MiR-223 promotes oral squamous cell carcinoma proliferation and migration by regulating FBXW7
Abstract
Abnormally expressed microRNAs (miRNAs) contribute widely to human cancer, including oral squamous cell carcinoma (OSCC), by regulating their downstream targets. MiR-223 has been proved to be up-regulated in both gastric cancer and ovarian cancer. However, the effect of miR-223 on OSCC is still unclear. Here, we showed that miR-223 was over-expressed in OSCC tissues using qRT-PCR. Next, we investigated the biological mechanism of miR-223 in OSCC. The results demonstrated that miR-223 facilitated the cell proliferation and migration of OSCC using MTT assay and Transwell assay. Furthermore, we stated that the FBXW7 expression was decreased in OSCC and re-expression of FBXW7 inhibited the proliferation and migration of OSCC. In addition, FBXW7 mimic inversed the promotion effect of miR-223 in regulating of OSCC cells. In short, miR-223 promoted OSCC cell proliferation and migration by downregulating FBXW7, which provided a novel therapeutic strategy for OSCC.
1.Introduction
Oral cancer is a common malignant tumor appearing in mouth, and oral squamous cell carcinoma (OSCC) accounts for about 90% of them [1]. Previous studies have shown that even after available treatment, over 50% of patients might die within five years due to the related complications, and the prognosis of OSCC remains dismal [2]. Therefore, it is necessary to carry out in-depth studies to reveal the internal mechanism of tumor occurrence and look for possible treatment methods of OSCC.
MicroRNAs (miRNA) are evolutionary conservative non-coding small molecules. They are encoded by the higher eukaryotic genome and regulate their expression by binding to the 3’ untranslatable region (3’UTR) of the target genes [3]. There were increasing evidences that abnormal expression of miRNAs caused a variety of cancers, including oral cancer. For example, the expression level of miR-381-3p was significantly decreased in oral squamous cell carcinoma and it suppressed OSCC cell proliferation [4]. MiR-155 acted as an oncogene in OSCC tissues, regulating cell cycle and apoptosis [5]. MiR-373-3p promoted tongue squamous cell carcinoma metastasis by targeting DKK1 [6]. Previous studies have demonstrated that miR-223 was up-regulated in oral cancer [7, 8], but the specific biological function of miR-223 in OSCC was not well understood.
FBXW7 was first discovered in drosophila as a regulatory protein for cell cycle and was named CDC4, acting as an important tumor suppressor gene found recently. FBXW7 is critical for the regulation of human cell cycle, cell growth and cell differentiation, and its deletion can lead to or accelerate the proliferation of cancer cells and increase poor prognosis [9, 10, 11]. Previously studies have shown that the absence of FBXW7 could lead to poor prognosis for colorectal cancer, esophageal cancer and gastric cancer [12, 13, 14]. Recently, some studies have shown that FBXW7 was a target gene of miRNAs in regulating different cancers. For instance, miR-223 regulated the acute lymphoblastic leukemia by inhibiting of FBXW7 [15]. MiR-27a promoted the growth of lung cancer cells via suppressing FBXW7, suggesting that FBXW7 played an important role in human tumorigenesis as a tumor suppressor [16]. Moreover, FBXW7 acted as a target of miR-23a in regulating of colorectal cancer cells proliferation [17]. Nevertheless, the regulation of miR-223 in OSCC by targeting FBXW7 is not reported ever.
The current study stated that miR-223 was over-expressed in OSCC and could regulate the cell proliferation and migration. Moreover, we first proved that miR-223 directly targeted FBXW7 in regulation of OSCC.
2.Material and method
2.1Tissue samples
Fifty paired cancerous (tumor center) and corresponding adjacent tongue epithelial tissue samples were obtained from OSCC patients who underwent surgery in the Yantai Yuhuangding Hospital. All patients signed informed consent before sample collection and were untreated before surgery. All samples were stored at
2.2Cell culture
SCC-15 (ATCCCRL-1623), SCC-4 (ATCC CRL-1624) and HSC-3 (BNCC341400) cell lines were purchased from American Type Culture Collection (ATCC). OECM1 cells were generous gifts from the National Defense Medical Center. CGHNC9 cells and normal keratinocyte cells CGHNK2 were obtained from Chang Gung Memorial Hospital. Normal keratinocytes CGHNK2 were cultured and maintained in Keratinocyte Serum Free Medium (K-SFM) (Life Technologies, USA). The oral cancer cell lines were cultured in RPMI-1640 with 10% fetal bovine serum (Gibco, USA), penicillin (100 U/ml) and streptomycin (100
2.3Cell transfection
The miR-223 mimic, miR-223 inhibitor or control mimic, control inhibitor used in this study was purchased from GenePharma (Shanghai, China). OSCC cells were transfected with miRNA mimic or miRNA inhibitor using the Lipofectamine 2000
2.4Western blot assay
Total proteins were extracted from cells by RIPA lysis containing proteinase inhibitors (Beyotime, China) and phenylmethanesulfonyl fluoride (PMSF). The protein concentration was detected by BCA reagent kit (Merck). Total proteins (50
2.5RNA isolation and qRT-PCR
TRIzol reagent (Invitrogen) was used to isolate total RNA from cells. All-in-One
2.6Methyl thiazolyl tetrazolium (MTT)
MTT assay was used to detect cell proliferation. The cells with different transfection were added into 96-well plates and cultured for 0, 24, 36, 48, 96 h, respectively. Then 20 ul MTT medium was added to each well. After incubation at 37
2.7Transwell assay
Cell migration ability was performed using transwell assay. The transwell chamber with 8
2.8Dual luciferase reporter assay
The recombinant pMIR-reportor luciferase vector was used for FBXW7 3’UTR luciferase assays. The wild-type and mut-type miR-223 putative targets on FBXW7 3’UTR were constructed into the pMIR-reporter luciferase vector, OECM1 cells were transfected with control mimic or miR-223 mimic using Lipofectamine 2000. Then, the Dual Luciferase Reporter Assay System (Promega, USA) was used to measure the luciferase activity after transfection for 48 h.
2.9Statistical analysis
All experiments were repeated in triplicate. SPSS v.19.0 software was used to perform statistical analyses and GraphPad Prism 5.02 Software was used to complete graph presentations. Results are represented as the mean
3.Results
3.1MiR-223 was frequently up-regulated and FBXW7 was down-regulated in OSCC cell lines and tissue specimens
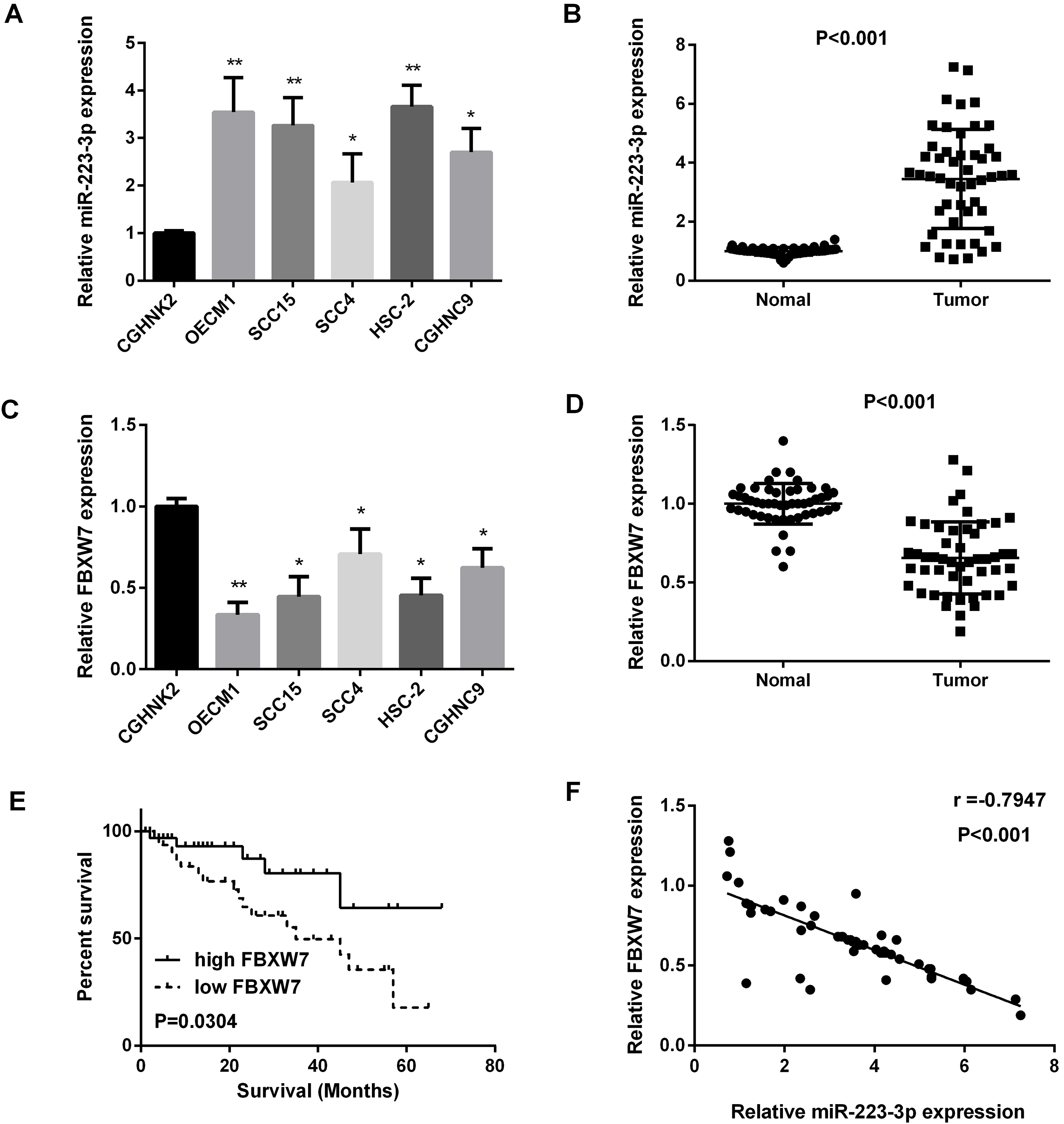
We used real-time PCR to quantify the miR-223 expression level in OSCC cell lines and 50 paired OSCC oral squamous cell carcinoma tissues, as results shown in Fig. 1A and B, miR-223 expression was increased in OSCC cell lines and OSCC tissues compared with the normal ones. We also found that, compared with the normal cells lines and normal tissues, FBXW7 expression levels were down-regulated in OSCC cell lines and OSCC tissues (Fig. 1C and D). Regression analysis showed the inverse correlation between FBXW7 and miR-223 expression level in 50 paired OSCC specimens (Fig. 1F). Furthermore, FBXW7 was correlated with the prognosis of patients (Fig. 1E). In addition, as we saw in Table 1, miR-223 was highly related to stage and tumor size. The results above indicated that miR-223 was correlated with OSCC progression.
Table 1
Relationship between miR-223 expression and their clinic-patho- logical characteristics of OSCC patients
| Item | Cases | miR-223 | ||
|---|---|---|---|---|
| High | Low | |||
| Age (years) | 0.786 | |||
| | 32 | 25 | 7 | |
| | 18 | 12 | 6 | |
| Gender | 0.230 | |||
| Female | 18 | 13 | 5 | |
| Male | 32 | 21 | 11 | |
| Clinical stage | 0.018 | |||
| I–II | 20 | 15 | 5 | |
| III–IV | 30 | 23 | 7 | |
| Tumor size | 0.004 | |||
| | 35 | 24 | 11 | |
| | 15 | 9 | 6 | |
Statistical analyses were performed by the
Figure 1.
Increased miR-223 expression and decreased FBXW7 expression in OSCC cell lines and tissues. (A and B) Detection of miR-223 expression in OSCC cell lines and tissue samples by qRT-PCR. (C and D) Detection of FBXW7 mRNA expression in OSCC cell lines and tissue samples by qRT-PCR. (E) The relationship between FBXW7 expression and survival. (F) Regression analysis of negatively correlation of FBXW7 and miR-223 expression in 50 OSCC samples (
3.2MiR-223 promoted OSCC cell proliferation and inhibited OSCC cell apoptosis
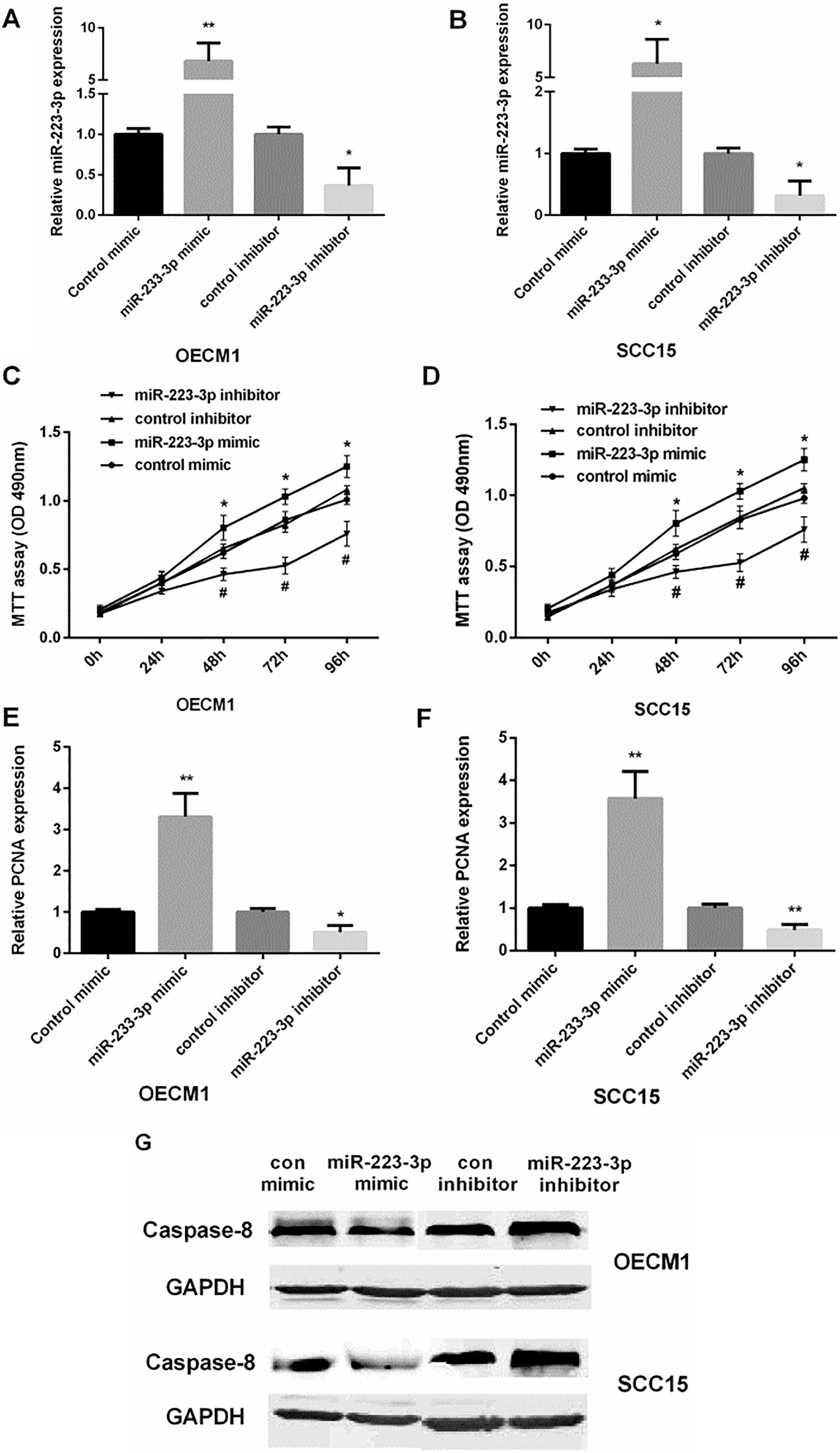
Control mimic/inhibitor and miR-223 mimic/ inhibitor were transfected into OECM1 and SCC15 cells. We used qRT-PCR to examine the miR-223 expression level in OSCC cells with different transfection, as shown in Fig. 2A and B. MTT assay showed that re-expression of miR-223 facilitated cell viability, while inhibiting miR-223 suppressed cell viability in both OECM1 and SCC15 cells (Fig. 2C and D). QRT-PCR results showed that miR-223 mimic increased PCNA expression, while miR-223 inhibitor decreased PCNA expression in both OECM1 and SCC15 cells (Fig. 2E and F). Moreover, Western blot showed that miR-223 mimic suppressed cell apoptosis while miR-223 inhibitor promoted cell apoptosis in both OECM1 and SCC15 cells (Fig. 2G). These results suggested that over-expression of miR-223 promoted OSCC cell proliferation, and inhibited cell apoptosis.
Figure 2.
The promotion of miR-223 in the proliferation and apoptosis of OSCC cells. (A and B) Detection of the relative miR-223 mRNA expression in OECM1 and SCC15 cell lines after transfected with control mimic/inhibitor or miR-223 mimic/inhibitor (
3.3MiR-223 promoted OSCC cell migration
We performed transwell assay to investigate the effect of miR-223 on OSCC cell migration, as shown in Fig. 3A and B, miR-223 mimic enhanced cell migration, while inhibiting miR-223 suppressed cell migration in both OECM1 and SCC15 cells.
Figure 3.
The promotion of miR-223 in the migration of OSCC cells. (A and B) Detection of relative cell migration in OECM1 and SCC15 cells lines after transfected with control mimic/control mimic, miR-223 mimic/inhibitor by transwell assay (
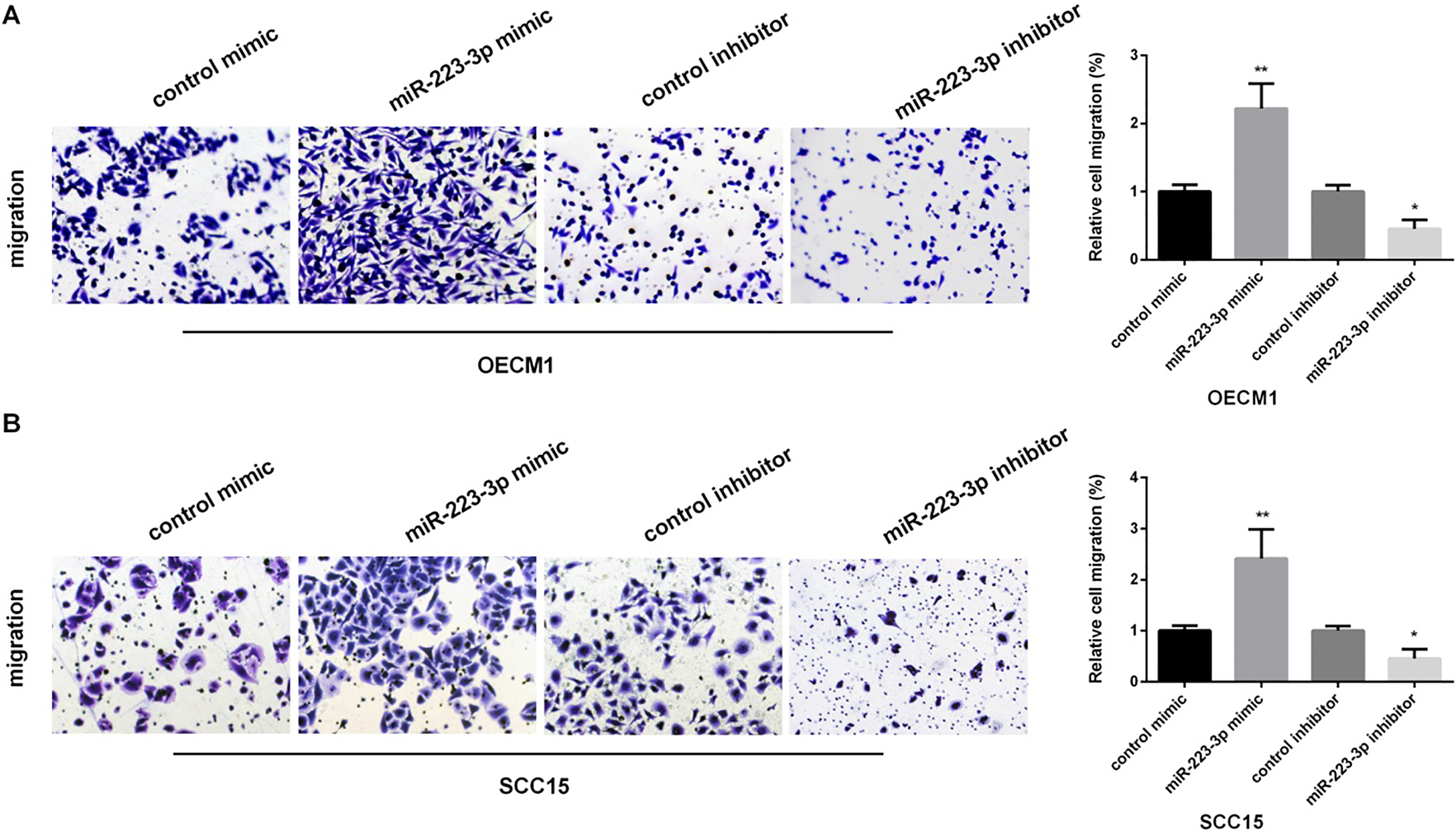
Figure 4.
FBXW7 as a target gene of miR-223 in regulating of OSCC cells. (A) The predicted sites of miR-223 in the 3’UTR of FBXW7. Between the 3’-UTR of FBXW7 and the complementary sites for the seed regions in miR-223 generated mutation. (B) Detection of the luciferase activity in OECM1 cells after transfected with miR-223 mimic/inhibitor (
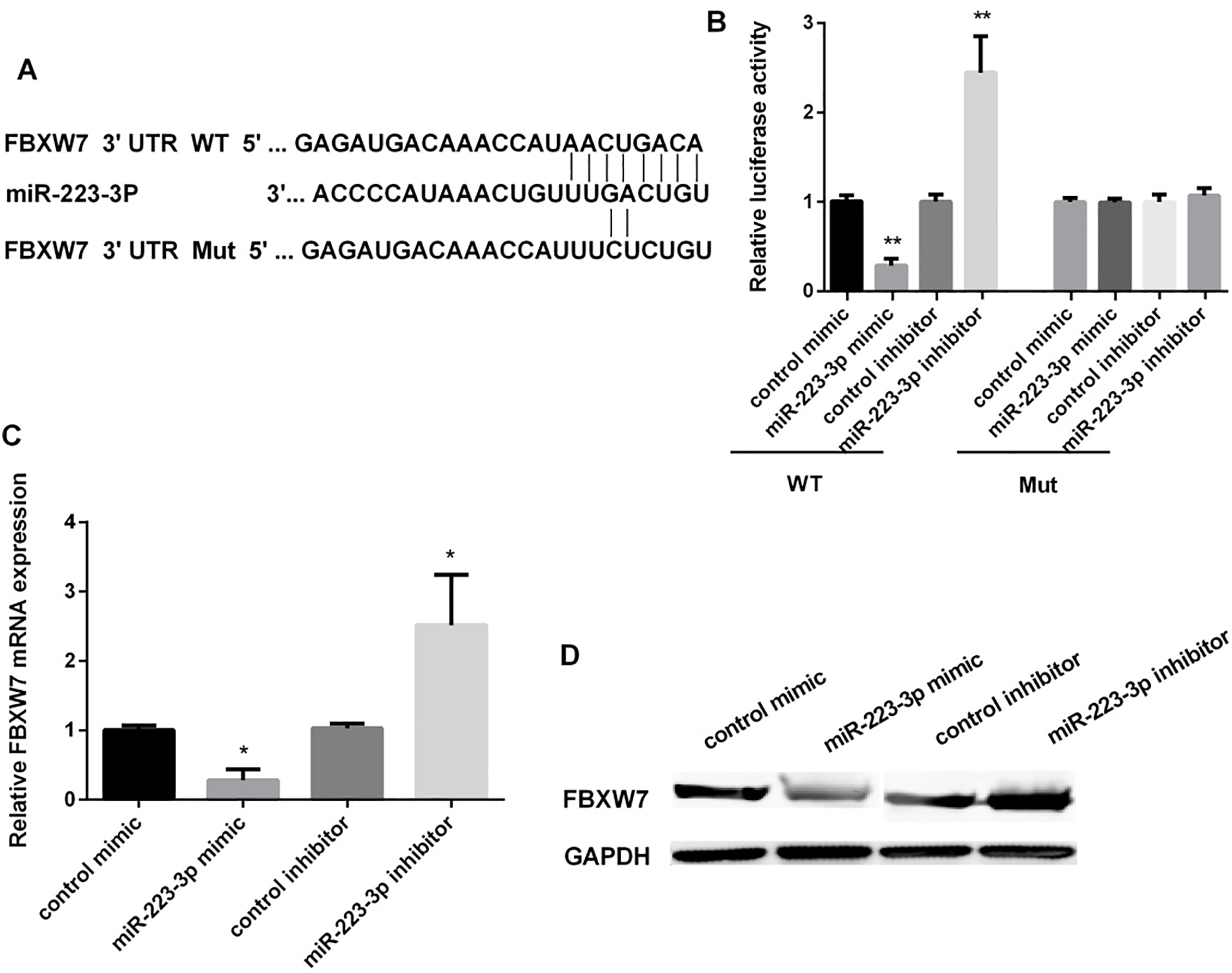
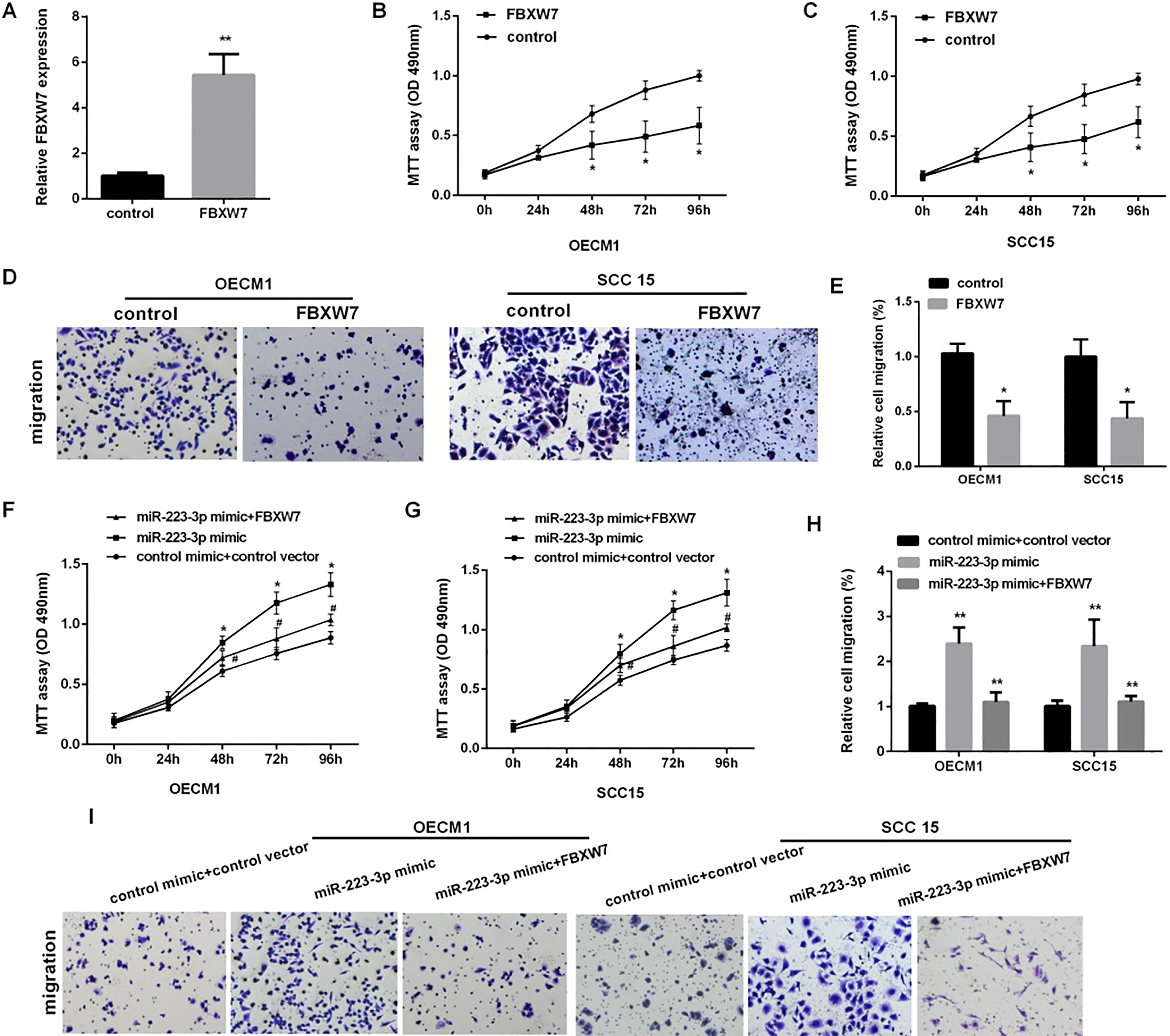
Figure 5.
FBXW7 reversed miR-223-mediated OSCC cells proliferation and migration. (A) Detection of the relative FBXW7 mRNA expression in OECM1 cells after overexpression of FBXW7 for 48 h (
3.4FBXW7 was the directly target of miR-223 in OSCC cells
Target Scan Human 7.1 was used to find the potential targets of miR-223, as the Fig. 4A showed, the FBXW7 might act as a target of miR-223. The effect of miR-223 mimic or inhibitor on the expression of FBXW7 in OECM1 cells was detected using luciferase reporter assay. MiR-223 mimic significantly inhibited the expression of FBXW7 (approximately decreased 71.5%), while miR-223 inhibitor promoted FBXW7 expression after OSCC cells transfected with wild-type FBXW7 3’-UTR. However, it has no effect on the expression of FBXW7 when transfected with mutant FBXW7 3’-UTR (Fig. 4B). Furthermore, miR-223 mimic could decrease the mRNA and FBXW7 protein expression. MiR-223 inhibitor increased the FBXW7 expression both in mRNA and protein levels (Fig. 4C and D).
3.5FBXW7 inversed the effect of miR-223 in regulating OSCC progression
To confirm the function of FBXW7 on the proliferation and migration of OSCC, we first over-expressed FBXW7 (Fig. 5A). As the Fig. 5B–D shown, increasing FBXW7 could inhibit the cells proliferation and migration in OECM1 and SCC15. Then, we detected cell proliferation and migration in OSCC co-transfected with FBXW7 vector and miR-223 mimic, the results showed that over-expression of FBXW7 markedly reversed miR-223-medicate promotion of cell proliferation in OECM1 and SCC15 cells (Fig. 5E), we also found that the migration cells number was decreased in OECM1 and SCC15 cells co-transfected with FBXW7 and miR-223 compared with the cells transfected with the miR-223 alone (Fig. 5F).
4.Discussion
Mounting researches suggest that miRNAs play important roles in progression of OSCC [18, 19]. Previous miRNA microarray analyses showed that miR-223 is down-regulated in OSCC tissues compared to adjacent normal tissues [20]. However, since then, there have no reports on further expression or functional data on miR-223 in OSCC. Here, we aimed to explore the biological function and underling mechanism of miR-223 in OSCC. The results showed that miR-223 expression was elevated in OSCC, whereas, FBXW7 expression was down-regulated. Furthermore, we also provided evidences that miR-223 and FBXW7 mRNA expression were negatively correlated, and FBXW7 could reverse the promotion effect of miR-223 in OSCC cell proliferation and migration.
Emerging studies have shown that miR-223 was abnormally expressed in cancer tissues, suggesting an essential role in tumorigenesis and tumor progression. For instance, miR-223 was up-regulated in gastric cancer, colorectal cancer, non-small cell lung cancer and pancreatic cancer [21, 22, 23, 24]. While, it was down-regulated in cervical cancer [25], prostate cancer [26], nasopharyngeal carcinoma [27] and acute myeloid leukemia [28]. Recent studies have reported that miR-223 expression in oral cancer was up-regulated [7, 8], which was consistent with our study that miR-223 expression was increased in OSCC cells compared with normal cells. In addition, Fang and his colleagues showed that miR-223 could promote ovarian cancer cells proliferation and invasion by targeting MAFB [29]. Also, Ma et al. found that miR-223 mimic could increase the gastric cancer cell proliferation and migration [30]. So we speculated that miR-223 might regulate cell proliferation, migration and invasion as an oncogene in OSCC based on the above researches. Our results showed that miR-223 mimic increased the cell proliferation and migration. Nevertheless, inhibiting miR-223 exerted the inverse effect in both OECM1 and SCC15 cell lines.
To better understand the tumor inhibitory effect of miR-223, bioinformatics analysis was used and FBXW7 was identified as a putative target of miR-223. FBXW7 was further confirmed as a direct target of miR-223 using the luciferase reporter assay. The expression of miR-223 was increased while FGFR2 was down-regulated in OSCC cells. FBXW7 was proved to be a tumor suppressor in regulating tumors progression. A study showed that FBXW7 was identified as a potential target of miR-367 in regulating tumor growth of NSCLC [31]. Also, miR-92a mimic promoted the tumor growth of osteosarcoma via suppressing FBXW7 [32]. Additionally, there was a study stated that FBXW7 level was remarkably reduced in breast cancer and it was a target of miR-223 in regulating the proliferation and migration of breast cancer cells [33]. Otherwise, miR-223 regulated the proliferation and apoptosis of testicular germ cells tumors by FBXW7 [34]. The researches above indicated that FBXW7 mainly function as a tumor suppressor in regulating tumor development, acting as a target gene of miRNA. The results in our study stated that FBXW7 expression level was down-regulated in OSCC and re-expression of FBXW7 inhibited cell proliferation and migration. Moreover, FBXW7 over-expression reversed the promotion effect of miR-223. In clinical tissues, FBXW7 expression was inversely correlated with that of miR-223. All of our data strongly support a role of FBXW7 as a downstream mediator of miR-223-dependent regulation of OSCC.
We summarize all our results and draw a conclusion that miR-223 expression was higher in OSCC and acted as an oncogene in the development of OSCC. We also found the target gene of miR-223 in regulating of OSCC cell proliferation and migration was FBXW7. The newly discovered miR-223/FBXW7 axis provided insight into the mechanisms of oral cancer and provided a potential therapeutic target for OSCC.
Conflict of interest
No conflicts declared.
References
[1] | P.L. Jornet, F.J. Garcia, M.L. Berdugo, F.P. Perez and A.P. Lopez, Mouth self-examination in a population at risk of oral cancer, Australian Dental Journal 60: ((2015) ), 59–64. |
[2] | W.L. Lo, S.Y. Kao, L.Y. Chi, Y.K. Wong and R.C. Chang, Outcomes of oral squamous cell carcinoma in Taiwan after surgical therapy: factors affecting survival, Journal of Oral and Maxillofacial Surgery: Official Journal of the American Association of Oral and Maxillofacial Surgeons 61: ((2003) ), 751–758. |
[3] | E. Huntzinger and E. Izaurralde, Gene silencing by microRNAs: contributions of translational repression and mRNA decay, Nature Reviews Genetics 12: ((2011) ), 99–110. |
[4] | X. Yang, H. Ruan, X. Hu, A. Cao and L. Song, miR-381-3p suppresses the proliferation of oral squamous cell carcinoma cells by directly targeting FGFR2, American Journal of Cancer Research 7: ((2017) ), 913–922. |
[5] | S. Fu, H.H. Chen, P. Cheng, C.B. Zhang and Y. Wu, MiR-155 regulates oral squamous cell carcinoma Tca8113 cell proliferation, cycle, and apoptosis via regulating p27Kip1, European Review for Medical and Pharmacological Sciences 21: ((2017) ), 937–944. |
[6] | J. Weng, H. Zhang, C. Wang, J. Liang, G. Chen, W. Li, H. Tang and J. Hou, MiR-373-3p targets DKK1 to promote EMT-induced metastasis via the wnt/beta-catenin pathway in tongue squamous cell carcinoma, BioMed Research International 2017: ((2017) ), 6010926. |
[7] | H. Tachibana, R. Sho, Y. Takeda, X. Zhang, Y. Yoshida, H. Narimatsu, K. Otani, S. Ishikawa, A. Fukao, H. Asao et al., Circulating miR-223 in oral cancer: its potential as a novel diagnostic biomarker and therapeutic target, PloS One 11: ((2016) ), e0159693. |
[8] | M. Manikandan, A.K. Deva Magendhra Rao, G. Arunkumar, M. Manickavasagam, K.S. Rajkumar, R. Rajaraman and A.K. Munirajan, Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism, Molecular Cancer 15: ((2016) ), 28. |
[9] | Y. Cheng and G. Li, Role of the ubiquitin ligase Fbw7 in cancer progression, Cancer Metastasis Reviews 31: ((2012) ), 75–87. |
[10] | H. Inuzuka, S. Shaik, I. Onoyama, D. Gao, A. Tseng, R.S. Maser, B. Zhai, L. Wan, A. Gutierrez, A.W. Lau et al., SCF (FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction, Nature 471: ((2011) ), 104–109. |
[11] | I.E. Wertz, S. Kusam, C. Lam, T. Okamoto, W. Sandoval, D.J. Anderson, E. Helgason, J.A. Ernst, M. Eby, J. Liu et al., Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7, Nature 471: ((2011) ), 110–114. |
[12] | M. Iwatsuki, K. Mimori, H. Ishii, T. Yokobori, Y. Takatsuno, T. Sato, H. Toh, I. Onoyama, K.I. Nakayama, H. Baba et al., Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance, International Journal of Cancer 126: ((2010) ), 1828–1837. |
[13] | T. Yokobori, K. Mimori, M. Iwatsuki, H. Ishii, F. Tanaka, T. Sato, H. Toh, T. Sudo, T. Iwaya, Y. Tanaka et al., Copy number loss of FBXW7 is related to gene expression and poor prognosis in esophageal squamous cell carcinoma, International Journal of Oncology 41: ((2012) ), 253–259. |
[14] | T. Yokobori, K. Mimori, M. Iwatsuki, H. Ishii, I. Onoyama, T. Fukagawa, H. Kuwano, K.I. Nakayama and M. Mori, p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases, Cancer Research 69: ((2009) ), 3788–3794. |
[15] | M.R. Mansour, T. Sanda, L.N. Lawton, X. Li, T. Kreslavsky, C.D. Novina, M. Brand, A. Gutierrez, M.A. Kelliher, C.H. Jamieson et al., The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia, The Journal of Experimental Medicine 210: ((2013) ), 1545–1557. |
[16] | Q. Wang, D.C. Li, Z.F. Li, C.X. Liu, Y.M. Xiao, B. Zhang, X.D. Li, J. Zhao, L.P. Chen, X.M. Xing et al., Upregulation of miR-27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen, Oncogene 30: ((2011) ), 3875–3886. |
[17] | S. Jahid, J. Sun, R.A. Edwards, D. Dizon, N.C. Panarelli, J.W. Milsom, S.S. Sikandar, Z.H. Gumus and S.M. Lipkin, miR-23a promotes the transition from indolent to invasive colorectal cancer, Cancer Discovery 2: ((2012) ), 540–553. |
[18] | H. Liu, W.-F. Chiang, T.-W. Chen, L.J. Chu, J.-S. Yu, S.-J. Chen, H.-C. Chen and B.C.-M. Tan, MiR-31-5p-ACOX1 axis enhances tumorigenic fitness in oral squamous cell carcinoma via the promigratory prostaglandin E2.%A lai YH, Theranostics 8: ((2018) ), 486–504. |
[19] | Y. CC, K. SY, L. CJ, L. SC and C. KW, MicroRNA-211 enhances the oncogenicity of carcinogen-induced oral carcinoma by repressing TCF12 and increasing antioxidant activity.%A chen YF, Cancer Research 76: ((2016) ), 4872–4886. |
[20] | A.K. Deva Magendhra Rao, G. Arunkumar, M. Manickavasagam, K.S. Rajkumar, R. Rajaraman and A.K. Munirajan, Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism.%A manikandan M, Molecular Cancer 15: ((2016) ), 28. |
[21] | X. Zhou, W. Jin, H. Jia, J. Yan and G. Zhang, MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7, Journal of Experimental & Clinical Cancer Research: CR 34: ((2015) ), 28. |
[22] | Z.W. Li, Y.M. Yang, L.T. Du, Z. Dong, L.L. Wang, X. Zhang, X.J. Zhou, G.X. Zheng, A.L. Qu and C.X. Wang, Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer, Medical Oncology 31: ((2014) ), 256. |
[23] | F.Y. Zhao, J. Han, X.W. Chen, J. Wang, X.D. Wang, J.G. Sun and Z.T. Chen, miR-223 enhances the sensitivity of non-small cell lung cancer cells to erlotinib by targeting the insulin-like growth factor-1 receptor, International Journal of Molecular Medicine 38: ((2016) ), 183–191. |
[24] | J. Ma, L. Cheng, H. Liu, J. Zhang, Y. Shi, F. Zeng, L. Miele, F.H. Sarkar, J. Xia and Z. Wang, Genistein down-regulates miR-223 expression in pancreatic cancer cells, Current Drug Targets 14: ((2013) ), 1150–1156. |
[25] | Y. Tang, Y. Wang, Q. Chen, N. Qiu, Y. Zhao and X. You, MiR-223 inhibited cell metastasis of human cervical cancer by modulating epithelial-mesenchymal transition, International Journal of Clinical and Experimental Pathology 8: ((2015) ), 11224–11229. |
[26] | A. Kurozumi, Y. Goto, R. Matsushita, I. Fukumoto, M. Kato, R. Nishikawa, S. Sakamoto, H. Enokida, M. Nakagawa, T. Ichikawa et al., Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer, Cancer Science 107: ((2016) ), 84–94. |
[27] | W. Yang, X. Lan, D. Li, T. Li and S. Lu, MiR-223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells, BMC Cancer 15: ((2015) ), 461. |
[28] | Y. Xiao, C. Su and T. Deng, miR-223 decreases cell proliferation and enhances cell apoptosis in acute myeloid leukemia via targeting FBXW7, Oncology Letters 12: ((2016) ), 3531–3536. |
[29] | G. Fang, J. Liu, Q. Wang, X. Huang, R. Yang, Y. Pang and M. Yang, MicroRNA-223-3p regulates ovarian cancer cell proliferation and invasion by targeting SOX11 expression, International Journal of Molecular Sciences 18: ((2017) ). |
[30] | L. Ma, Y. Chen, B. Zhang and G. Liu, Increased microRNA-223 in Helicobacter pylori-associated gastric cancer contributed to cancer cell proliferation and migration, Bioscience, Biotechnology, and Biochemistry 78: ((2014) ), 602–608. |
[31] | G. Xiao, X. Gao, X. Sun, C. Yang, B. Zhang, R. Sun, G. Huang, X. Li, J. Liu, N. Du et al., miR-367 promotes tumor growth by inhibiting FBXW7 in NSCLC, Oncology Reports 38: ((2017) ), 1190–1198. |
[32] | X. Jiang, X. Li, F. Wu, H. Gao, G. Wang, H. Zheng, H. Wang, J. Li and C. Chen, Overexpression of miR-92a promotes the tumor growth of osteosarcoma by suppressing F-box and WD repeat-containing protein 7, Gene 606: ((2017) ), 10–16. |
[33] | W. Xia, J. Zhou, H. Luo, Y. Liu, C. Peng, W. Zheng and W. Ma, MicroRNA-32 promotes cell proliferation, migration and suppresses apoptosis in breast cancer cells by targeting FBXW7, Cancer Cell International 17: ((2017) ), 14. |
[34] | J. Liu, H. Shi, X. Li, G. Chen, C. Larsson and W.O. Lui, miR2233p regulates cell growth and apoptosis via FBXW7 suggesting an oncogenic role in human testicular germ cell tumors, International Journal of Oncology 50: ((2017) ), 356–364. |




