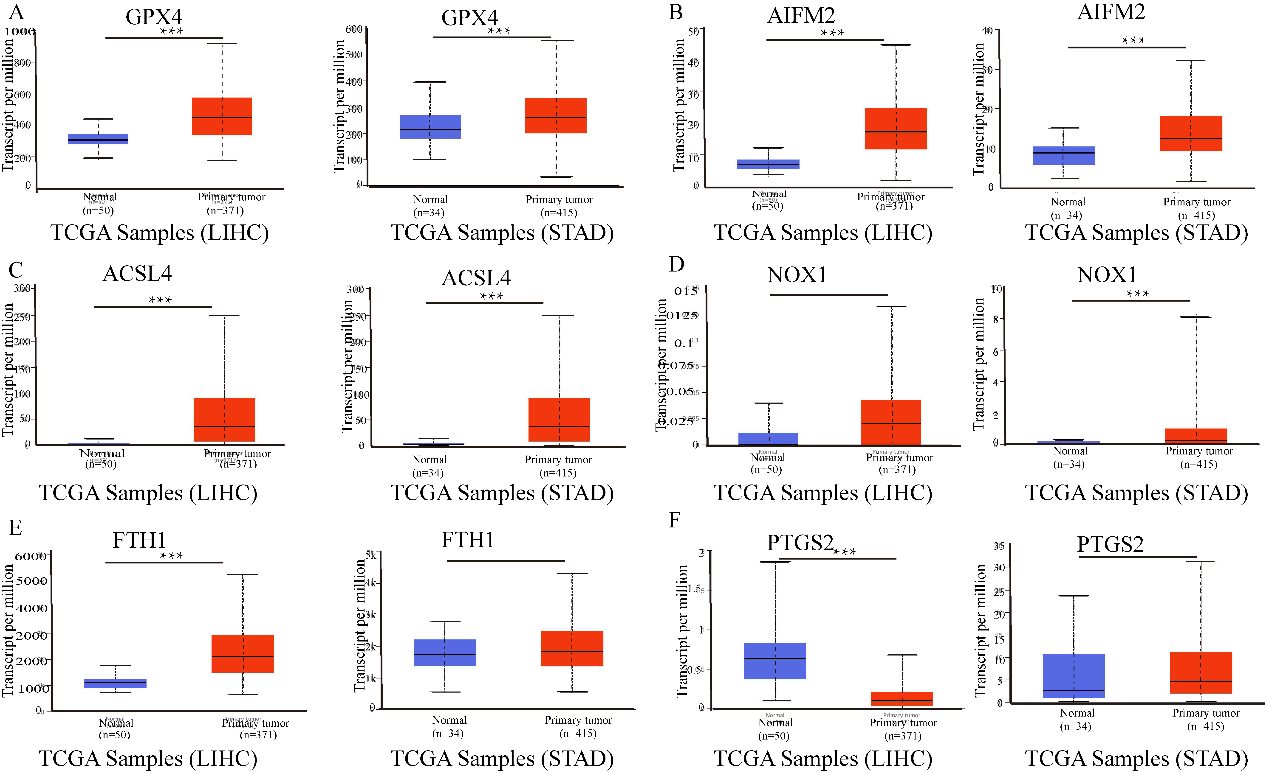
The results in Fig. 1 show that GPX4, AIFM2, and ACSL4 levels were higher in LIHC and STAD tissues than in normal tissues. Moreover, FTH1 expression was higher in LIHC than in adjacent tissues. However, the expression level of PTGS2 was lower in LIHC tissues than in adjacent normal tissues. NOX1 expression was higher in STAD than in adjacent tissues.
Moreover, PTGS2 expression was higher in STAD than in normal tissues. However, the expression level of PTGS2 was lower in LIHC tissues than in normal tissues. In addition, the relative expression levels of ferroptosis-related genes in LIHC and STAD tissues are shown in Fig. 2. FTH1 was the most highly expressed gene in LIHC and STAD. In addition, GPX4, AIFM2, and ACSL4 were also overexpressed in LIHC and STAD (Fig. 2).
Figure 2.
The relative levels of ferroptosis-related genes in LIHC and STAD. FTH1 was the most highly expressed gene in LIHC and STAD.

3.2The correlation between mRNA expression of ferroptosis-related genes and tumour stages in LIHC and STAD patients
Figure 3.
The correlation between ferroptosis-related genes and tumour stage in STAD patients (GEPIA). The mRNA expression levels of these genes were not correlated with patients’ individual tumour stage (Pr (
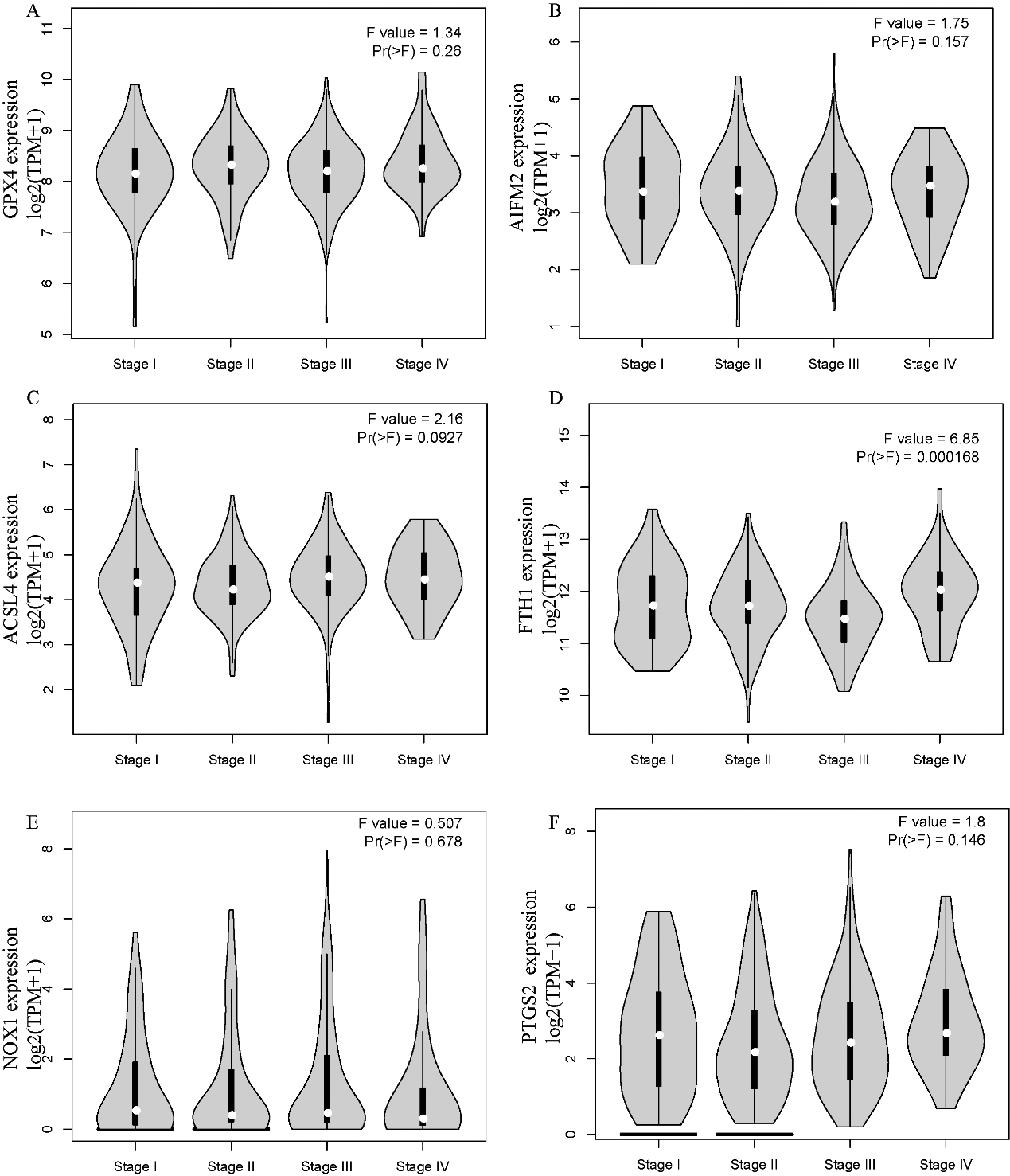
Figure 4.
Prognostic value of the mRNA expression of ferroptosis-related genes in LIHC and STAD patients (UALCAN). The survival curves comparing patients with high (red) and low (blue) risk. Ferroptosis-related gene expression in LIHC and STAD was plotted using UALCAN at the threshold of
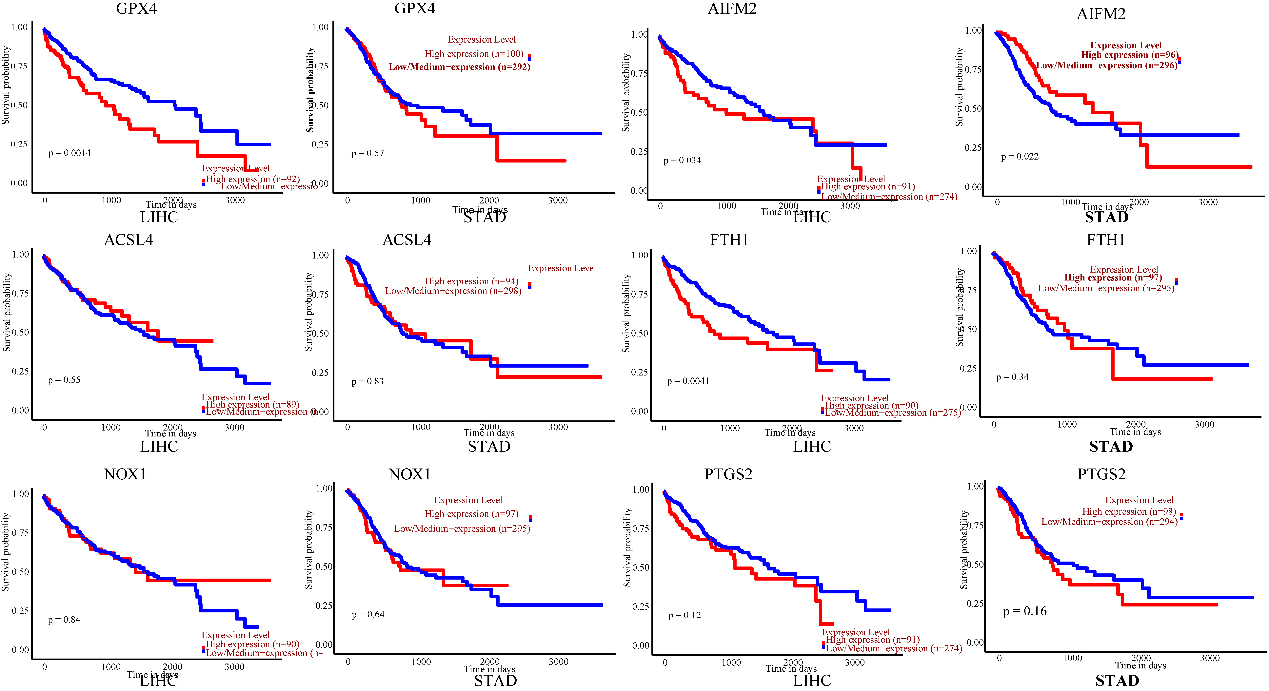
Figure 5.
Genetic mutations in ferroptosis-related genes and their correlation with the OS of LIHC patients (cBioPortal). (A) Outline of alterations in ferroptosis-related genes in LIHC. (B) Correlations of different ferroptosis-related genes with each other in LIHC. (C) Genetic alterations in ferroptosis-related genes were associated with shortened OS in LIHC. (D) Genetic alterations in ferroptosis-related genes and OS in LIHC. (
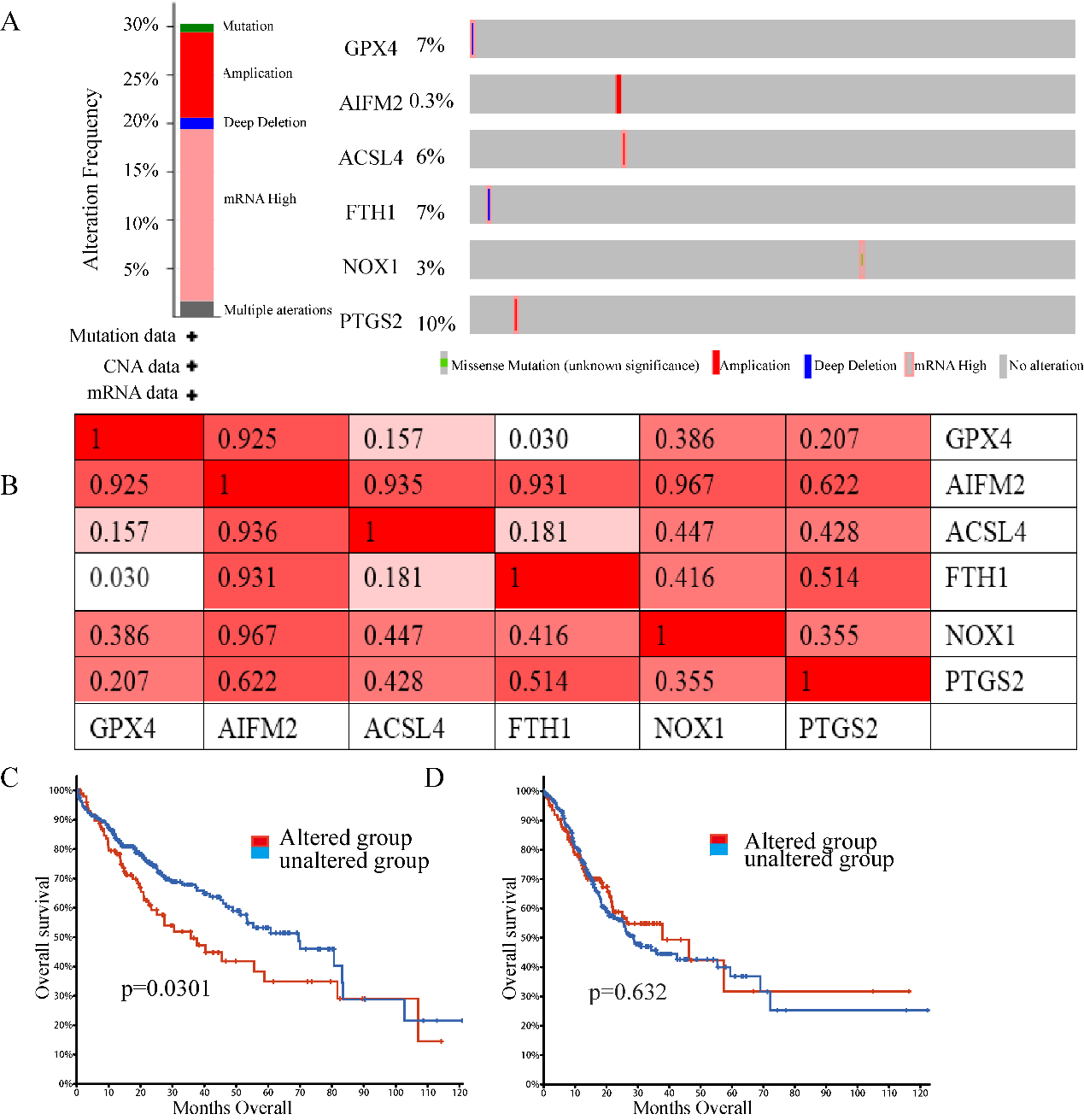
Figure 6.
GO and KEGG enrichment analyses of ferroptosis-related genes and their coexpressed genes in LIHC. (STRING and DAVID). (A) PPI network. The nodes indicate proteins; the edges indicate the interaction of proteins. (B) BP. (C) CC. (D) MF. (E) KEGG. Systematic Analysis of Expression Profiles and Prognostic Significance for Ferroptosis-related Genes in LIHC.
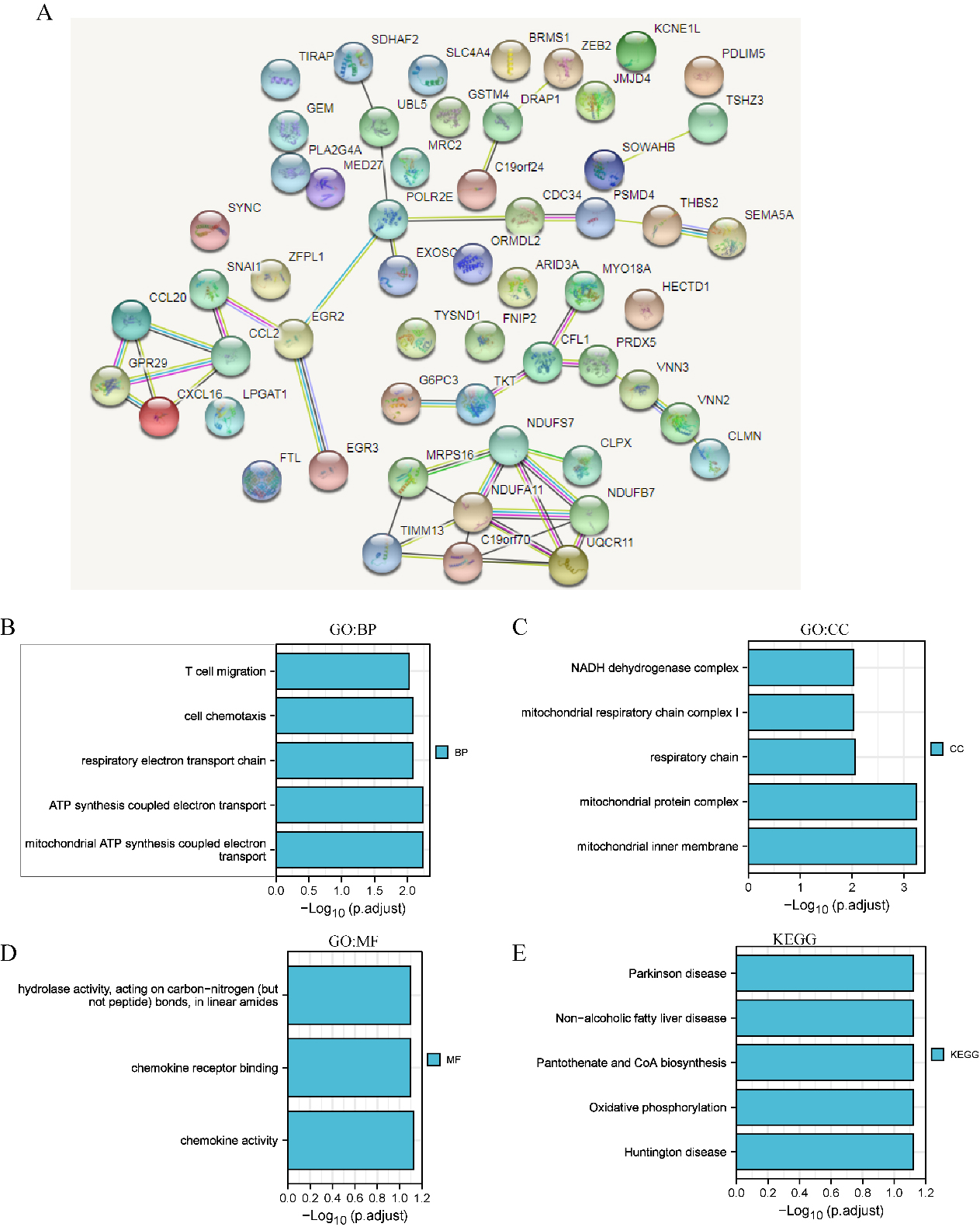
Figure 7.
The correlation of ASCL4 and PTGS2 expression with tumour-infiltrating lymphocytes in LIHC and STAD. A–D. The relationship of ASCL4 and PTGS2 with immune infiltration levels in LIHC and STAD. E–H. The coexpression levels of CD274 with ASCL4 and PTGS2 in LIHC and STAD.
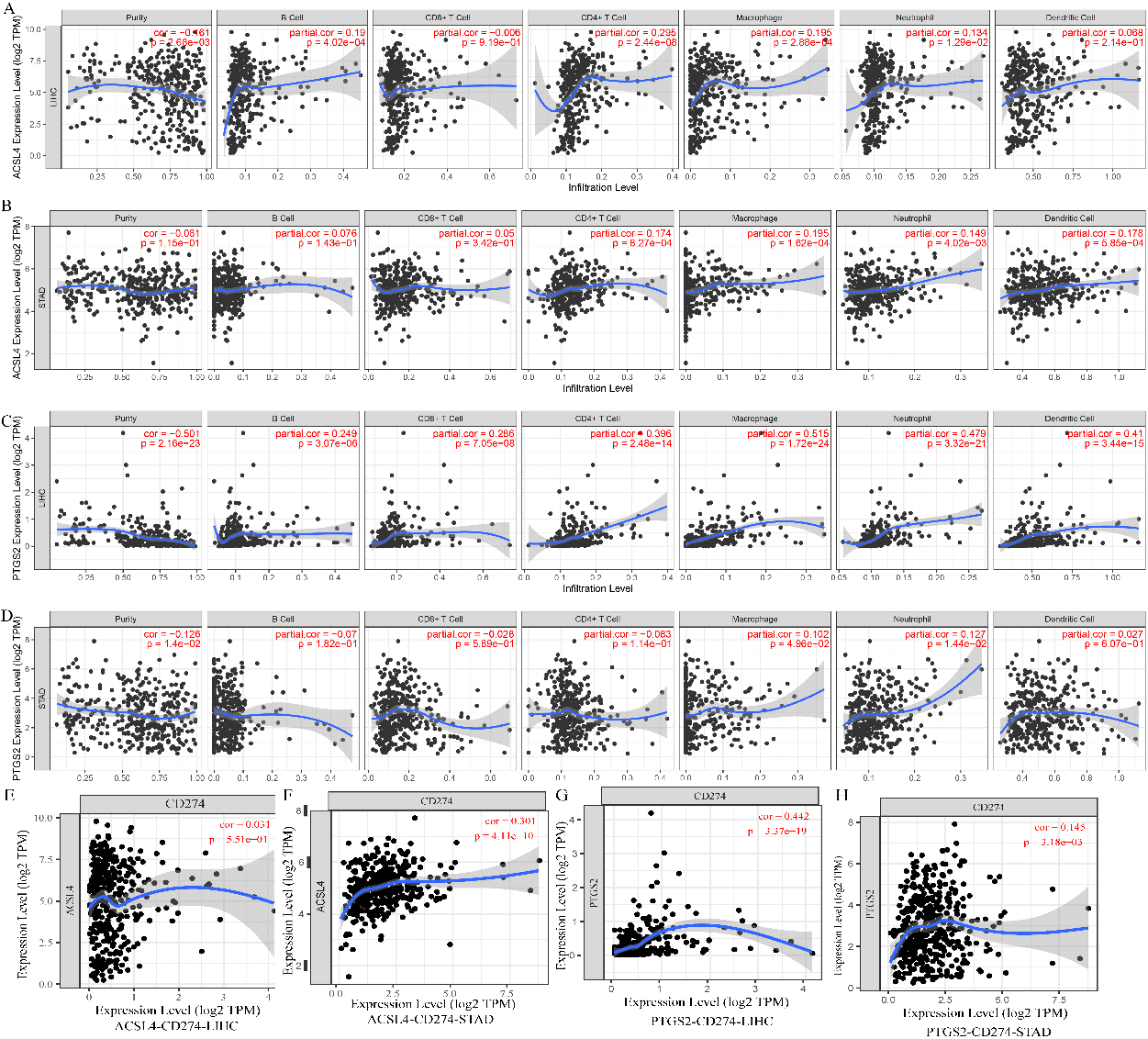
Figure 8.
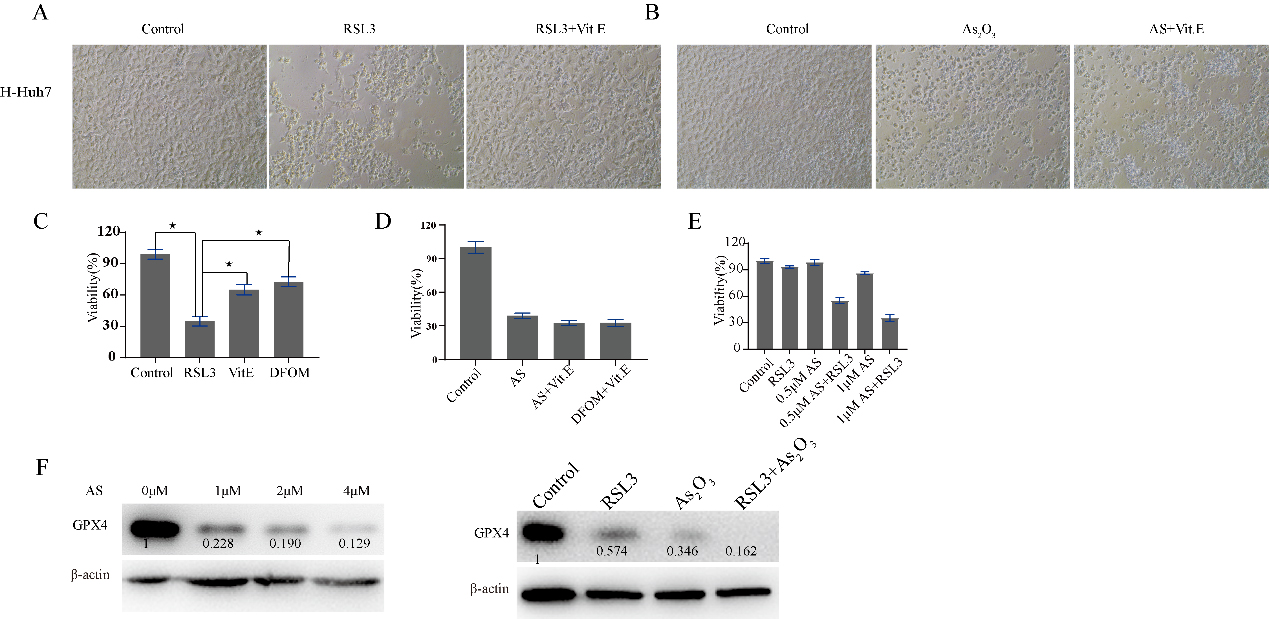
RSL3 and As2O3 suppress LIHC cell proliferation. Huh7 cells were treated with RSL3 or As2O3 for 48 hours with or without Vit. E and DFOM. A. Microscopic image of Huh7 cells treated with PBS, RLS3, or RLS3
Next, we explored the correlation of the mRNA expression of different ferroptosis-related genes to patients’ individual tumour stage in LIHC and STAD patients by GEPIA. The FTH1 groups prominently varied in STAD, whereas the GPX4, AIFM2, ACSL4, NOX1, and PTGS2 groups did not significantly differ (Fig. 3). The mRNA expression of ferroptosis-related genes in different tumour stages of LIHC did not significantly differ. In summary, the results above indicated that the mRNA expression of FTH1 was appreciably correlated with patients’ individual cancer stages in STAD, and STAD in more advanced stages tended to have higher mRNA expression levels of FTH1
3.3Prognostic values of ferroptosis-related genes in patients with LIHC and STAD
The prognostic values of ferroptosis-related genes for LIHC and STAD, including OS, were explored using UALCAN. Patients were divided into low- and high-risk groups based on a cut-off value (Fig. 4). Negative correlations were displayed between OS and the mRNA levels of GPX4, AIFM2, and FTH1 in LIHC. Nevertheless, there was no prominent correlation in LIHC between OS and ACSL4, NOX1, and PTGS2 expression. Increased AIFM2 mRNA expression levels were positively associated with OS in STAD. The AIFM2 expression level in STAD was negatively associated with the infiltration of CD4
3.4Genetic mutations in ferroptosis-related genes and their associations with OS in LIHC and STAD
Epigenetic alteration exerts an important role in early malignancies. Therefore, we explored ferroptosis-related gene alterations and correlations using the cBioPortal online tool for LIHC (TCGA, Firehose Legacy), in which ferroptosis-related genes varied in 110 samples out of 369 patients with LIHC (35.74%) (Fig. 5A). PTGS2, GPX4, FTH1, and ACSL4 were the top four genes with genetic alterations, and their mutation rates were 10, 7, 7, and 6%, respectively. In addition, we also analysed the correlations between ferroptosis-related genes using PTGS2, GPX4, PTH1, and ACSL4 mRNA expression in LIHC using the cBioPortal online tool (TCGA, Firehose Legacy), which contained Pearson’s correction. We also found that AIFM2, ACSL4, NOS1 and PTGS2 were highly coexpressed with the other genes, but other ferroptosis-related genes were partly coexpressed (Fig. 5B). In addition, we analysed the association of genetic alterations in ferroptosis-related genes with the OS of LIHC patients. Results from the Kaplan-Meier plot and log-rank test showed that genetic alterations in ferroptosis-related genes were associated with shortened OS (Fig. 5C,
3.5GO and KEGG enrichment analyses of ferroptosis-related genes and their 60 coexpressed genes in LIHC and STAD patients
GO and KEGG enrichment analyses of ferroptosis-related genes and their 60 coexpressed genes in LIHC patients (Fig. 6). As shown in in Fig. 6B–E, we found that BP terms such as GO: 0042775 (mitochondrial ATP synthesis coupled electron transport), GO: 0042773 (ATP synthesis coupled electron transport), GO: 0022904 (respiratory electron transport chain), GO: 0060326 (cell chemotaxis), and GO: 0072678 (T-cell migration) were prominently regulated by ferroptosis-related genes in LIHC (Fig. 6B). Moreover, CC terms, including GO: 0005743 (mitochondrial inner membrane), GO: 0098798 (mitochondrial protein complex), GO: 0070469 (respiratory chain), and GO: 0005747 (mitochondrial respiratory chain complex I), were appreciably correlated with ferroptosis-related genes (Fig. 6C). Furthermore, ferroptosis-related genes also significantly influenced MFs, such as GO: 0008009 (chemokine activity), GO: 0042379 (chemokine receptor binding), and GO: 0016811 (hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides). In the KEGG analysis, hsa00190 (oxidative phosphorylation) was closely connected to the functions of ferroptosis-related genes in LIHC (Fig. 6E).
In addition, in STAD patients, we found that BPs, such as T-cell cytokine production, regulation of the neuroinflammatory response and cellular response to lipopolysaccharide, were prominently regulated by ferroptosis-related genes in STAD (S3). Moreover, CC terms, including cell division site, microvilli, and glial cell projection, were significantly associated with ferroptosis-related genes. Furthermore, ferroptosis-related genes also significantly influenced MF terms, such as cytokine activity, receptor ligand activity, and growth factor receptor binding. In the KEGG analysis, rheumatoid arthritis, the IL-17 signalling pathway and transcriptional misregulation in cancer were strongly related to the functions of ferroptosis-related genes in STAD (S3).
3.6The correlation between the expression levels of PTGS2 and ACSL4 and immune infiltration in LIHC and STAD
ASCL4 and PTGS2 coexpressed with cytokines that may be associated with immune cell attraction. ASCL4 coexpressed with CCL20, CCR6, and CXCL16 in LIHC. PTGS2 coexpressed with CCL2 in LIHC. PTGS2 coexpressed with CXCL8, IL11, IL24, CSF2, IL6, and IL1B in STAD.
We further analysed the correlation between the expression levels of ASCL4 and PTGS2 and immune infiltration in LIHC. ASCL4 and PTGS2 were overexpressed in LIHC, while ASCL4 was downregulated in STAD. In LIHC, the level of ASCL4 was significantly correlated with the infiltration levels of B cells, CD4
3.7A GPX4 inhibitor in combination with As2O3 enhances antitumor therapy
GPX4, ACSL4, FTH1, NOS1, PTGS2 and AIFM2 play important roles in ferroptosis. GPX4 is the most widely explored among them. GPX4 plays a key role in ferroptosis resistance, and RSL3 is usually used as a ferroptosis inducer via the reduction of GPX4 expression. Ferroptosis induced by RSL3 in Huh7 cells can be reversed by Vit. E or DFOM, there was no difference in cell viability between Vit. E and DFOM; however, the cell death induced by arsenic trioxide could not be reversed by Vit. E. Compared to RSL3 or As2O3 alone, As2O3 plus RSL3 significantly inhibited the growth of Huh7 cells. As2O3 downregulated the expression of GPX4 with increasing concentrations Fig. 8F, Supplementary Fig. 5). Moreover, GPX4 was downregulated to an undetectable level when in combination with RSL3 (Fig. 8).
4.Discussion
In this study, we intended to explore the relationships between ferroptosis-related genes and OS in digestive system neoplasms such as STAD and LIHC. The association of OS and the expression levels of ferroptosis-related genes in LIHC, STAD and normal tissues was comprehensively explored. The analysis of genetic mutations, functional enrichment, and tumour microenvironment indicated that the ferroptosis-related gene signature can effectively predict the prognosis and clinical status of LIHC and STAD patients.
Ferroptosis is a form of regulated cell death dependent on iron and reactive oxygen species and is characterized by the accumulation of lipid peroxides [8]. Ferroptosis, a novel form of regulated cell death induced by iron-dependent lipid peroxidation, plays a key role in the development and drug resistance of tumours. It is reported that ferroptosis has close relation with tumorigenesis and drug resistance [19, 20, 21]. Functional analysis revealed that ferroptosis was closely related to cell cycle, cell metabolism, and immune pathways. Ferroptosis related genes were critical regulators modulating drug resistance, tumor microenvironment infiltration, and cancer stemness. Recent studies have reported that therapy-resistant tumour cells are vulnerable to ferroptosis. Therefore, ferroptosis induction is a potential therapeutic method for treating tumours, especially drug-resistant ones. Our study found that GPX4, AIFM2, ACSL4, and FTH1 were highly expressed in LIHC and STAD by analysing UALCAN. Downregulation or inactivation of GPX4 by RSL3 could induce ferroptosis in LIHC. Higher expression of GPX4 was related to unfavourable OS in LIHC, while GPX4 had no effects on OS in STAD. GPX4 inhibitors, such as RSL3, loaded by nanomaterials have been used in many studies in different tumours [22, 23, 24, 25, 26, 27]. Given that GPX4 expression was higher in LIHC than in normal tissues, nanomaterials loaded with RSL3 may have antitumor effects in LIHC by accumulating in the liver region with less toxicity. In future research, we may perform a study of RSL3 nanoparticles in the treatment of LIHC. AIFM2 is another ferroptosis resistance-related gene screened using CRISPR methods [13, 28]. Apart from GPX4, AIFM2 could also exert a function in ferroptosis resistance. The relationship of AIFM2 to OS was different in diverse cancer types. The OS of AIFM2 in LIHC and STAD had opposite trends, and we analysed the immune infiltration levels by using TIMER data to explain the potential mechanisms involved We found that the AIFM2 expression level in LIHC was not related to the infiltration of B cells, CD4
Furthermore, the expression of FTH1 was increased in human LIHC compared with normal tissues, while there was no significant difference in FTH1 level in STAD tissues compared with normal tissues. There was no obvious correlation in STAD with OS, but FTH1 expression was related to tumour stage in patients with STAD. However, there was no obvious correlation with tumour stage in patients with LIHC, but FTH1 expression was related to OS in LIHC.
Immune checkpoints have been a hot topic in the antitumor area. A ferroptosis-related gene signature was related to overall survival prediction and immune infiltration in lung squamous cell carcinoma. Tianfang Wang found that ferroptosis genes was related to immune infiltration in thyroid papillary carcinoma and associated with overall survival [29, 30]. In this study, we attempted to explain the undiscovered mechanisms of immune cell infiltration. In addition to ferroptosis-related characteristics, including mitochondrial ATP synthesis coupled electron transport and the respiratory electron transport chain, GO and KEGG enrichment analyses of ferroptosis-related genes and their 60 coexpressed genes in LIHC revealed the functions of cell chemotaxis and T-cell migration. Among them, ASCL4 and PTGS2 were coexpressed with cytokines that may be associated with immune cell attraction. Although ASCL4 and PTGS2 may promote immune cell infiltration, ASCL4 and PTGS2 were not related to OS in either tumour type. PTGS2 was coexpressed with CD274 in LIHC. PTGS2 and ACSL4 were coexpressed with CD274 in STAD. The interaction of CD274 with PD-1 hampers antitumor immunity. The higher expression of PTGS2 and ACSL4 in tumour tissues may be used as an indicator of sensitivity to immune checkpoint inhibitors. However, the correlation of ferroptosis-related genes to the sensitivity of immune checkpoint inhibitors may be not exact in some tumor types, so ferroptosis in combination to apoptosis and pyroptosis related genes may be better to predict the therapeutic efficacy.
The ferroptosis related protein was dysregulated after drug treatment, and we usually define it as the key indicator of ferroptosis. In addition, GPX4 inhibitors in combination with other drugs enhanced antitumor therapy [31, 32]. Arsenic trioxide downregulated the protein expression of GPX4. However, the death of Huh7 cells induced by arsenic trioxide treatment could not be reversed by Vit. E. According to previous reports, apoptosis in LIHC could be induced by arsenic trioxide. The above phenomenon indicates that the downregulation of GPX4 may not exert a dominant role in the cell death induced by arsenic trioxide. The higher expression of GPX4 was significantly correlated with overall survival in LIHC. Moreover, GPX4 was higher in LIHC than in normal tissue. Arsenic trioxide may be beneficial in LIHC in the clinic.
5.Conclusion
In this study, we analysed the expression and prognostic value of ferroptosis genes in STAD and LIHC. Our results suggest that GPX4, AIFM2, and FTH1 are dysregulated in LIHC and are candidate prognostic biomarkers in LIHC. The higher expression of PTGS2 and ACSL4 in tumour tissues may be used as an indicator of sensitivity to immune checkpoint inhibitors. Future studies should focus on the regulation of ferroptosis in tumour cells and immune cells in different types of cancers.
Authors’ contributions
Conception: WCP, JHS, JH. Interpretation or analysis of data: WCP, MRJ, HYL. Preparation of the manuscript: WCP, JH. Revision for important intellectual content: WCP, MRJ, HYL, JHS, JH. Supervision: JH.
Funding
This work was supported by the Changning District medical and health research special (CNKW2022Y58). Shenzhen’s Natural Science Foundation-Basic Research Project (JCYJ20210324113811032, 202103181320250 02).
Availability of data and materials
All remaining data are available within the article and supplementary files, or available from the authors upon request.
Consent for publication
All authors have agreed to publish this manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no confict of interests.
Supplementary data
The supplementary files are available to download from http://dx.doi.org/10.3233/CBM-230114.
Acknowledgments
Not applicable.
References
[1] | S. Chuah, J. Lee, Y. Song, H.D. Kim, M. Wasser, N.A. Kaya et al., Uncoupling immune trajectories of response and adverse events from anti-PD-1 immunotherapy in hepatocellular carcinoma, Journal of Hepatology 3: (77) ((2022) ), 683–694. |
[2] | Y. Yang, J. Ouyang, Y. Zhou, J. Zhou and H. Zhao, The CRAFITY score: A promising prognostic predictor for patients with hepatocellular carcinoma treated with tyrosine kinase inhibitor and immunotherapy combinations, Journal of Hepatology 77: (2) ((2022) ), 574–576. |
[3] | Z. Ning, X. Guo, X. Liu, C. Lu, A. Wang, X. Wang et al., USP22 regulates lipidome accumulation by stabilizing PPARγ in hepatocellular carcinoma, Nature Communications 13: (1) ((2022) ), 2187. |
[4] | C.J. Allen, D.T. Pointer Jr., A.N. Blumenthaler, R.J. Mehta, S.E. Hoffe, B.D. Minsky et al., Chemotherapy versus chemotherapy plus chemoradiation as neoadjuvant therapy for resectable gastric adenocarcinoma: A multi-institutional analysis, Annals of Surgery 274: (4) ((2021) ), 544–548. |
[5] | C. Röcken, A. Amallraja, C. Halske, L. Opasic, A. Traulsen, H.M. Behrens et al., Multiscale heterogeneity in gastric adenocarcinoma evolution is an obstacle to precision medicine, Genome Medicine 13: (1) ((2021) ), 177. |
[6] | C. Liang, X. Zhang, M. Yang and X. Dong, Recent progress in ferroptosis inducers for cancer therapy, Advanced Materials (Deerfield Beach, Fla) 31: (51) ((2019) ), e1904197. |
[7] | H. Wang, Y. Cheng, C. Mao, S. Liu, D. Xiao, J. Huang et al., Emerging mechanisms and targeted therapy of ferroptosis in cancer, Molecular Therapy: The Journal of the American Society of Gene Therapy 29: (7) ((2021) ), 2185–2208. |
[8] | T. Xu, W. Ding, X. Ji, X. Ao, Y. Liu, W. Yu et al., Molecular mechanisms of ferroptosis and its role in cancer therapy, Journal of Cellular and Molecular Medicine 23: (8) ((2019) ), 4900–4912. |
[9] | H. Wang, P. Wang and B.T. Zhu, Mechanism of Erastin-Induced Ferroptosis in MDA-MB-231 Human Breast Cancer Cells: Evidence for a Critical Role of Protein Disulfide Isomerase, Molecular and cellular biology, (2022) , e0052221. |
[10] | Y. Su, B. Zhao, L. Zhou, Z. Zhang, Y. Shen, H. Lv et al., Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs, Cancer letters 483: ((2020) ), 127–136. |
[11] | Y. Zou and S.L. Schreiber, Progress in understanding ferroptosis and challenges in its targeting for therapeutic benefit, Cell Chemical Biology 27: (4) ((2020) ), 463–471. |
[12] | J. Ni, K. Chen, J. Zhang and X. Zhang, Inhibition of GPX4 or mTOR overcomes resistance to Lapatinib via promoting ferroptosis in NSCLC cells, Biochemical and Biophysical Research Communications 567: ((2021) ), 154–160. |
[13] | S. Doll, F.P. Freitas, R. Shah, M. Aldrovandi, M.C. da Silva, I. Ingold et al., FSP1 is a glutathione-independent ferroptosis suppressor, Nature 575: (7784) ((2019) ), 693–698. |
[14] | J. Cheng, Y.Q. Fan, B.H. Liu, H. Zhou, J.M. Wang and Q.X. Chen, ACSL4 suppresses glioma cells proliferation via activating ferroptosis, Oncology reports 43: (1) ((2020) ), 147–158. |
[15] | X. Sun, Z. Ou, R. Chen, X. Niu, D. Chen, R. Kang et al., Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells, Hepatology (Baltimore, Md) 63: (1) ((2016) ), 173–184. |
[16] | A. Vandierendonck, H. Degroote, B. Vanderborght, X. Verhelst, A. Geerts, L. Devisscher et al., NOX1 inhibition attenuates the development of a pro-tumorigenic environment in experimental hepatocellular carcinoma, Journal of Experimental & Clinical Cancer Research: CR 40: (1) ((2021) ), 40. |
[17] | N. Yamada, T. Karasawa, H. Kimura, S. Watanabe, T. Komada, R. Kamata et al., Ferroptosis driven by radical oxidation of n-6 polyunsaturated fatty acids mediates acetaminophen-induced acute liver failure, Cell Death & Disease 11: (2) ((2020) ), 144. |
[18] | F. Gao, Y. Zhao, B. Zhang, C. Xiao, Z. Sun, Y. Gao et al., Suppression of lncRNA Gm47283 attenuates myocardial infarction via miR-706/Ptgs2/ferroptosis axis, Bioengineered 13: (4) ((2022) ), 10786–10802. |
[19] | Z. Ren, M. Hu, Z. Wang, J. Ge, X. Zhou, G. Zhang et al., Ferroptosis-related genes in lung adenocarcinoma: Prognostic signature and immune, drug resistance, mutation analysis, Frontiers in Genetics 12: ((2021) ), 672904. |
[20] | W. Wang, F. Pan, X. Lin, J. Yuan, C. Tao and R. Wang, Ferroptosis-related hub genes in hepatocellular carcinoma: Prognostic signature, immune-related, and drug resistance analysis, Frontiers in Genetics 13: ((2022) ), 907331. |
[21] | X. Sheng, Z. Xia, H. Yang and R. Hu, The ubiquitin codes in cellular stress responses, Protein & cell, (2023) . |
[22] | H. Zheng, J. Jiang, S. Xu, W. Liu, Q. Xie, X. Cai et al., Nanoparticle-induced ferroptosis: Detection methods, mechanisms and applications, Nanoscale 13: (4) ((2021) ), 2266–2285. |
[23] | X. Zhang, Y. Ma, J. Wan, J. Yuan, D. Wang, W. Wang et al., Biomimetic nanomaterials triggered ferroptosis for cancer theranostics, Frontiers in Chemistry 9: ((2021) ), 768248. |
[24] | L. Luo, H. Wang, W. Tian, X. Li, Z. Zhu, R. Huang et al., Targeting ferroptosis-based cancer therapy using nanomaterials: Strategies and applications, Theranostics 11: (20) ((2021) ), 9937–9952. |
[25] | W. Li, X. Liu, X. Cheng, W. Zhang, C. Gong, C. Gao et al., Effect of Malt-PEG-Abz@RSL3 micelles on HepG2 cells based on NADPH depletion and GPX4 inhibition in ferroptosis, Journal of Drug Targeting 30: (2) ((2022) ), 208–218. |
[26] | Y. Li, M. Li, L. Liu, C. Xue, Y. Fei, X. Wang et al., Cell-specific metabolic reprogramming of tumors for bioactivatable ferroptosis therapy, ACS Nano 16: (3) ((2022) ), 3965–3984. |
[27] | K. Li, C. Lin, M. Li, K. Xu, Y. He, Y. Mao et al., Multienzyme-like reactivity cooperatively impairs glutathione peroxidase 4 and ferroptosis suppressor protein 1 pathways in triple-negative breast cancer for sensitized ferroptosis therapy, ACS Nano 16: (2) ((2022) ), 2381–2398. |
[28] | P. Koppula, G. Lei, Y. Zhang, Y. Yan, C. Mao, L. Kondiparthi et al., A targetable CoQ-FSP1 axis drives ferroptosis- and radiation-resistance in KEAP1 inactive lung cancers, Nature Communications 13: (1) ((2022) ), 2206. |
[29] | T.W. Miao, D.Q. Yang, F.Y. Chen, Q. Zhu and X. Chen, A ferroptosis-related gene signature for overall survival prediction and immune infiltration in lung squamous cell carcinoma, Bioscience Reports 42: (8) ((2022) ), BSR20212835. |
[30] | R. Lin, C.E. Fogarty, B. Ma, H. Li, G. Ni, X. Liu et al., Identification of ferroptosis genes in immune infiltration and prognosis in thyroid papillary carcinoma using network analysis, BMC Genomics 22: (1) ((2021) ), 576. |
[31] | Q. Wang, C. Bin, Q. Xue, Q. Gao, A. Huang, K. Wang et al., GSTZ1 sensitizes hepatocellular carcinoma cells to sorafenib-induced ferroptosis via inhibition of NRF2/GPX4 axis, Cell Death & Disease 12: (5) ((2021) ), 426. |
[32] | M. Asperti, S. Bellini, E. Grillo, M. Gryzik, L. Cantamessa, R. Ronca et al., H-ferritin suppression and pronounced mitochondrial respiration make Hepatocellular Carcinoma cells sensitive to RSL3-induced ferroptosis, Free Radical Biology & Medicine 169: ((2021) ), 294–303. |




