MiR-511 mimic transfection inhibits the proliferation, invasion of osteosarcoma cells and reduces metastatic osteosarcoma tumor burden in nude mice via targeting MAPK1
Abstract
Osteosarcoma, a highly aggressive cancer, can rapidly metastasize to distant organs such as lung, liver, brain. Despite much progress in the therapeutic regime has been made, the prognosis of osteosarcoma remains poor. In present study, microRNA-511 (miR-511) is lowly expressed in osteosarcoma cells, including MG63, U-2 OS, Saos-2 cells, while mitogen activated protein kinase1 (MAPK1) is highly expressed in osteosarcoma cells. Interestingly, MAPK1 might be a target of miR-511. We found that overexpression of miR-511 by miR-511 mimic transfection may result to low expression of MAPK1. Further study showed that miR-511 mimic inhibits the development of osteosarcoma MG63 cell, including proliferation and invasion. Moreover, miR-511 mimic transfection reduces metastatic osteosarcoma tumor burden in nude mice. These activities are mediated by targeting MAPK1. Our study provides a new sight for the molecular pathogenesis of osteosarcoma.
1.Introduction
Osteosarcoma, one of the most common forms of solid bone cancer in children and adolescents, often attack proximal humerus, proximal tibia and the metaphyseal regions of distal femur with high morbidity and mortality [1]. Typical symptoms of osteosarcoma include joint movement limitation, trabecular fracture, local swelling and pain [2]. The combination of surgery, post-operative radiotherapy and chemotherapy are the traditional therapies of osteosarcoma. Unfortunately, radiotherapy and chemotherapy usually result in some life-threatening side effects such as nephrotoxicity, infertility and cardiotoxicity. Therefore, it is important to understand the underlying molecular mechanism of tumorigenesis and explore a new therapy regime to prevent further deterioration of osteosarcoma.
More and more researches have demonstrated that microRNAs (miRNAs) are key predictors of the severity and prognosis of cancer. Studies over the last decades have indicated that a variety of abnormal miRNAs were existed in the pathogenesis of osteosarcoma [3]. microRNA-511(miR-511) has been demonstrated to be dyregulated in diverse human cancer types including breast cancer [4], colorectal cancer [5], lung adenocarcinoma [6]. However, whether miR-511 influences the progression of osteosarcoma remains unknown yet. The mitogen activated protein kinase (MAPK) signaling cascade are membrane-to-nucleus signaling modules and play important roles in multiple physiological processes [7, 8]. MAPK1 works as a downstream oncogene of MAPK signaling pathway. Furthermore, some previous studies suggested that MAPK1 was significantly up-regulated in various types of human cancers, such as ovarian cancer [7], cervical cancer [9] and gastric cancer [10]. It is likely that MAPK1 also contributes to the development of osteosarcoma.
In Targetscan Website, we can see that the 3’UTR of MAPK1 is perfectly complementary with miR-511-3p. Therefore, we speculate that MAPK1 might be a target of miR-511-3p and restoring miR-511-3p expression may suppress osteosarcoma via targeting MAPK signaling pathway. In our research, we investigated the impact of miR-511 mimic on the proliferation and invasion of osteosarcoma and the potential molecular mechanisms in vitro and in vivo.
2.Material and methods
2.1Cell lines and cell culture
Osteosarcoma cell lines MG63, U-2 OS, Saos-2 and osteoblast hFOB1.19 cell lines were purchased from American Type Culture Collection (ATCC; Rockville, MD, USA). MG63 cells were cultured in Eagle’s Minimum Essential Medium (EMEM; Gibco, Rockville, MD, USA) supplemented with 10% FBS. U-2 OS cells were cultured in McCoy’s 5a Medium Modified supplemented with 10% FBS. Saos-2 cells were cultured in McCoy’s 5a Medium Modified supplemented with 15% FBS. hFOB1.19 cells were kept in 1:1 mixture of Ham’s F12 Medium Dulbecco’s Modified Eagle’s Medium, with 2.5 mM L-glutamine added 0.3 mg/ml G418 and 10% FBS. All cells except hFOB1.19 cells (34
2.2Animals and ethics
Nude mice (6–8 weeks old) were obtained from North Sichuan Medical College Affiliated Hospital. The experimental protocol was supported by the Committee on the Ethics of Animal Experiments of North Sichuan Medical College Affiliated Hospital.
2.3Cell transfection
The interaction probability between miR-511 and MAPK1 was predicted by TargetScan. MiR-511 mimic wild type (wt), miR-511 mutant (mut) (Fig. 1A), mimic control, pcDNA-MAPK1 and pcDNA vector were obtained from GenePharma Co., Ltd (Shanghai, China). MG63 cells were transfected with pcDNA-MAPK1 (20
2.4Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells cell lines (MG63, U2OS, SaOS2 and hFOB1.19) using TRIzol reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. Equal amounts of RNA were reversely-transcribed to cDNA with SuperScript Reverse Transcriptase Kit (Vazyme, Nanjing, China). Then mRNA levels of miR-511 and MAPK1 were analyzed by SYBR Green PCR Master Mix (Vazyme) in a Fast Real-time PCR 7300 System (Applied Biosystems, Foster City, USA). Primers for MAPK1, miR-511-3p, snRNA U6 were synthesized by Sangon Bioteh (Shanghai, China). Data were analyzed according to the comparative Ct method also referred to as the 2
2.5Transwell assay
MG63 cells were seeded (5
2.6Matrigel tube formation assay
Matrigel tube formation assay was conducted as previous reported [11]. When MG63 cell density reached to 90%, the growth medium was replaced by serum-free medium, and cells were incubated for 24 h at 37
2.7BrdU staining
The transfected cells were seeded into 24-well upper chamber (Corning, NY, USA) pre-coated with matrigel (BD, New Jersey, USA). The lower chamber was filled with RPMI-1640 medium containing 10% FBS. 24 h later, the invasive cells were stained with crystal violet (Sigma, MO, USA) for 15 min. Photos were taken under optical microscope (Olympus).
2.8Western-blot assay
After transfection for 48 h, the expressions of Ki67, cleaved caspase-3, vascular endothelial growth factor (VEGF),
2.9TOP/FOP-FLASH luciferase reporter assay
TOP/FOP-FLASH assay was performed as reported elsewhere [13]. Briefly, transfected MG63 cells in each group were incubated with mixture containing serum-free DMEM (20
2.10Osteosarcoma model and groups
Nude mice were randomly divided into two groups (
2.11Immunohistochemical staining
3
2.12HE staining
HE staining was performed for observation of tumor focal in lung and liver tissue of mice. Briefly, the lung and liver tissues were fixed in 10% neutral-buffered formalin, paraffin embedded, and sectioned at 4 mm thickness according to the standard procedure. The sections were deparaffinized and hydrated gradually, then examined by H&E staining.
2.13Statistical analysis
All statistical analyses were conducted using the SPSS statistical software program (version 17.0; SPSS, Inc., Chicago, IL, USA). The data presented in this study were expressed as means
3.Results
3.1The expressions of miR-511 and MAPK1 were dysregulated in osteosarcoma cells
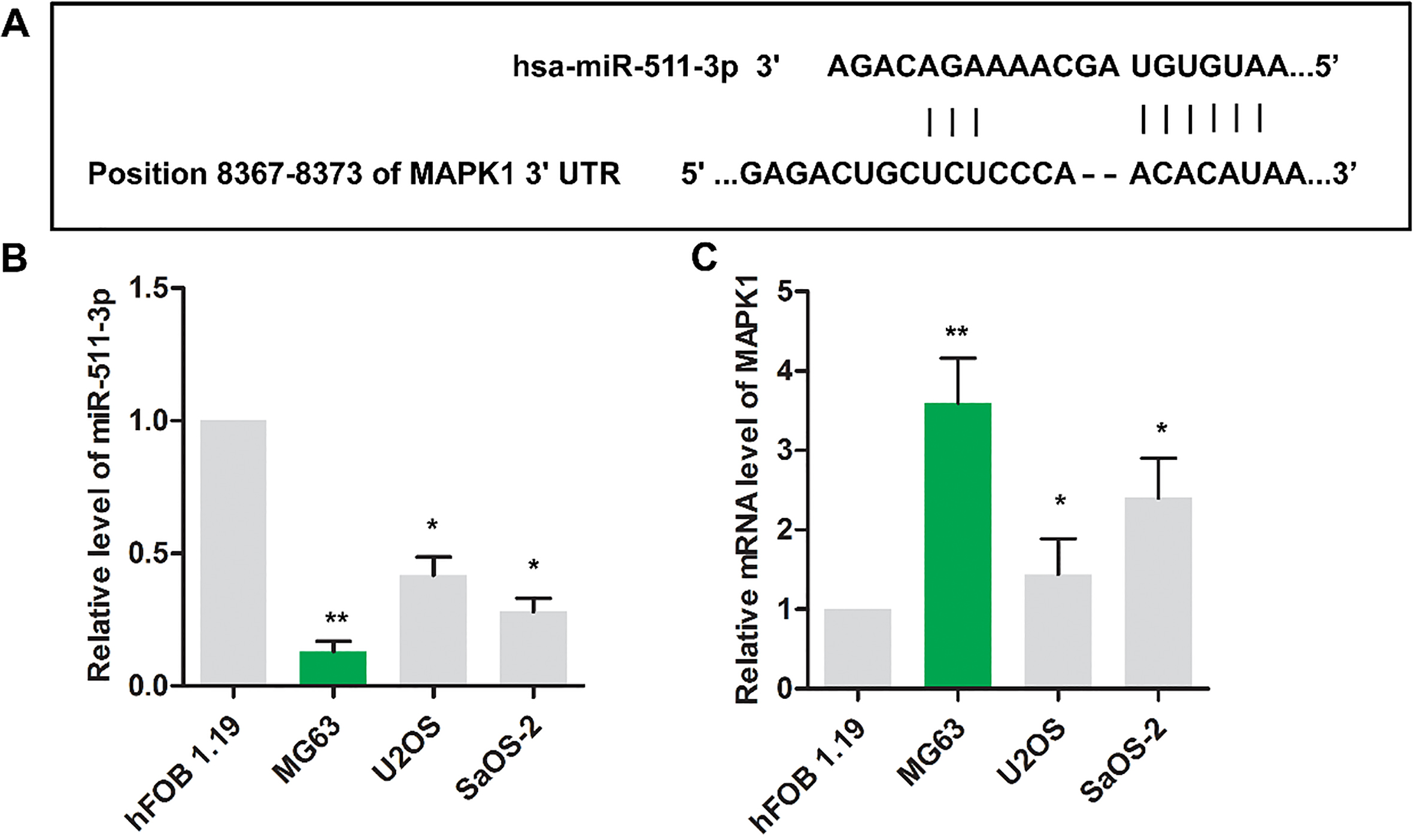
The binding sites between miR-511 and MAPK1 were shown in Fig. 1A via Targetscan website. As shown in Fig. 1B, the level of miR-511 was decreased in osteosarcoma cells (MG63, U2OS and SaOS-2) compared with that in nonneoplastic cell (hFOB1.19) (
Figure 1.
The expression of miR-511 and MAPK1 were dysregulated in osteosarcoma cells. (A) The binding sites between miR-511 and MAPK1 were predicted via Targetscan website. The relative expressions miR-511(B) and MAPK1(C) were detected using RT-PCR in HFOB1.19, MG63, Saos2 and U2OS cell lines.
3.2MAPK1 is a target of miR-511
As the results shown in Fig. 2A, miR-511 mimic transfection led to high expression level of miR-511, indicating the high transfection efficiency. Interestingly, miR-511 mimic transfection significantly reduced the mRNA level (Fig. 2B) and protein level (Fig. 2C and D) of MAPK1, which indicated that a reverse relationship was found in miR-511 and MAPK1. Luciferase reporter assay result shown in Fig. 2E suggested that miR-511 might target on MAPK1 and negatively regulate the expression of MAPK1.
Figure 2.
MAPK1 is a target of miR-511. MG63 cells were randomly divided into 3 groups. Control group: normal MG63 cells; mimic NC group: MG63 cells were transfected with scramble mimics; miR-511 mimic group: MG63 cells were transfected with miR-511 mimic; The relative expressions miR-511(A) and MAPK1(B) were detected using RT-PCR in MG63 cells.
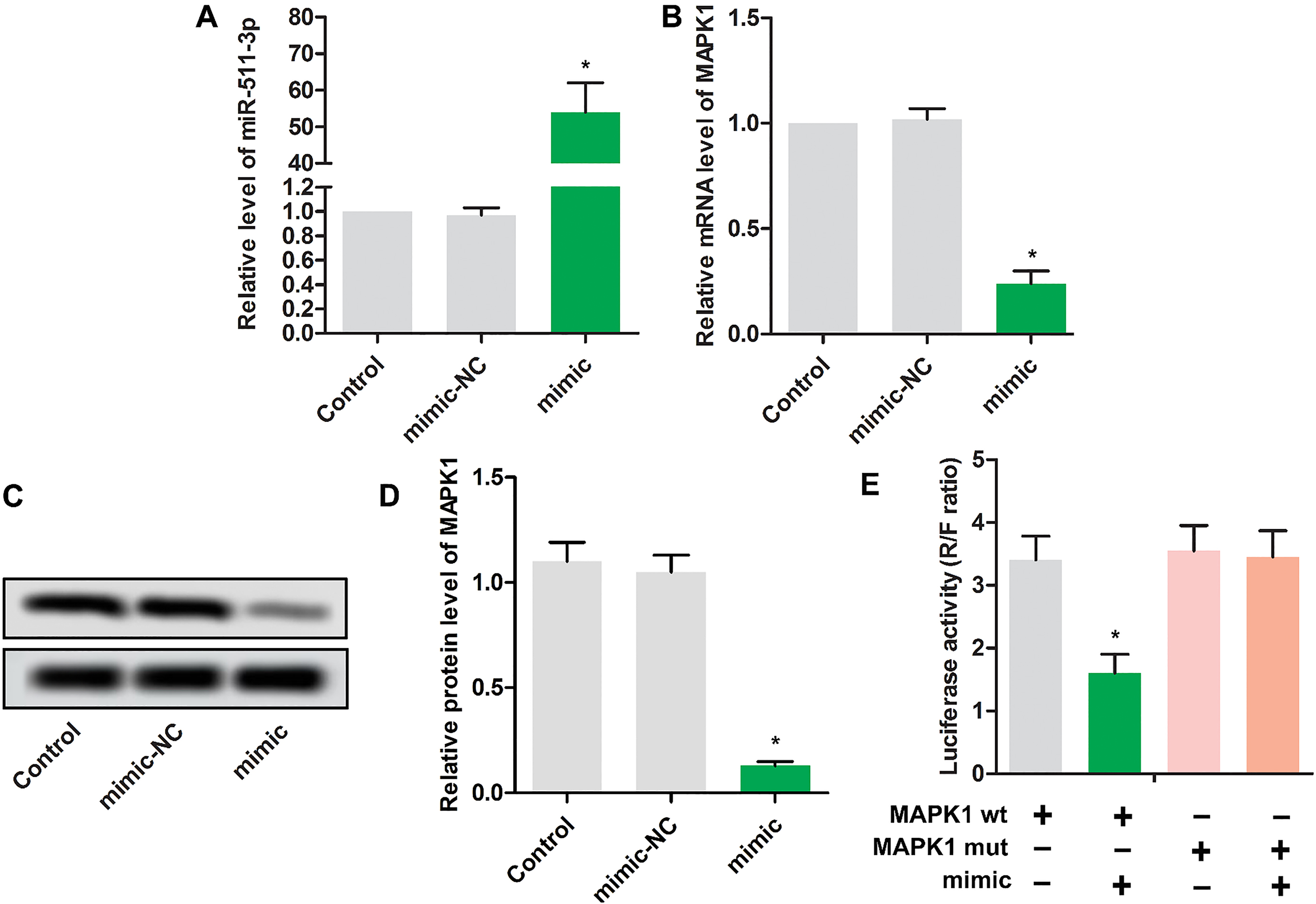
Figure 3.
Up-regulated miR-511 inhibited the proliferation of MG63 cells. MG63 cells were randomly divided into 3 groups. Control group: normal MG63 cells; pcDNA-MAPK1 group: MG63 cells were transfected with pcDNA-MAPK1; vector group: MG63 cells were transfected with pcDNA. (A) The relative mRNA expression MAPK1 was detected using RT-PCR in MG63 cells.
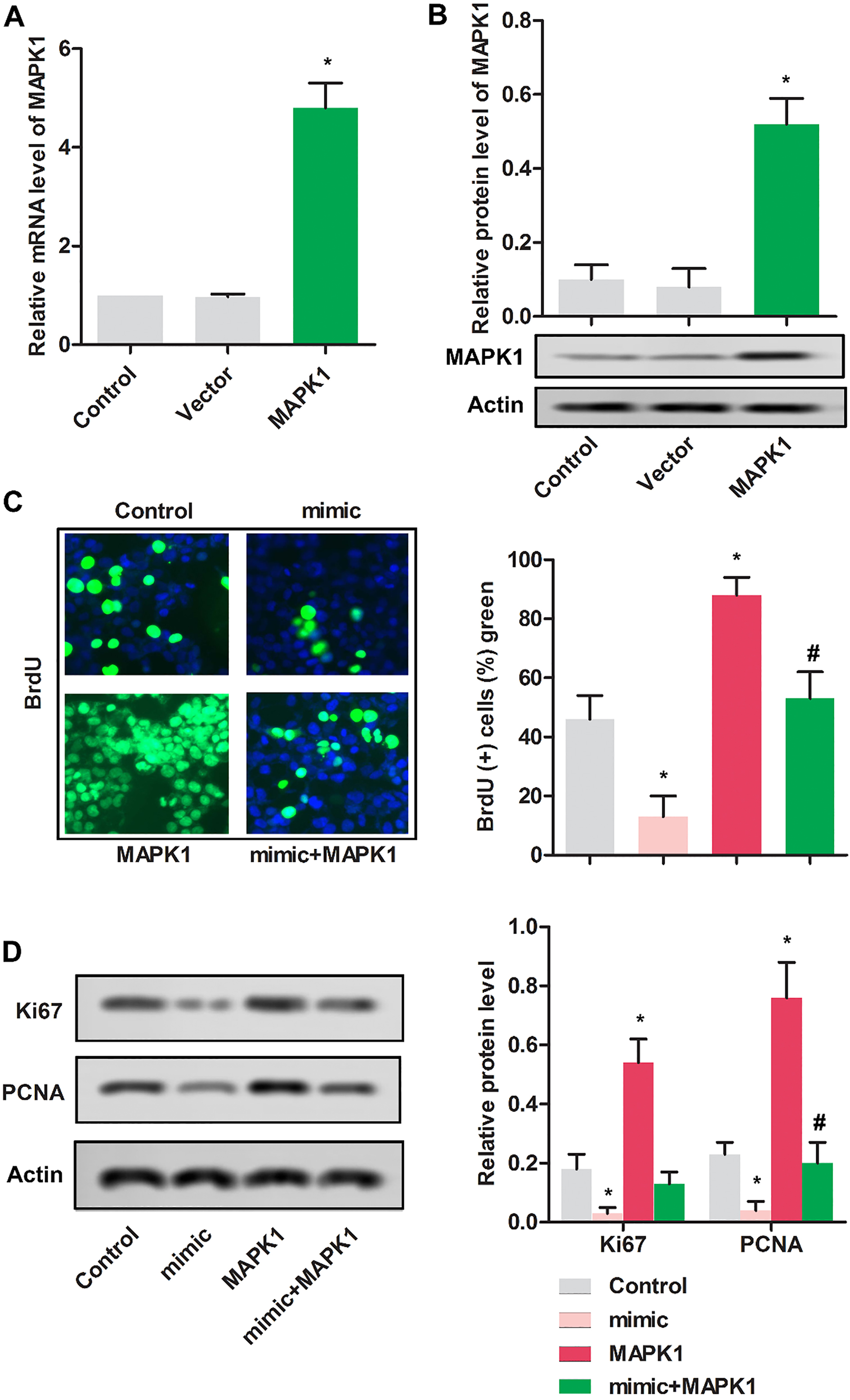
Figure 4.
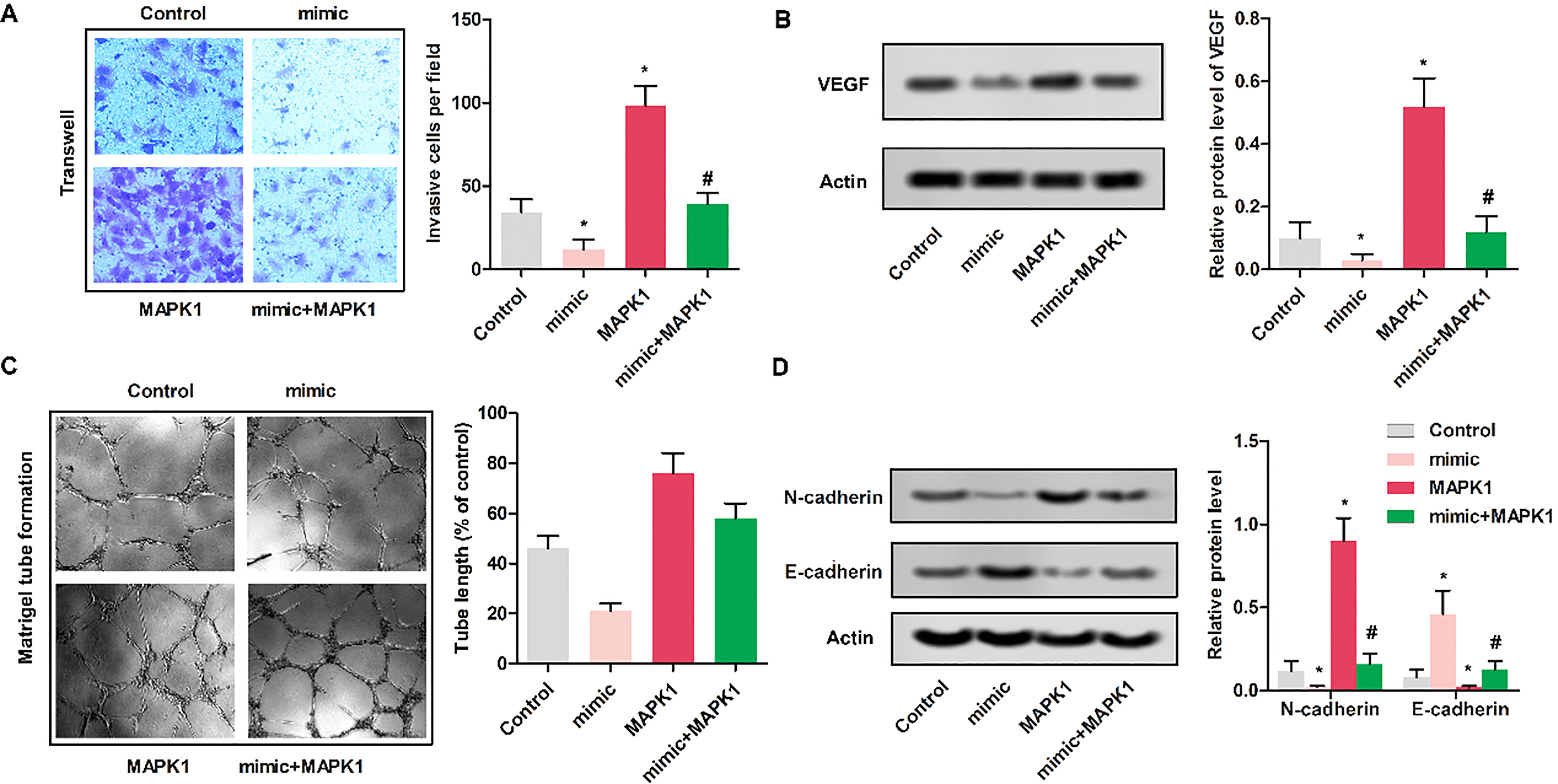
Up-regulated miR-511 inhibited the invasion of MG63 cells. MG63 cells were randomly divided into 4 groups. Control group: normal MG63 cells; mimic: MG63 cells were transfected with miR-511 mimic; MAPK1 group: MG63 cells were transfected with pcDNA-MAPK1; MAPK1
3.3Up-regulated miR-511 inhibited the proliferation of MG63 cells
As shown in Fig. 3A and B, relative mRNA and protein expressions were remarkably increased in MG63 cells transfected with pcDNA-MAPK1 compared with cells transfected with vector.
To determine the impact of miR-511 on the proliferation of MG63 cells, BrdU assay was used. As shown in Fig. 3C, cells in mimic group showed the reduced proliferative cells compared with control group (
Figure 5.
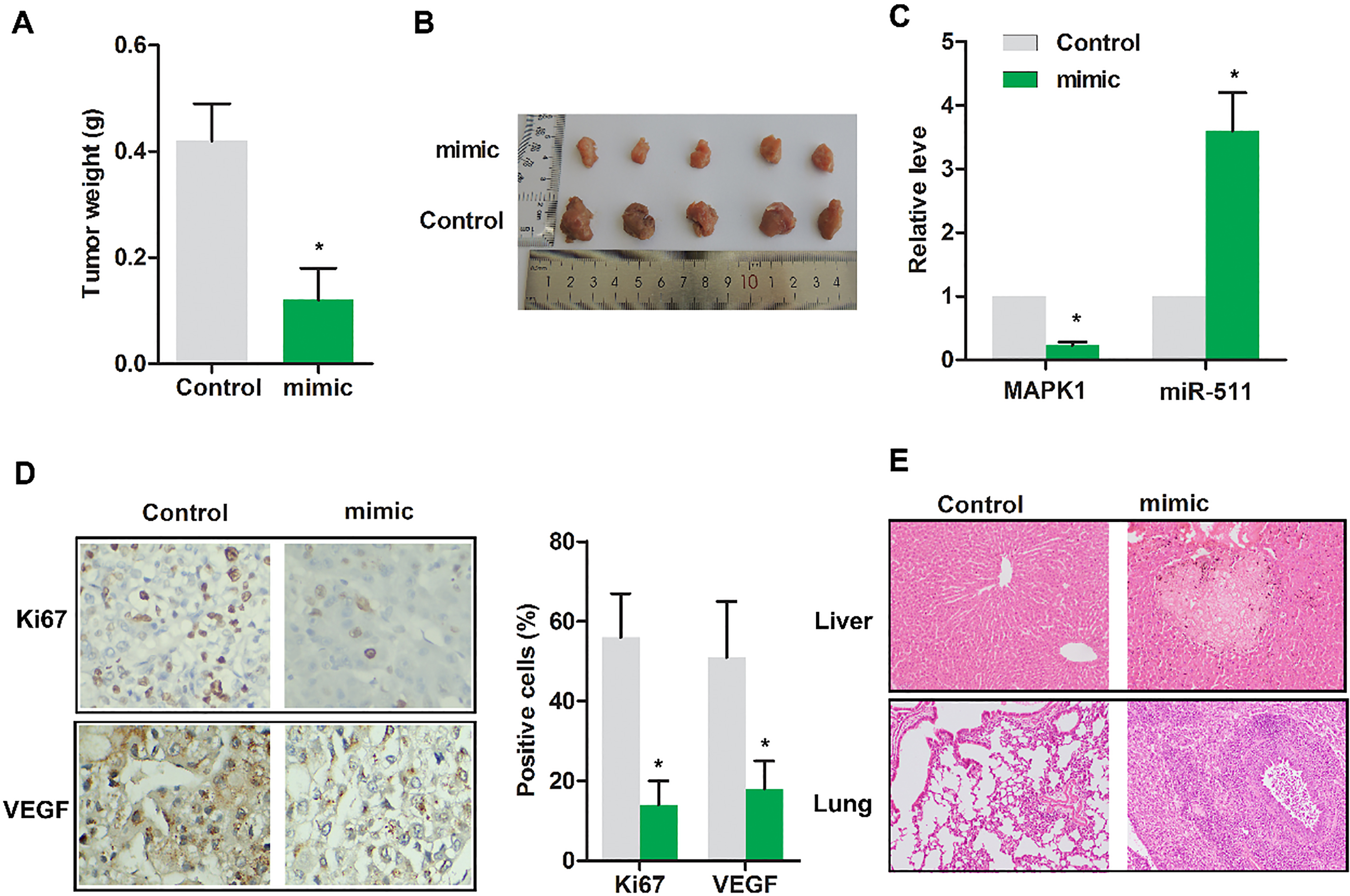
Up-regulated miR-511 suppressed the progression of osteosarcoma in vivo. Nude mice were randomly divided into 2 groups. Control group: tumor extracted from nude mice receiving subcutaneous injection of MG63 cells transfected with miR-NC; mimic group: tumor extracted from nude mice receiving subcutaneous injection of MG63 cells transfected with miR-511 mimic. (A) Gross tumor weight was detected. (B) The representative tumor images were exhibited. (C) The expressions of MAPK1 and miR-511 were evaluated using RT-PCR. (D) The levels of Ki67 and VEGF were detected by immunohistochemical assay. (E) The tumor focal lesions in liver/lung were detected by HE staining. The representative tumor images were exhibited. The experiments were repeated at least 3 times, and error bars represent
3.4Up-regulated miR-511 inhibited the invasion of MG63 cells
To determine the impact of miR-511 on the invasion of MG63 cells, Transwell assay was performed. As shown in Fig. 4A, cells in mimic group showed the reduced invasive cells compared with control group (
3.5Up-regulated miR-511 suppressed the progression of osteosarcoma in vivo
To validate miR-511 indeed play an anti-cancer role in vivo, tumor weight was detected. As displayed in Fig. 5A, the tumor weight was remarkably reduced in mimic group compared with control (
4.Discussion
Osteosarcoma, a kind of highly aggressive and rapidly metastasizes cancer, arises from metaphysis of the long bones and produces immature bone [14]. Despites many efforts have been taken in the available treatment of osteosarcoma, the effects are limited. The key to overcome osteosarcoma is to explore the molecules and signaling pathways associated with this disease.
MiRNAs is implicated in the regulation of gene expression a posttranscriptional level. A lot of studies have demonstrated the importance of miRNA in multiple human cancers [15]. miR-511 is considered as a tumor suppressor in multiple cancers including lung cancer, prostate cancer [16], gastric cancer [17], gallbladder cancer [18, 19], hepatocellular carcinoma [20] and colorectal cancer [5] and only few reports showed that miR-511 served as a carcinogenic factor in other cancers, such as HCV-associated diffuse large B-cell lymphoma as well as in human blood monocyte-derived dendritic cells and macrophages [21]. Interestingly, our study revealed that miR-511 was remarkably down-regulated in osteosarcoma MG63, SaOS2 and U2OS cells. Importantly, miR-511 was observed to inhibit cell proliferation, invasion of osteosarcoma cells and in vivo tumor growth by restoring miR-511 level (mimic). These results suggest that miR-511 acts as a tumor suppressor in osteosarcoma.
Researches revealed different targets of miR-511, such as AKT1, Insulin-like growth factor-1, Hepatoma-derived growth factor, TLR4, NO synthase 1 and TRIM24 [5, 18, 22, 23, 24, 25]. In present study, the interaction has been predicted in targetscan (http://www. targetscan.org/cgi-bin/targetscan/vert_71/targetscan. cgi?species=Human&mir_nc=miR-511-3p). In addition, the interaction between miR-511 and MAPK1 was further validated by luciferase reporter assay result in our study. we found that MAPK1 is up-regulated in osteosarcoma MG63, SaOS2 and U2OS cells, which is dramatically contrary with miR-511. Besides, the overexpression of MAPK1 counteracts the inhibitory effect of miR-511, which further improves the interaction between MAPK1 and miR-511. Furthermore, some previous studies suggested that MAPK1 was significantly up-regulated in various types of human cancer, such as breast cancer [26], lung cancer [27], and ovarian cancer [28]. Herein, MAPK1 was demonstrated to be highly expressed and could be regulated by miR-511 in osteosarcoma. Also, MAPK1 is thought as a oncogene in osteosarcoma in this study. The carcinogenesis of MAPK1 can be confirmed by previous study [26, 27, 28].
In vivo, we checked the role of miR-511 on tumor growth, MAPK1 expression, Ki67 and VEGF expression as well as in vivo metastasis in mice. The results show that restoring miR-511 level by mimic can significantly inhibit tumor growth and the expression of MAPK1, Ki67 and VEGF. Since osteosarcoma easily transfers to liver and lung. We checked whether the tumor transferred to lung or liver, consequently, miR-511 mimic reduced the metastasis of xenograft tumor.
In summary, this study demonstrated that over-expressed miR-511 obstructed tumorigenesis by targeting MAPK1 in vitro and in vivo. Considering the carcinogenic effect of MAPK1, it may be an effective therapeutic target for osteosarcoma. Besides that, miR-511 may be a potential biomarker for the diagnosis and prognosis of osteosarcoma.
Conflict of interest
None.
References
[1] | H.J. Kim, P.N. Chalmers and C.D. Morris, Pediatric osteogenic sarcoma, Curr Opin Pediatr 22: ((2010) ), 61–6. |
[2] | D.D. Moore and H.H. Luu, Osteosarcoma, Cancer Treat Res 162: ((2014) ), 65–92. |
[3] | A.L. Leichter, M.J. Sullivan, M.R. Eccles and A. Chatterjee, MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours, Molecular Cancer 16: ((2017) ), 15. |
[4] | Y. Zhao, W. Pang, N. Yang, L. Hao and L. Wang, MicroRNA-511 inhibits malignant behaviors of breast cancer by directly targeting SOX9 and regulating the PI3K/Akt pathway, International Journal of Oncology 53: ((2018) ), 2715–2726. |
[5] | S. He, G. Wang, J. Ni, J. Zhuang, S. Zhuang and G. Wang, MicroRNA-511 inhibits cellular proliferation and invasion in colorectal cancer by directly targeting hepatoma-derived growth factor, Oncology Research 26: ((2018) ), 1355–1363. |
[6] | H.H. Zhang, M. Pang, W. Dong, J.X. Xin, Y.J. Li, Z.C. Zhang, L. Yu, P.Y. Wang, B.S. Li and S.Y. Xie, miR-511 induces the apoptosis of radioresistant lung adenocarcinoma cells by triggering BAX, Oncology Reports 31: ((2014) ), 1473–9. |
[7] | M. Xu, K. Zhou, Y. Wu, L. Wang and S. Lu, Linc00161 regulated the drug resistance of ovarian cancer by sponging microRNA-128 and modulating MAPK1, Molecular Carcinogenesis 58: ((2019) ), 577–587. |
[8] | C.M Johannessen, J.S. Boehm, S.Y. Kim et al., COT drives resistance to RAF inhibition through MAP kinase pathway reactivation, Nature 468: ((2010) ), 968–72. |
[9] | W. Li, J. Liang, Z. Zhang et al., MicroRNA-329-3p targets MAPK1 to suppress cell proliferation, migration and invasion in cervical cancer, Oncology Reports 37: ((2017) ), 2743–2750. |
[10] | B. Fei and H. Wu, MiR-378 inhibits progression of human gastric cancer MGC-803 cells by targeting MAPK1 in vitro, Oncology Research 20: ((2012) ), 557–64. |
[11] | T. Zhang, J. Li, F. Yin, B. Lin, Z. Wang, J. Xu, H. Wang, D. Zuo, G. Wang, Y. Hua and Z. Cai, Toosendanin demonstrates promising antitumor efficacy in osteosarcoma by targeting STAT3, Oncogene 36: ((2017) ), 6627–6639. |
[12] | G. Wang, S. Wang and C. Li, MiR-183 overexpression inhibits tumorigenesis and enhances DDP-induced cytotoxicity by targeting MTA1 in nasopharyngeal carcinoma, Tumour Biol 39: ((2017) ), 1010428317703825. |
[13] | K. Zhuang, Y. Yan, X. Zhang, J. Zhang, L. Zhang and K. Han, Gastrin promotes the metastasis of gastric carcinoma through the beta-catenin/TCF-4 pathway, Oncol Rep 36: ((2016) ), 1369–76. |
[14] | Z.Q. Wen, X.G. Li, Y.J. Zhang, Z.H. Ling and X.J. Lin, Osteosarcoma cell-intrinsic colony stimulating factor-1 receptor functions to promote tumor cell metastasis through JAG1 signaling, Am J Cancer Res 7: ((2017) ), 801–815. |
[15] | A. Lujambio and S.W. Lowe, The microcosmos of cancer, Nature 482: ((2012) ), 347–55. |
[16] | F. Zhang and Z. Wu, Significantly altered expression of miR-511-3p and its target AKT3 has negative prognostic value in human prostate cancer, Biochimie 140: ((2017) ), 66–72. |
[17] | Z. Fang, L. Zhang, Q. Liao, Y. Wang, F. Yu, M. Feng, X. Xiang and J. Xiong, Regulation of TRIM24 by miR-511 modulates cell proliferation in gastric cancer, J Exp Clin Canc Res 36: ((2017) ), 17. |
[18] | L.-X. Li, Y.-J. Li and J.-X. He, Long noncoding RNA PAGBC contributes to nitric oxide (NO) production by sponging miR-511 in airway hyperresponsiveness upon intubation, Journal of Cell Biochemistry 26: ((2018) ), 1–12. |
[19] | X.S. Wu, F. Wang, H.F. Li et al., LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis, 18: ((2017) ), 1837–1853. |
[20] | G.G. Cao, G.W. Dong, G.X. Meng, G.H. Liu, G.H. Liao and G.S. Liu, Biology and Medicine, MiR-511 inhibits growth and metastasis of human hepatocellular carcinoma cells by targeting PIK3R3, Tumour Biology 36: ((2015) ), 4453–9. |
[21] | C. Augello, U. Gianelli, F. Savi et al., MicroRNA as potential biomarker in HCV-associated diffuse large B-cell lymphoma, 67: ((2014) ), 697–701. |
[22] | X. Yang, L. Liu, H. Zou, Y.W. Zheng and K.P. Digestive, l.d.o.j.o.t.I.S.o. Gastroenterology and t.I.A.f.t.S.o.t. Liver, circZFR promotes cell proliferation and migration by regulating miR-511/AKT1 axis in hepatocellular carcinoma, Digestive and Liver Disease S1590-8658(19)30552-3. |
[23] | W. Wei, G. Wang, Y. Cheng, R. Yang, J. Song, S. Huang, H. Li, H. Geng, H. Yu, S. Liu and L. Hao, Cellular and d.b. Animal, A miR-511-binding site SNP in the 3’UTR of IGF-1 gene is associated with proliferation and apoptosis of PK-15 cells, In Vitro Cell Dev Biol Anim 55: ((2019) ), 323–330. |
[24] | G. Curtale, T.A. Renzi, L. Drufuca et al., Glucocorticoids downregulate TLR4 signaling activity via its direct targeting by miR-511-5p, Eur J Immunol 47: ((2017) ), 2080–2089. |
[25] | Z. Fang, L. Zhang, Q. Liao et al., Regulation of TRIM24 by miR-511 modulates cell proliferation in gastric cancer, J Exp Clin Cancer Res 36: ((2017) ), 17. |
[26] | L. Niu, Y. Zhou, W. Zhang and Y. Ren, Pathology and practice, Long noncoding RNA LINC00473 functions as a competing endogenous RNA to regulate MAPK1 expression by sponging miR-198 in breast cancer, Pathol Res Pract 215: ((2019) ), 152470. |
[27] | T. Jun, F.S. Zheng, K.M. Ren, H.Y. Zhang, J.G. Zhao and J.Z. Zhao, Medical and p. sciences, Long non-coding RNA UCA1 regulates the proliferation, migration and invasion of human lung cancer cells by modulating the expression of microRNA-143, Eur Rev Med Pharmacol Sci 22: ((2018) ), 8343–8352. |
[28] | M. Xu et al., Linc00161 regulated the drug resistance of ovarian cancer by sponging microRNA-128 and modulating MAPK1, Mol Carcinog 58: ((2019) ), 577–587. |




