LncRNA BCAR4 expression predicts the clinical response to neoadjuvant chemotherapy in patients with locally advanced breast cancer
Abstract
BACKGROUND:
Neoadjuvant chemotherapy (NAC) is an important treatment for locally advanced breast cancer (LABC). However, there are no effective biomarkers to predict the efficacy. Therefore, there is an urgent need for new biomarkers to predict the response of LABC to NAC. LncRNA BCAR4 has been detected in a variety of malignant tumor tissues and used as a new biomarker for diagnosis and prognosis. However, LncRNA BCAR4 predicts the response of LABC to NAC is unclear.
OBJECTIVE:
Explore the predictive effect of LncRNA BCAR4 on the efficacy of NAC for LABC in three different evaluation systems.
METHODS:
First, the TCGA database was used to analyze the expression of LncRNA BCAR4 in 33 kinds of malignant tumors, and further explore its expression in breast cancer and its impact on the survival and prognosis of breast cancer. Furthermore, quantitative methods were used to measure the expression level of LncRNA BCAR4 in cancer tissues of 48 LABC patients, and the correlation between LncRNA BCAR4 and clinicopathological status and response to NAC under the evaluation system of 3, RECIST1.1, Miller-Payne (MP) score and whether it reaches pCR,was analyzed.
RESULTS:
TCGA data analysis found that LncRNA is highly expressed in a variety of malignant tumor tissues, including breast cancer. And relatively low expression, the shorter the overall survival time of high expression patients. The high expression of LncRNA BCAR4 is related to the size of the tumor, and there are differences in expression between stage I and other stages, but there is no obvious correlation with the positive lymph node and hormone receptor status. Among the three evaluation systems, only in the RECIST 1.1 evaluation system LncRNA BCAR4 has a predictive effect on NAC for LABC. The expression of LncRNA BCAR4 has no significant correlation with clinical stage, Ki-67% and hormone receptor status, and has no significant correlation with whether patients with locally advanced breast cancer obtain pCR during neoadjuvant chemotherapy.
CONCLUSION:
LncRNA BCAR4 is highly expressed in LABC tissues and may be an effective marker for predicting the efficacy of NAC for LABC.
1.Introduction
Breast cancer is the most common malignant tumor in women worldwide [1]. About 8.5% of Americans and 4% of Europeans have been diagnosed with locally advanced breast cancer [2]. Neoadjuvant chemotherapy (NAC), as the first-line treatment for locally advanced breast cancer (LABC), and studies have confirmed that the pathological complete remission of breast cancer patients in NAC can significantly improve the prognosis, and longer disease-free survival and overall survival in advance [3]. This may be because some patients who fail to obtain pCR (pathological complete response) have developed drug resistance to NAC, which will not only fail to benefit from NAC, but also lead to further progress of the disease and miss the best opportunity for surgery [4, 5]. Therefore, in order to make breast cancer patients better benefit from NAC, it is necessary to find biomarkers that can effectively predict patients’ sensitivity to NAC.
Long Non-coding RNA (LncRNA) is a type of non-coding RNA with a length greater than 200 base pairs. It is different from coding RNA and does not have a typical start codon, promoter conserved region and open reading region, and contains a large number of stop codons. According to their location on the genome, they can be divided into: intergenic LncRNA, intron LncRNA, antisense LncRNA, promoter-related lncRNA, enhancer lncRNA, and untranslated lncRNA [6]. According to their different modes of action, they can be divided into four types of molecules: Signal, Decoy, Guide and Scaffold [7]. LncRNA can participate in complex gene expression regulation through epigenetic modification, transcription and post-transcription [8]. LncRNA is involved in a variety of physiological and pathological processes, especially in malignant tumors [9, 10, 11]. Some LncRNA can promote the invasion and migration of malignant tumors [12], and can be used as a predictive marker for the prognosis of patients [13, 14], or be used as a marker to predict the efficacy of chemotherapy for malignant tumors [15, 16, 17]. LncRNA H19 is highly expressed in the peripheral circulation of breast cancer patients, and compared with high-expressing patients, low-expressing LncRNA H19 in the peripheral circulation can obtain pCR, suggesting that it may be a marker for predicting the efficacy of breast cancer on neoadjuvant chemotherapy [11]. The expression of LncRNA H19 and LnCRNA UCA1 in rectal cancer cells and tissues before and after chemotherapy is significantly different, and can predict the efficacy of rectal cancer on 5FU neoadjuvant chemotherapy [18]. A prediction model constructed from 1 LncRNA and 2 coding genes can predict whether patients with triple-negative breast cancer will get pCR during neoadjuvant chemotherapy [19]. LncRNA HOTAIR in serum can predict the response of breast cancer patients to neoadjuvant chemotherapy [17]. A new signature composed of 36 LncRNAs can predict the efficacy of neoadjuvant chemotherapy and predict whether pCR can be obtained [16]. LncRNA H19 can be used as a marker for the efficacy of NAC for breast cancer [20].
LncRNA BCAR4 (long non-coding RNA breast cancer anti-estrogen resistance 4) was first discovered in the screening of anti-estrogen resistance genes in breast cancer cells, and it was located on chromosome 16p13.13 [21]. LncRNA BCAR4 is highly expressed in a variety of malignant tumor cells or tissues, and because of its high expression, the survival prognosis of patients becomes worse [22, 23, 24], such as osteosarcoma [24], cervical cancer [25], colon cancer [26] and so on. Up-regulation of LncRNA BCAR4 expression promotes tumor cell proliferation, migration and apoptosis, as well as tumor cell resistance [27, 28]. The ErbB2/ErbB3 signaling pathway leads to breast cancer cell resistance to tamoxifen [27], and the activation of Wnt/
The overexpression of LncRNA BCAR4 in gastric cancer can lead to cisplatin resistance, and the abnormal expression of other LncRNA in breast cancer can be used as markers to predict the efficacy of NAC. However, it is not clear whether LncRNA BCAR4 expression is related to the efficacy of NAC and whether it can be used as a marker to predict the efficacy of NAC in LABC. In this study, we investigated the relative expression of LncRNA BCAR4 in LABC and its correlation with the efficacy of NAC under different efficacy evaluation systems, and verified its predictive role in the efficacy of NAC in LABC.
2.Material and methods
2.1Bioinformation analysis
The clinical data and LncRNA sequencing expression data of breast cancer patients were downloaded from TCGA on July 28, 2020. (https://cancer-genome.nih.gov/). They covered raw data on LncRNA-seq of 1109 samples of BC tissues and 113 samples of paracancerous tissues as well as corresponding clinical information.
The expression levels of LncRNA BCAR4 in 33 cancers were analyzed based on TCGA data. LncRNA BCAR4 expression levels and overall survival data in the TCGA database were extracted from starBase (http://starbase.sysu.edu.cn/). Experimen data were divided into high and low groups based on the median level of LncRNA BCAR4 expression.
2.2Patients
Breast cancer tissue specimens were collected from the Department of Pathology, Affiliated Hospital of Zunyi Medical University between January 1, 2018 and January 1, 2019. The inclusion criteria were as follows: 1) Female; 2) Biopsy proven primary invasive breast cancer without distant metastasis; 3) LABC, stage IIB-IIIB; 4) No history of other cancers; 5) Complete NAC with no any prior treatment; 6) Surgery followed by a pathologic examination performed after completion of NAC. The exclusion criteria were as follows:1) Male; 2) The biopsy was diagnosed as carcinoma in situ and early breast cancer; 3) Bilateral breast cancer; 4) Combined with a history of other cancers; 5) NAC treatment not completed; 6) surgery not performed at our hospital or no postoperative pathologic assessment. The included patients were divided into different subgroups according to the chemotherapy response.
The study was approved by the Ethics Committee of Zunyi Medical University.
2.3Pathology prior to neoadjuvant chemotherapy
Breast cancer tissue is core-needle biopsies before NAC, and embedded in paraffin for preservation. Based on nuclear staining of estrogen receptor (ER) and progesterone receptor (PR), we defined
2.4Evaluation of the efficacy of neoadjuvant chemotherapy
The pCR was defined as no residual invasive breast cancer in the histopathology specimen of the breast and axillary lymph nodes (ypT0/ypTisN0). Any remaining positive lymph nodes or residual disease in the breast due to partial tumor response was defined as pathological non-complete response (non-pCR).
The Miller-Payne (MP) grading system was used to evaluate the pathological response in the breast [33]: no change or some alteration in individual malignant cells but no reduction in overall cellularity was considered as score 1; up to a 30% reduction of tumor cells was considered as score 2; between an estimated 30% and 90% reduction of tumor cells was considered as score 3; more than a 90% reduction of tumor cells, such that only small clusters or widely dispersed individual cells remained, was considered as score 4; and no remaining invasive malignant cells was considered as score 5. MP score
The clinical efficacy evaluation were evaluated according to the revised Response Evaluation Criteria in Solid Tumors (RECIST) guidelines version 1.1. At least a 30% decrease in the sum of the diameters of the tumor was considered a partial response (PR). Progressive disease (PD) was defined as an increase of
Table 1
Primer sequence
| Gene | Bidirectional primer sequence |
|---|---|
| LncRNA | Forward: 5 |
| BCAR4 | Reverse: 5 |
| Reverse: 5 | |
| Reverse: 5 |
Figure 1.
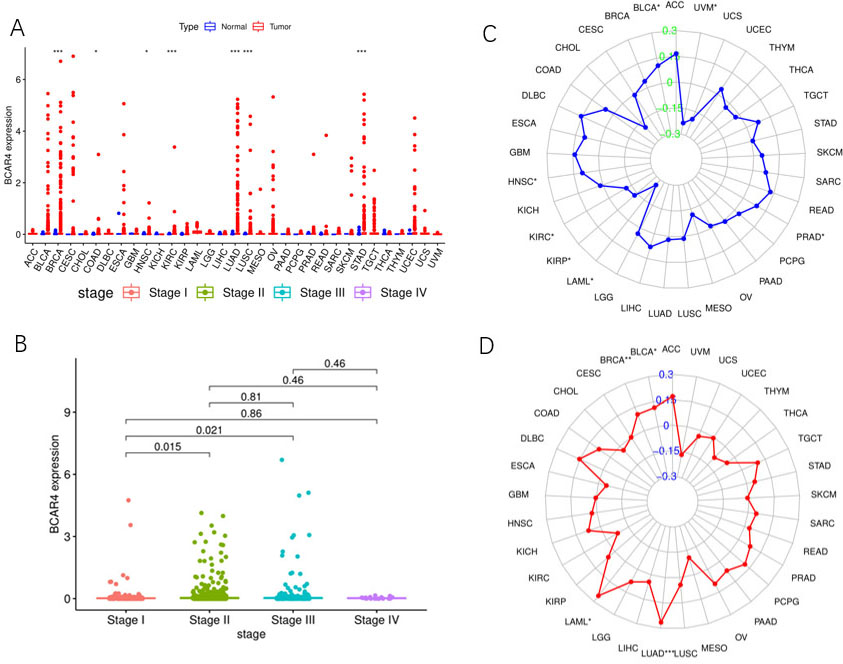
TCGA database pan cancer analysis results. A. The relative expression of LncRNA BCAR4 in 33 kinds of malignant tumor tissues and relative normal tissues. B. The relative expression of LncRNA BCAR4 in different stages of invasive breast cancer. C. MSI correlation of LncRNA BCAR4 in 33 malignant tumors. D. The TMB correlation of LncRNA BCAR4 in 33 malignant tumors.
2.5Neoadjuvant chemotherapy regimen
All patients received 6–8 cycles of NAC with the ‘TAC/TAC(H)’ chemotherapy regimen (T: docetaxel, 75 mg/m
2.6RNA extraction and quantitative real-time PCR analysis (qPCR)
According to the standard guidance of TRIzol reagent (Invitrogen life technologies), total RNAs were extracted. Reverse Transcriptase Kit (SuperScriptTM III Reverse TranscriptaseInvitrogen) was obtained for synthesizing cDNA. qPCR was performed with 2X PCR master mix90 (Arraystar).
2.7Statistical analysis
The edge R package in R was used to analyze the differential expression of LncRNA in the TCGA data obtained from breast cancer. Get relevant survival data through R’s survival package and draw a survival graph. The expression levels of potential LncRNAs were extracted from the downloaded LncRNA information. Statistical analysis between two groups was performed using Student’s
Figure 2.
TCGA database analyzes LncRNA that has differences expressed (DE) in breast cancer tissues. A. Heat map of DE-LncRNA. B. Volcano map of DE-LncRNA. C. Survival curve of differential expression of LncRNA BCAR4 in breast cancer: there is a significant difference in survival between the expression level of LncRNA BCAR4, and the survival prognosis of those with high expression is worse (
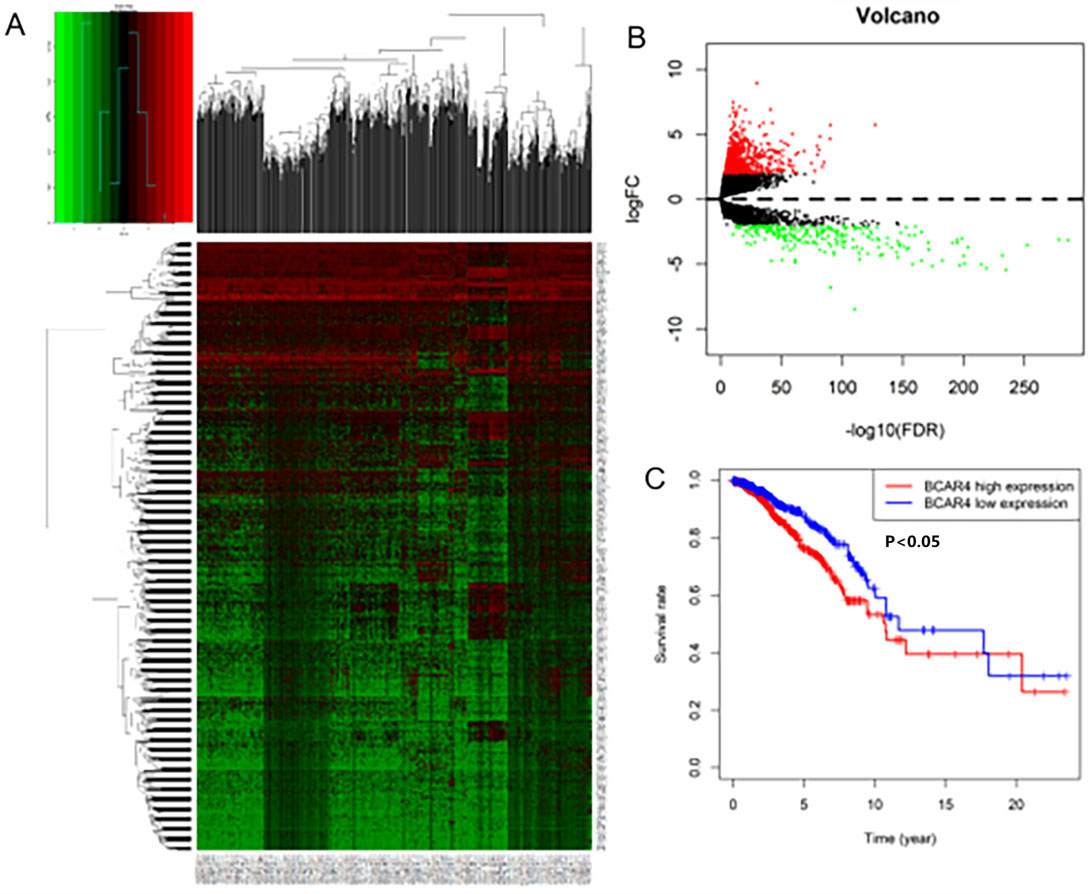
Figure 3.
The relative expression of LncRNA BCAR4. A. The relative expression of LncRNA BCAR4 in breast cancer tissues and normal breast tissues. B. The relative expression of LncRNA BCAR4 in different breast cancer subtypes.
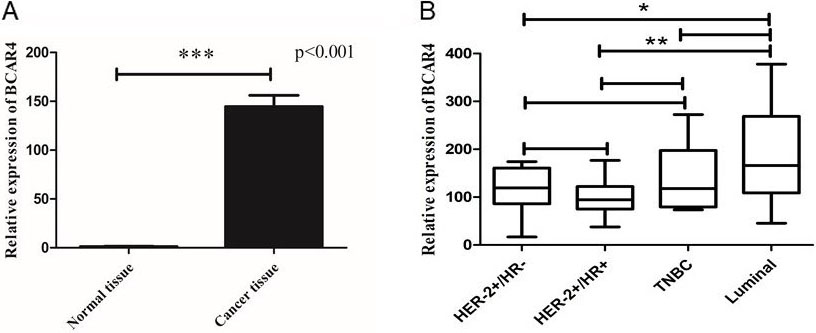
Table 2
Demographic and clinicopathological characteristics of BC patients
| Characteristics | |
|---|---|
| Age (years) | |
| | 28(58.3) |
| | 20(41.7) |
| Grade | |
| G1 | 7(14.6) |
| G2 | 37(77.1) |
| G3 | 4(8.3) |
| Molecular classification | |
| HER2 positive and HR positive type | 10(20.8) |
| HER2 positive and HR negative type | 11(12.9) |
| Luminal type | 17(35.4) |
| TNBC type | 10(20.8) |
| ER status | |
| ER negative | 24(50) |
| ER positive | 24(50) |
| PR status | |
| PR negative | 31(64.6) |
| PR positive | 17(35.4) |
| HER2 status | |
| HER2 negative | 27(56.3) |
| HER2 positive | 21(43.8) |
| Ki67 | |
| | 25(52.1) |
| | 23(47.9) |
| Tumor size | |
| | 29(60.4) |
| | 19(39.6) |
| Clinical stage | |
| II | 33(68.8) |
| III | 15(31.3) |
| Clinical response | |
| cCR | 24(50) |
| cSD | 24(50) |
| Pathological response | |
| pCR | 20(41.7) |
| non-pCR | 28(58.3) |
| Miller-Payne (MP) score | |
| | 19(39.6) |
| | 29(60.4) |
Table 3
Relationship between expression level of lncRNA BCAR4 and clinicopathological characteristics in breast cancer patients undergoing neoadjuvant chemotherapy
| Characteristics | LncRNA BCAR4 level | |
| |
| High level% | Low level% | |||
| Age at entry (years) | 0.196 | 0.658 | ||
| | 13(46.4) | 15(53.6) | ||
| | 8(40.0) | 12(60.0) | ||
| Tumor size (cm) | 7.778 | 0.005 | ||
| | 8(27.6) | 21(72.4) | ||
| | 13(68.4) | 6(31.6) | ||
| Clinical stage | 0.962 | 0.327 | ||
| II | 16(48.5) | 17(51.5) | ||
| III | 5(33.3) | 10(66.7) | ||
| Clinical lymph node stage | 1.011 | 0.603 | ||
| N0 | 3(30.0) | 7(70.0) | ||
| N1 | 14(48.3) | 15(51.7) | ||
| N2 | 4(44.4) | 5(55.6) | ||
| Estrogen receptor (ER) status | 2.116 | 0.146 | ||
| ER positive | 13(54.2) | 11(45.8) | ||
| ER negative | 8(33.3) | 16(66.7) | ||
| Progesterone receptor (PR) status | 2.43 | 0.119 | ||
| PR positive | 10(58.8) | 7(41.2) | ||
| PR negative | 11(35.5) | 20(64.5) | ||
| HER2 status | 6.032 | 0.014 | ||
| HER-2 positive | 6(28.6) | 15(71.4) | ||
| HER-2 negative | 16(59.3) | 11(40.7) | ||
| Ki-67% | 2.98 | 0.585 | ||
| | 11(47.8) | 12(52.2) | ||
| | 10(40.0) | 15(60.0) | ||
| Grade | 2.623 | 0.269 | ||
| G1 | 4(57.1) | 3(42.9) | ||
| G2 | 14(37.8) | 23(62.2) | ||
| G3 | 3(75.0) | 1(25.0) | ||
| Molecular classification | 8.538 | 0.036 | ||
| HER2 positive and HR negative type | 3(27.3) | 8(72.7) | ||
| HER2 positive and HR positive type | 2(20.0) | 8(80.0) | ||
| TNBC | 4(40.0) | 6(60.0) | ||
| Luminal type | 12(70.6) | 5(29.4) | ||
3.Results
3.1Pan-cancer analysis based on 33 cancer types
Through the pan cancer analysis of TCGA database, it was found that LncRNA BCAR4 was significantly higher in various cancer tissues than in matched normal tissues: 7 of 33 tumor tissues (BRCA/COAD/HNSGC/ KIRC/LUAD/LUSC/STAD) (Fig. 1A). And further study found that LncRNA BCAR4 is differentially expressed in different stages of breast cancer. Stage I and Stage II have differential expression,
Further analysis of the MSI and TMB analysis of LncRNA BCAR4 in 33 kinds of malignant tumors found that LncRNA BCAR4 was significantly correlated with MSI in HNSC, KIRC, KIRP, LAML and BLCA, but there was no significant correlation in BRCA (Fig. 1C). In BRCA, BLCA, LGG and other malignant tumors, LncRNA BCAR4 has a significant correlation with TMB (Fig. 1D) (For the abbreviations of 33 types of malignant tumors, see Appendix 1).
3.2lncRNA BCAR4 is highly expressed in locally advanced breast cancer tissues, and it is correlated with poor prognosis in breast cancer patients treated with neoadjuvant chemotherapy
LncRNAs data comes from the TCGA database. A total of 1109 breast cancer tissues and 113 unmatched normal breast tissues were obtained. Through Biotype labeling, 14447 LncRNAs were screened out. Using
3.3The association between LncRNA BCAR4 expression and the clinicopathological characteristics of 48 breast cancer patients treated with neoadjuvant chemotherapy
In order to verify the expression of LncRNA BCAR4 in breast cancer, we used the inclusion and exclusion criteria to include a total of 48 NAC punctured tissues of breast cancer patients before chemotherapy and 10 unpaired normal breast tissues for quantitative real-time PCR (qPCR) (Demographic and clinicopathological characteristics of BC patients shown in Table 2). It was confirmed that LncRNA BCAR4 level was significantly increased in breast cancer patients compared with control group (
The expression levels of LncRNA BCAR4 were categorized into high level group (above the cut-off value) and low level group (below the cut-off value) using the median value as the cut-off value, and the cut-off value is 1.28. Next, we will analyze the correlation between the expression level of LncRNA BCAR4 and the clinicopathological conditions of 48 breast cancer patients. The expression levels of LncRNA BCAR4 were associated with larger tumor size (
3.4The correlation of LncRNA BCAR4 with the response to neoadjuvant chemotherapy
According to breast cancer diagnosis and treatment guidelines, most of the current NAC regimens for breast cancer are composed of anthracyclines, paclitaxel combined with cyclophosphamide. To evaluate the relationship between LncRNA BCAR4 expression level and chemotherapy efficacy, we analyze through 3 different evaluation systems. The clinical responses were evaluated according to the RECIST1.1. In the present study, patients with CR and PR were categorized as responders, and patients with SD and PD as non-responders. The study found that the expression of LncRNA BCAR4 was correlated with the efficacy of RECIST1.1 criteria (
Table 4
Response to neoadjuvant chemotherapy in patients with high or low LncRNA BCAR4 expression No. (%)
| Characteristics | LncRNA BCAR4 level |
|
| |
|---|---|---|---|---|
| High level% | Low level% | |||
| Clinical Response | 10.243 | 0.001 | ||
| cCR | 5(20.8) | 19(79.2) | ||
| cSD | 16(66.7) | 8(33.3) | ||
| Pathological response | 0.196 | 0.658 | ||
| PCR | 8(40.0) | 12(60.0) | ||
| non-PCR | 13(46.4) | 15(53.6) | ||
| Miller-Payne (MP) score | 0.167 | 0.683 | ||
| | 9(47.4) | 10(52.6) | ||
| | 12(41.4) | 17(58.6) | ||
Figure 4.
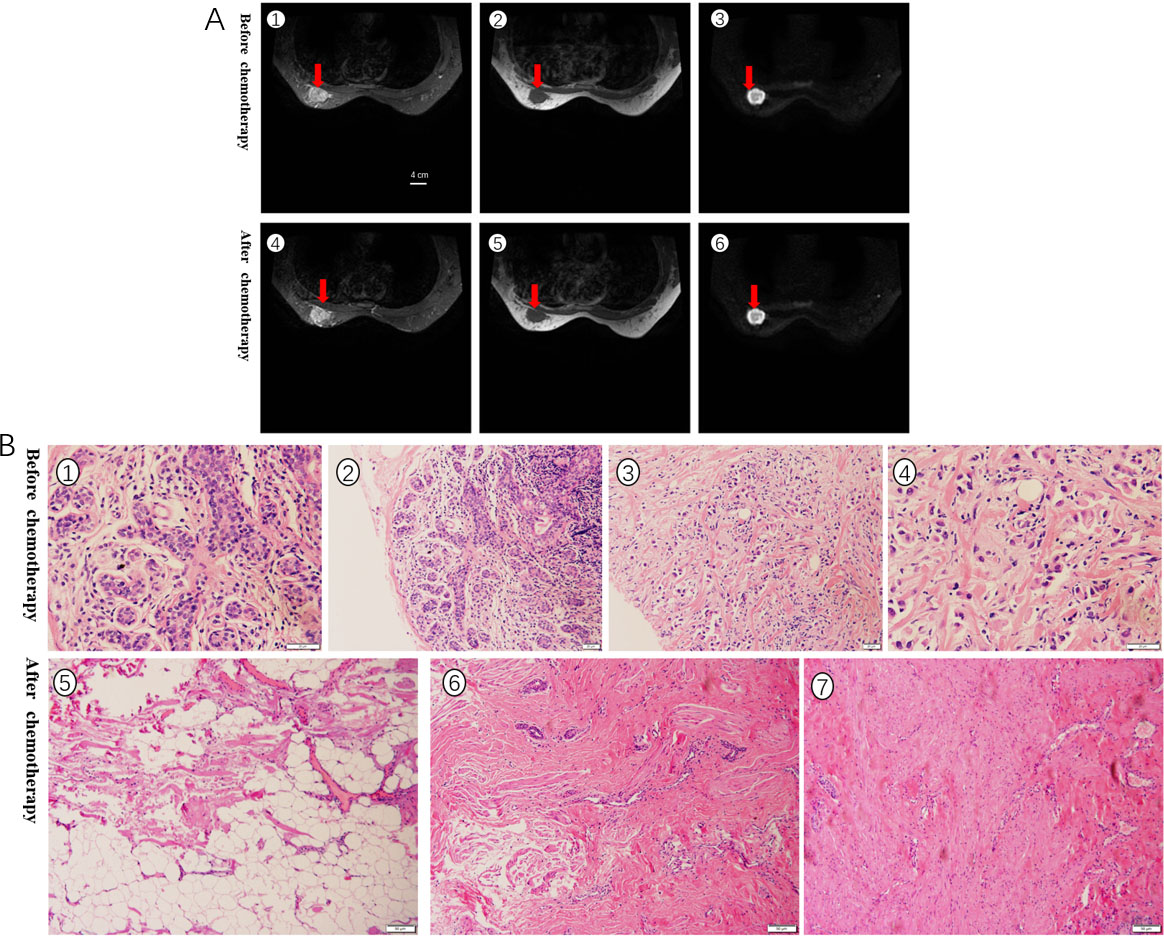
A. RECIST1.1 evaluated the curative effect, the size of the tumor was basically unchanged before and after chemotherapy, and it was evaluated as SD. The red arrow indicates the location of the tumor. B. Pathological evaluation of HE stained specimens, no cancer cells were seen after chemotherapy, and the grade was PCR.
3.5Univariate analysis of the correlation between clinical indicators and neoadjuvant chemotherapy efficacy
In order to verify the expression of LncRNA BCAR4 in breast cancer tissues, this study conducted further in-depth studies in human breast cancer tissues. A total of 48 patients with breast cancer who received NAC were enrolled in this study. According to RECIST1.1, 24 (50%) patients achieved clinical effective (cCR
Table 5
Factors influencing the efficacy of neoadjuvant chemotherapy in breast cancer patients
| Characteristics | cCR | cSD |
|
| PCR | non-PCR |
|
| MP | MP |
|
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age at entry (years) | 0.343 | 0.558 | 2.508 | 0.113 | 3.049 | 0.081 | ||||||
| | 13(46.4) | 15(53.6) | 9(32.1) | 19(67.9) | 14(50.0) | 14(50.0) | ||||||
| | 11(55.0) | 9(45.0) | 11(55.0) | 9(45.0) | 5(25.0) | 15(75.0) | ||||||
| Clinical stage | 0.873 | 0.350 | 1.222 | 0.155 | 6.692 | 0.010 | ||||||
| II | 1854.4 | 1545.5 | 16(48.5) | 17(51.5) | 9(27.3) | 24(72.7) | ||||||
| III | 640.0 | 960.0 | 4(26.7) | 11(73.3) | 10(66.7) | 5(33.3) | ||||||
| Estrogen receptor (ER) status | 1.333 | 0.248 | 0.343 | 0.558 | 0.087 | 0.768 | ||||||
| ER negative | 14(58.3) | 10(41.7) | 11(45.8) | 13(54.2) | 9(37.5) | 15(62.5) | ||||||
| ER positive | 10(41.7) | 14(58.3) | 9(37.5) | 15(62.5) | 10(41.7) | 14(58.3) | ||||||
| Progesterone receptor (PR) status | 0.091 | 0.763 | 1.626 | 0.202 | 4.075 | 0.044 | ||||||
| PR negative | 15(48.4) | 16(51.6) | 15(48.4) | 16(51.6) | 9(29.0) | 22(71.0) | ||||||
| PR positive | 9(52.9) | 8(47.1) | 5(29.4) | 12(70.6) | 10(58.8) | 7(41.2) | ||||||
| HER2 status | 6.857 | 0.009 | 6.291 | 0.012 | 0.035 | 0.853 | ||||||
| HER2 negative | 9(33.3) | 18(66.7) | 7(25.9) | 20(74.1) | 11(40.7) | 16(59.3) | ||||||
| HER2 positive | 15(71.4) | 6(28.6) | 13(61.9) | 8(38.1) | 8(38.1) | 13(61.9) | ||||||
| BCAR4 expression level | 10.243 | 0.001 | 0.196 | 0.658 | 0.167 | 0.683 | ||||||
| High level | 5(23.8) | 16(76.2) | 8(38.1) | 13(61.9) | 9(42.9) | 12(57.1) | ||||||
| Low level | 19(70.4) | 8(29.6) | 12(44.4) | 15(55.6) | 10(37.0) | 17(63.0) | ||||||
| Molecular classification | 9.183 | 0.027 | 6.554 | 0.093 | 3.552 | 0.314 | ||||||
| HER2 positive and HR negative type | 7(63.3) | 4(36.4) | 7(63.6) | 4(36.4) | 5(45.5) | 6(54.5) | ||||||
| HER2 positive and HR positive type | 8(80.0) | 2(20.0) | 6(60.0) | 4(40.0) | 3(30.0) | 7(70.0) | ||||||
| TNBC | 5(50.0) | 5(50.0) | 3(30.0) | 7(70.0) | 2(20.0) | 8(80.0) | ||||||
| Luminal type | 4(23.5) | 13(76.5) | 4(23.5) | 13(76.5) | 9(52.9) | 8(47.1) | ||||||
| Grade | 0.170 | 0.919 | 2.062 | 0.357 | 3.154 | 0.240 | ||||||
| G1 | 3(42.9) | 4(57.1) | 3(42.9) | 4(57.1) | 1(14.3) | 6(85.7) | ||||||
| G2 | 19(51.4) | 18(48.6) | 14(37.8) | 23(62.2) | 17(45.9) | 20(54.1) | ||||||
| G3 | 2(50.0) | 2(50.0) | 3(75.0) | 1(25.0) | 1(25.0) | 3(75.0) | ||||||
| Tumor size (cm) | 4.269 | 0.039 | 0.002 | 0.960 | 0.042 | 0.772 | ||||||
| | 18(62.1) | 11(37.9) | 12(41.4) | 17(58.6) | 11(37.9) | 18(62.1) | ||||||
| | 6(31.6) | 13(68.4) | 8(42.1) | 11(57.9) | 8(42.1) | 11(57.9) | ||||||
| Ki-67 stage (%) | 0.083 | 0.773 | 0.600 | 0.807 | 0.004 | 0.951 | ||||||
| | 13(52.0) | 12(48.0) | 10(40.0) | 15(60.0) | 10(40.0) | 15(60.0) | ||||||
| | 11(47.8) | 12(52.2) | 10(43.5) | 13(56.5) | 9(39.1) | 14(60.9) | ||||||
According to the evaluation of pathological efficacy, 20 of the 48 patients obtained pCR, accounting for 41.7%. Among the many indicators, only HER-2 status has a significant correlation with whether to obtain pCR,
According to the MP score, the score of 0–3 represents the failure of NAC, and the score of 4–5 represents the effectiveness of chemotherapy. Among 48 patients, 29 patients were effective in NAC, accounting for 60.4%. The clinical stage (
The logistic multivariate analysis of clinical parameters related to the clinical efficacy of NAC for breast cancer obtained from the univariate analysis and other literature supports found that HER-2 negative (
Table 6
Multivariate Logistic analysis on the correlation between LncRNA BCAR4 expression and the clinical response (cCR
| Characteristics |
|
| OR | 95%CI |
|---|---|---|---|---|
| HER2 status (positive vs negative) | 1.561 | 0.043 | 4.765 | 1.049–21.657 |
| LncRNA BCAR4 level (low vs high) | 2.091 | 0.008 | 8.091 | 1.729–37.856 |
| Ki-67 stage ( | 0.351 | 0.627 | 1.420 | 0.345–5.841 |
| Clinical stage (II vs III) | 1.657 | 0.068 | 5.241 | 0.883–31.128 |
4.Discussion
NAC has a very important position in the overall management of breast cancer patients, especially for LABC. Due to the lack of sensitive and specific biomarkers, we cannot predict at an early stage whether patients will benefit the most from NAC. Therefore, it is urgent to find an effective molecular marker to predict the efficacy of NAC for breast cancer. Some biomarkers in tumor tissue or body fluid may help to predict the response of breast cancer patients to NAC. For example, the expression of circulating LncRNA H19 is related to the pCR of breast cancer [20]. A study [19] developed a “response score” for NAC for triple negative breast cancer through Gene Expression Omnibus database, and the prediction model consisting of 1 LncRNA and 2 coding genes showed good predictive ability, AUC
In this study, we first performed pan cancer data analysis and single cancer analysis using TCGA database, and found that LncRNA BCAR4 expression was abnormal in a variety of malignant tumor tissues. This has also been confirmed by other studies, such as high expression in breast [34], gastric [35], colon [36], and cervical cancer [25], and its expression is also related to the degree of malignancy of the tumor and has independent prognostic value [37]. Further analysis found that the expression of LncRNA BCAR4 has significant differences between breast cancer stage I and stage II (
Chemotherapy response evaluation can be assessed through a variety of evaluation systems, mainly the RISIST1.1 system and the Miller-Payne score system, as well as whether the pCR is achieved after chemotherapy is frequently used. At present, the most commonly used is whether to achieve pCR after chemotherapy to determine the efficacy of breast cancer chemotherapy, and most studies have found that compared with breast cancer patients who have not obtained pCR, breast cancer patients who have obtained pCR have a longer OS. Compared with the pathological evaluation method of pCR, the Miller-Payne score system can more accurately judge the residual status of breast cancer cells after NAC, and more intuitively reflect the response of breast cancer patients to NAC. Although the clinical evaluation of tumors is subjective, and physicians may not be accurate enough in evaluating tumor size changes, related studies and this study have found that there is a certain correlation between clinical evaluation and pathological evaluation. The most important point is that clinical evaluation can predict the patient’s response to chemotherapy in the first few cycles of chemotherapy for breast cancer patients, and does not need to wait until the entire chemotherapy cycle is completed before surgery to evaluate the efficacy of chemotherapy. This reminds us that the value of the simplest and most intuitive clinical evaluation cannot be ignored. In order to more comprehensively evaluate whether LncRNA BCAR4 can be used as a biomarker of NAC for breast cancer, research and analysis were conducted under these three evaluation systems. This study found that LncRNA BCAR4 has only significant expression differences in the RISISI1.1 evaluation system: low expression levels can obtain effective clinical responses (cCR and cPR) (
How does LncRNA BCAR4 affect the efficacy of chemotherapy? The expression of LncRNA BCAR4 in gastric cancer tissues is higher than that in adjacent tissues [28]; and it regulates Wnt signaling pathway to make gastric cancer cells resistant to cisplatin. And LncRNA BCAR4 also highly expresse in breast cancer tissues, and promotes the aggressiveness of tumor cells, and drives resistance to tamoxifen through the RBB2/ERBB3 signaling pathway [27]. The above research shows that LncRNA BCAR4 is highly expressed in malignant tumors and mediates drug resistance through different signaling pathways. The above research shows that LncRNA BCAR4 is highly expressed in malignant tumors and mediates drug resistance through different signaling pathways. This provides a new research idea for this study to explain the correlation between the expression of LncRNA BCAR4 and the clinical efficacy of NAC for breast cancer: perhaps by changing the sensitivity of cells to drugs, breast cancer cells are resistant to chemotherapy. Therefore, the effect of chemotherapy is different. TMB is an important indicator of the prognosis of immunotherapy. In the CheckMate-026 [39] clinical study, the researchers used Nivolumab and platinum-based chemotherapy respectively for the first-line treatment of advanced NSCLC. At the same time, WES was used to measure TMB, and the patients were divided into three groups (
This study also has many shortcomings. First of all, this study included a small sample size, only 48 breast cancer patients were included, which could not fully reflect the response of LncRNA BCAR4 to the efficacy of NAC for breast cancer. The study specimens need to be expanded for further study. Secondly, this study initially found that the expression of LncRNA BCAR4 is related to the clinical efficacy of NAC for breast cancer. However, there is no further research and analysis of its mechanism, so further research is still needed, and it will lay a solid foundation for LncRNA BCAR4 as a biomarker for the treatment of breast cancer in the future.
In conclusion, this study confirmed that LncRNA BCAR4 is significantly higher expressed in locally advanced breast cancer tissues than in normal breast tissues, and its expression status has a significant correlation with tumor size, molecular typing, and HER-2 status. And further research and analysis found that the level of its expression has a significant correlation with the clinical efficacy of neoadjuvant chemotherapy for locally advanced breast cancer, which can be used as a targeted predictor.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China, No. 81860469; the Zunyi Science and Technology Bureau, China, No. Grant Zunshi kehe HZ (2019) No. 85; the Honghuagang District Science and Technology Bureau of Zunyi City, China, No. Grant Zunhong Kehe Shezi (2018) No. 12.
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
Conception: Fengjiao gan, Qing Luo and Suhong Sun
Interpretation or analysis of data: Fengjiao Gan and Yi Li
Preparation of the manuscript: Fengjiao Gan, Kang Hu, Yan Li Mengxi Xu, Tie Zhou and Shun Wu
Revision for important intellectual content: Fengjiao Gan, Suhong Sun and Qing Luo
Supervision: Qing Luo and Suhong Sun
References
[1] | Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J Clin 70: ((2020) ), 313. |
[2] | EBCTCG (Early Breast Cancer Trialists’ Collaborative Group), P. McGale, C. Taylor, C. Correa, D. Cutter, F. Duane, M. Ewertz, R. Gray, G. Mannu, R. Peto, T. Whelan, Y. Wang, Z. Wang and S. Darby, Effect of radiotherapy after mastectomy and axillary surgery on 10-year recurrence and 20-year breast cancer mortality: Meta-analysis of individual patient data for 8135 women in 22 randomised trials, Lancet 383: ((2014) ), 2127–2135. |
[3] | P. Cortazar, L. Zhang, M. Untch, K. Mehta, J.P. Costantino, N. Wolmark, H. Bonnefoi, D. Cameron, L. Gianni, P. Valagussa, S.M. Swain, T. Prowell, S. Loibl, D.L. Wickerham, J. Bogaerts, J. Baselga, C. Perou, G. Blumenthal, J. Blohmer, E.P. Mamounas, J. Bergh, V. Semiglazov, R. Justice, H. Eidtmann, S. Paik, M. Piccart, R. Sridhara, P.A. Fasching, L. Slaets, S. Tang, B. Gerber, C.E. Geyer, Jr, R. Pazdur, N. Ditsch, P. Rastogi, W. Eiermann and G. von Minckwitz, Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis, Lancet 384: ((2014) ), 164–172. |
[4] | M. Tanioka, C. Fan, J.S. Parker, K.A. Hoadley, Z. Hu, Y. Li, T.M. Hyslop, B.N. Pitcher, M.G. Soloway, P.A. Spears, L.N. Henry, S. Tolaney, C.T. Dang, I.E. Krop, L.N. Harris, D.A. Berry, E.R. Mardis, E.P. Winer, C.A. Hudis, L.A. Carey and C.M. Perou, Integrated analysis of RNA and DNA from the phase III trial CALGB 40601 identifies predictors of response to trastuzumab-based neoadjuvant chemotherapy in HER2-Positive breast cancer, Clin Cancer Res 24: ((2018) ), 5292–5304. |
[5] | D. Fumagalli, D. Venet, M. Ignatiadis, H.A. Azim Jr, M. Maetens, F. Roth, R. Salgado, I. Bradbury, L. Pusztai, N. Harbeck, H. Gomez, T.W. Chang, M.A. Coccia-Portugal, S. Di Cosimo, E. de Azambuja, L. de la Pea, P. Nuciforo, J.C. Brase, J. Huober, J. Baselga, M. Piccart, S. Loi and C. Sotiriou, RNA sequencing to predict response to neoadjuvant Anti-HER2 therapy: A secondary analysis of the NeoALTTO randomized clinical trial, JAMA Oncol 3: ((2017) ), 227–234. |
[6] | L. Ma, V.B. Bajic and Z. Zhang, On the classification of long non-coding RNAs, RNA Biol 10: ((2013) ), 925–933. |
[7] | K.C. Wang and H.Y. Chang, Molecular mechanisms of long noncoding RNAs, Mol Cell 43: ((2011) ), 904–914. |
[8] | J. Zhu, H. Fu, Y. Wu and X. Zheng, Function of lncRNAs and approaches to lncRNA-protein interactions, Sci China Life Sci 56: ((2013) ), 876–885. |
[9] | M.N. Cabili, C. Trapnell, L. Goff, M. Koziol, B. Tazon-Vega, A. Regev and J.L. Rinn, Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses, Genes Dev 25: ((2011) ), 1915–1927. |
[10] | S. Geisler and J. Coller, RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts, Nat Rev Mol Cell Biol 14: ((2013) ), 699–712. |
[11] | J.R. Prensner and A.M. Chinnaiyan, The emergence of lncRNAs in cancer biology, Cancer Discov 1: ((2011) ), 391–407. |
[12] | W.X. Peng, R.Z. He, Z. Zhang, L. Yang and Y.Y. Mo, LINC00346 promotes pancreatic cancer progression through the CTCF-mediated Myc transcription, Oncogene 38: ((2019) ), 6770–6780. |
[13] | J.J. Qiu, X.J. Lin, X.Y. Tang, T.T. Zheng, X.Y. Zhang and K.Q. Hua, Long noncoding RNA TC0101441 induces epithelial-mesenchymal transition in epithelial ovarian cancer metastasis by downregulating KiSS1, Int J Cancer 146: ((2020) ), 2588–2598. |
[14] | C. Chen, Y. Feng, J. Wang, Y. Liang and W. Zou, Long non-coding RNA SNHG15 in various cancers: A meta and bioinformatic analysis, BMC Cancer 20: ((2020) ), 1156. |
[15] | W. Han, X. Du, M. Liu, J. Wang, L. Sun and Y. Li, Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer, Int J Biol Macromol 121: ((2019) ), 270–278. |
[16] | G. Wang, X. Chen, Y. Liang, W. Wang and K. Shen, A long noncoding RNA signature that predicts pathological complete remission rate sensitively in neoadjuvant treatment of breast cancer, Transl Oncol 10: ((2017) ), 988–997. |
[17] | R. Lu, J. Zhang, W. Zhang, Y. Huang, N. Wang, Q. Zhang and S. Qu, Circulating HOTAIR expression predicts the clinical response to neoadjuvant chemotherapy in patients with breast cancer, Cancer Biomark 22: ((2018) ), 249–256. |
[18] | Y. Yokoyama, T. Sakatani, R. Wada, K. Ishino, M. Kudo, M. Koizumi, T. Yamada, H. Yoshida and Z. Naito, In vitro and in vivo studies on the association of long non-coding RNAs H19 and urothelial cancer associated 1 with the susceptibility to 5-fluorouracil in rectal cancer, Int J Oncol 55: ((2019) ), 1361–1371. |
[19] | Q. Wang, C. Li, P. Tang, R. Ji, S. Chen and J. Wen, A minimal lncRNA-mRNA signature predicts sensitivity to neoadjuvant chemotherapy in triple-negative breast cancer, Cell Physiol Biochem 48: ((2018) ), 2539–2548. |
[20] | E. Özgür, F. Ferhatolu, F. en, P. Saip and U. Gezer, Circulating lncRNA H19 may be a useful marker of response to neoadjuvant chemotherapy in breast cancer, Cancer Biomark 27: ((2020) ), 11–17. |
[21] | D. Meijer, T. van Agthoven, P.T. Bosma, K. Nooter and L.C. Dorssers, Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells, Mol Cancer Res 4: ((2006) ), 379–386. |
[22] | A. Wang, J. Meng, H. Liu, C. Li and Z. Zhou, Long non-coding RNA BCAR4 promotes liver cancer progression by regulating proliferation, migration and invasion, Oncol Lett 20: ((2020) ), 2779–2787. |
[23] | W. Zhao, Z. Wang, X. Fang, N. Li and J. Fang, Long noncoding RNA Breast cancer antiestrogen resistance 4 is associated with cancer progression and its significant prognostic value, J Cell Physiol 234: ((2019) ), 12956–12963. |
[24] | L. Ju, Y.M. Zhou and G.S. Yang, Up-regulation of long non-coding RNA BCAR4 predicts a poor prognosis in patients with osteosarcoma, and promotes cell invasion and metastasis, Eur Rev Med Pharmacol Sci 20: ((2016) ), 4445–4451. |
[25] | R. Zou, X. Chen, X. Jin, S. Li, R. Ou, J. Xue, X. Yan, L. Chen, Y. Hu and H. Zhu, Up-regulated BCAR4 contributes to proliferation and migration of cervical cancer cells, Surg Oncol 27: ((2018) ), 306–313. |
[26] | S. Ouyang, X. Zheng, X. Zhou, Z. Chen, X. Yang and M. Xie, LncRNA BCAR4 promotes colon cancer progression via activating Wnt/β-catenin signaling, Oncotarget 8: ((2017) ), 92815–92826. |
[27] | M.F. Godinho, A.M. Sieuwerts, M.P. Look, D. Meijer, J.A. Foekens, L.C. Dorssers and T. van Agthoven, Relevance of BCAR4 in tamoxifen resistance and tumour aggressiveness of human breast cancer, Br J Cancer 103: ((2010) ), 1284–1291. |
[28] | L. Wang, Q. Chunyan, Y. Zhou, Q. He, Y. Ma, Y. Ga and X. Wang, BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients, Eur Rev Med Pharmacol Sci 21: ((2017) ), 4064–4070. |
[29] | F. Chen, J. Mo and L. Zhang, Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription, Tumour Biol 37: ((2016) ), 13403–13412. |
[30] | N. Li, W.J. Gao and N.S. Liu, LncRNA BCAR4 promotes proliferation, invasion and metastasis of non-small cell lung cancer cells by affecting epithelial-mesenchymal transition, Eur Rev Med Pharmacol Sci 21: ((2017) ), 2075–2086. |
[31] | J. Gong, H. Zhang, L. He and L. Wang, J. Wang, Increased expression of long non-coding rna bcar4 is predictive of poor prognosis in patients with non-small cell lung cancer, Tohoku J Exp Med 241: ((2017) ), 29–34. |
[32] | T. Fujii, T. Kogawa, W. Dong, A.A. Sahin, S. Moulder, J.K. Litton, D. Tripathy, T. Iwamoto, K.K. Hunt, L. Pusztai, B. Lim, Y. Shen and N.T. Ueno, Revisiting the definition of estrogen receptor positivity in HER2-negative primary breast cancer, Ann Oncol 28: ((2017) ), 2420–2428. |
[33] | K.N. Ogston, I.D. Miller, S. Payne, A.W. Hutcheon, T.K. Sarkar, I. Smith, A. Schofield and S.D. Heys, A new histological grading system to assess response of breast cancers to primary chemotherapy: Prognostic significance and survival, Breast 12: ((2003) ), 320–327. |
[34] | M.F. Godinho, J.D. Wulfkuhle, M.P. Look, A.M. Sieuwerts, S. Sleijfer, J.A. Foekens, E.F. Petricoin 3rd, L.C. Dorssers and T. van Agthoven, BCAR4 induces antioestrogen resistance but sensitises breast cancer to lapatinib, Br J Cancer 107: ((2012) ), 947–955. |
[35] | G.R. Zhou, D.P. Huang, Z.F. Sun and X.F. Zhang, Long non-coding RNA BCAR4 accelerates cell proliferation and suppresses cell apoptosis in gastric cancer via regulating MAPK/ERK signaling, Eur Rev Med Pharmacol Sci 24: ((2020) ), 3657–3664. |
[36] | H. Siddique, A. Al-Ghafari, H. Choudhry, S. AlTurki, H. Alshaibi, H. Al Doghaither and H. Alsufiani, Long noncoding RNAs as prognostic markers for colorectal cancer in saudi patients, Genet Test Mol Biomarkers 23: ((2019) ), 509–514. |
[37] | C. Tu, X. Ren, J. He, C. Zhang, R. Chen, W. Wang and Z. Li, The value of LncRNA BCAR4 as a prognostic biomarker on clinical outcomes in human cancers, J Cancer 10: ((2019) ), 5992–6002. |
[38] | X. Li, D. Dai, B. Chen, H. Tang and W. Wei, Oncological outcome of complete response after neoadjuvant chemotherapy for breast conserving surgery: A systematic review and meta-analysis, World J Surg Oncol 15: ((2017) ), 210. |
[39] | E. Nadal, B. Massuti, M. Dmine, R. Garca-Campelo, M. Cobo and E. Felip, Immunotherapy with checkpoint inhibitors in non-small cell lung cancer: Insights from long-term survivors, Cancer Immunol Immunother 68: ((2019) ), 341–352. |
Appendices
Appendix
Appendix
| Abbreviation | Full name | Abbreviation | Full name | Abbreviation | Full name |
|---|---|---|---|---|---|
| ACC | Adrenocortical Carcinoma | KIRC | Kidney Renal Clear Cell Carcinoma | PD | Progressive Disease |
| BCAR4 | Breast Cancer Anti-Estrogen Resistance 4 | KIRP | Kidney Renal Papillary Cell Carcinoma | PR | Partial Response |
| BLCA | Bladder Urothelial Carcinoma | LABC | Locally Advanced Breast Cancer | PRAD | Prostate Adenocarcinoma |
| BRCA | Breast Invasive Carcinoma | LAML | Acute Myeloid Leukemia | READ | Rectum Adenocarcinoma |
| CESC | Cervical Squamous Cell Carcinoma And Endocervical Adenocarcinoma | LGG | Brain Lower Grade Glioma | SARC | Sarcoma |
| CHOL | Cholangiocarcinoma | LIHC | Liver Hepatocellular Carcinoma | SD | Progressive Disease |
| COAD | Colon Adenocarcinoma | LUAD | Lung Adenocarcinoma | SKCM | Skin Cutaneous Melanoma |
| CR | Complete Response | LUSC | Lung Squamous Cell Carcinoma | STAD | Stomach Adenocarcinoma |
| DLBC | Diffuse Large B-Cell Lymphoma | MESO | Mesothelioma | TGCT | Testicular Germ Cell Tumors |
| ESCA | Esophageal Carcinoma | MP | Miller-Payne | THCA | Thyroid Carcinoma |
| GBM | Glioblastoma Multiforme | MSI | Microsatellite Instability | THYM | Thymoma |
| HNSC | Head And Neck Squamous Cell Carcinoma | NAC | Neoadjuvant Chemotherapy | TMB | Tumor Mutational Burden |
| IHC | Immunohistochemistry | OV | Ovarian Serous Cystadenocarcinoma | UCEC | Uterine Corpus Endometrial Carcinoma |
| ISH | In Situ Hybridization | PAAD | Pancreatic Adenocarcinoma | UCS | Uterine Carcinosarcoma |
| KICH | Kidney Chromophobe | PCPG | Pheochromocytoma And Paraganglioma | UVM | Uveal Melanoma |
Abbreviation list




