A novel PIK3CA hot-spot mutation in breast cancer patients detected by HRM-COLD-PCR analysis
Abstract
BACKGROUND:
The PI3K protein is involved in the PI3K/AKT/mTOR pathway. Deregulation of this pathway through PIK3CA mutation is common in various tumors. The aim of this work is to identify hotspot mutation at exons 9 and 20 in Tunisian patients with sporadic or hereditary breast cancer.
METHODS:
Hotspot mutations in exon 9 and exon 20 of the PIK3CA gene were identified by QPCR-High Resolution Melting followed by COLD-PCR and sequencing in 63 (42 sporadic cases and 21 hereditary cases) tumor tissues collected from Tunisian patient with breast cancer. MCF7, and BT20 breast cancer cell lines harboring the PIK3CA hotspot mutations E545K and H1047R in exon 9 and exon 20 respectively, were used as controls in HRM experiments.
RESULTS:
PIK3CA hotspot mutations were detected in 66.7% (28 out of 42) of sporadic BC cases, and in 14.3% (3 out of 21) of hereditary BC. The E545K and the H1048Y were the most prevalent mutations identified in patients with sporadic and hereditary BC, whereas the H1047R hotspot mutation was not found in our patients. Statistical analysis showed that PIK3CA mutation associated with an aggressive behavior in patients with sporadic BC, while it’s correlated with age, tumor stage and tumor size in the group patients with hereditary breast cancer.
CONCLUSIONS:
Our results showed a novel PIK3CA hotspot mutation in Tunisian breast cancer patients detected by HRM-COLD-PCR. Moreover, the absence of PIK3CA hotspot mutation associated with good prognosis.
1.Introduction
Breast cancer (BC) is a complex disease resulting from genetic and epigenetic alterations affecting several genes such as oncogenes, tumor suppressor, etc. [1]. BC is the most frequent cancer and 1/10 woman has a risk to develop this malignancy during her life [2]. In Tunisia, the incidence of BC is approximately 30/100.000/per year [3]. In Tunisian patients, BC appears more frequently in young women (<35 years) with aggressive clinical behavior such as Triple Negative (ER/PR/Her2 negative) profile [3,4]. PIK3CA is among the most commonly mutated gene in various human cancer including BC [5,6]. PIK3CA mediates different processes such as cell transformation, tumor initiation and proliferation, resistance to apoptosis, etc. Its activity is stimulated by extracellular growth factors and hormones [7]. The dysregulation of PI3K initiates activity of the serine/threonine kinase AKT which modulate proteins that promote uncontrolled cellular and tumor growth [8]. There are three classes of PI3Ks according to their primary structures, substrate preferences and regulation: Class I (Ia, Ib), Class II and Class III. Most relevant for cellular regulation are the PI3Ks of class Ia which act as heterodimers of regulatory and catalytic subunits [7]. The catalytic subunit of the class I PI3-kinase p110α is encoded by the PIK3CA gene. The p110α protein has five domains: an adaptor-binding-domain for linking the regulatory subunit, a Ras-binding-domain, a C2-domain for binding PIP2 and PIP3, a helical domain and a kinase domain [8]. Genetic alterations of PIK3CA contributing to carcinogenesis are somatic mutations and/or gene amplification leading to a gain of function [9–12]. Most of PIK3CA mutations are clustered at the coding region of the helical and kinase domains of the protein and more than 80% of mutations are located in exon 9 such as the E542K, and E545K, and in exon 20 such as the H1047R [10,11]. The percentage of PIK3CA gene mutations varied widely among cancer types with 26.8% in breast cancer, 32% in colon cancer, 27% in brain cancer, 12% in ovarian cancer, 25% in stomach cancer, and 4% in lung cancer [13–17].
The present work aims to identify hotspot mutations in exons 9 and 20 of the PIK3CA gene in sporadic and hereditary BC patients from the south of Tunisia. To address this study, we opted for the PCR-HRM [18]. Mutation screening by High Resolution Melting (HRM) is based on the dissociation behavior of DNA in decreasing temperature step. Signal generated is modified in the presence of fluorescent dyes that actively intercalating double-stranded DNA when it translate from a double to single strand [18]. The HRM melting profile gives a specific sequence-related pattern allowing discrimination between wild-type sequences and homozygote-heterozygote variants [19]. Actually, HRM analysis requires the use of standard DNA representing wild-type and/or mutated sequences. In the present study, DNA extracted from breast cancer cell lines (wild-type and containing PIK3CA hotspot mutations) was used as standards. Due to its high sensitivity, HRM has been used to screen different oncogenes or tumor suppressor to detect the minimal fraction of mutated cells in tumor tissues [20–22].
To identify and validate the PCR-HRM results, we used the Fast-COLD-PCR ‘CO-amplification at Lower Denaturation temperature’ [23]. This technique allows selective amplification of the minority mutant allele to enrich it prior to DNA sequencing.
2.Materials and methods
2.1.Patients and tumor samples
Sixty three patients with BC (21 hereditary cases, and 42 sporadic cases) were included in this study. Frozen tissues were collected at the Department of Anatomo-Pathology from Habib Bourguiba Hospital of Sfax (Tunisia) from 2007 to 2009. All patients were ranked through the Scarff-Bloom-Richardson system and the clinical stage was obtained by the TNM classification. Informed consents were obtained from all patients prior to collect samples according to institutional guidelines. The mean age of patients with hereditary breast cancer was 52.05 years (33 to 74 years), and those with sporadic breast cancer was 48.36 years (25 s to 83 years). Table 1 summarizes the clinic pathological features of patients included in this study.
Table 1
Clinico pathological features of patients with sporadic or hereditary breast cancer (BC)
| Variables | Sporadic BC (%) | Hereditary BC (%) |
| Age | ||
| ≤45 | 15 (38.5%) | 10 (47.6) |
| >45 | 24 (61.4%) | 11 (52.4) |
| SBR grade | ||
| I | 8 (20.5) | 2 (9.5) |
| II | 19 (48.7) | 12 (57.1) |
| III | 12 (30.8) | 7 (33.3) |
| T-stage | ||
| 1 | 5 (20) | 3 (14.3) |
| 2 | 5 (20) | 8 (38.1) |
| 3 | 1 (4) | 4 (19) |
| 4 | 14 (56) | 6 (28.6) |
| Tumor size | ||
| ≤3 cm | 20 (51.3) | 10 (47.6) |
| >3 cm | 19 (48.7) | 11 (52.4) |
| N | ||
| N0 | 15 (38.5) | 11 (52.4) |
| N1 | 24 (61.5) | 10 (47.6) |
| M | ||
| M0 | 7 (43.8) | 13 (61.9) |
| M1 | 9 (56.3) | 8 (38.1) |
| ER | ||
| Positive | 23 (59) | 10 (47.6) |
| Negative | 16 (41) | 11 (52.4) |
| PR | ||
| Positive | 25 (64.1) | 13 (61.9) |
| Negative | 14 (35.9) | 8 (38.1) |
| HER-2 | ||
| Positive | 7 (31.8) | 4 (20) |
| Negative | 15 (68.2) | 16 (80) |
2.2.Cell lines
Breast cancer cell lines (MCF7, and BT20) were used to obtain the mutated DNA fragments corresponding to the E545K and H1047R mutations located in exon 9 and exon 20 respectively. To obtain the wild type sequence for exon 9 and exon 20, we used the T47D, and MCF7 cell lines respectively. Cells were cultured in DMEN (10% fetal calf serum, 1% ampicillin/streptomycin, non essential amino acid) medium at 37 °C in a controlled atmosphere (5%
2.3.DNA extraction
Briefly, cell pellets were collected, dissolved in buffer containing proteinase K (20 ng/ml) and incubated 55 °C for 3 hours. DNA was extracted by adding phenol/chloroform (v/v), and after centrifugation for 15 min at 13.000 rpm, DNA was precipitated by adding 2.5 v ethanol and 1/10 v 3M sodium acetate pH 5.2 and incubated overnight at −20 °C. The DNA pellet was washed with 75% ethanol, dissolved in sterile water and quantified by NanoDrop spectrophotometer (Thermofisher).
For fresh frozen tissues, 60 μm sections were used for DNA extraction using the TRIzol reagent according to the manufacturer’s instructions (Invitrogene).
2.4.Standards DNA
DNA extracted from breast cancer cell lines were used as template to amplify by PCR DNA fragment containing the PIK3CA hotspot mutation in exon 9 and exon 20. Reactions were performed in 25 μL mixture containing 20 pmol of each primer, 1 unit of Taq DNA polymerase (Fermentas), 200 mM dNTP, 1× PCR buffer and 100 ng of genomic DNA for 35 cycles as follow: 30 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. The amplicons were inserted into the pGEM-T Easy (Promega), and the ligation mixture served to transform the E. coli. Top10F’ strain. DNA extracted from recombinant plasmids were quantified by NanoDrop and used as standards in HRM experiments. Samples showing profiles that are different from those obtained with wild-type, mutated in exon 9 (E545K), or in exon 20 (H1047Y) were retained for further analysis.
Primers sequences using for exon 9 and exon 20 amplification were previously reported and presented in Table 2.
Table 2
Sequence of primers used for PCR, real-time PCR and sequencing
| Exon 9 | Sequence 5′ —3′ | Lenght (pb) | % GC | Tm (°C) |
| Forward | ctagctagagacaatgaattaagggaaa | 130 | 35.7 | 63.6 |
| Reverse | cattttagcacttacctgtgactcca | 42.3 | 66 | |
| Exon 20 | ||||
| Forward | tgagcaagaggctttggagt | 134 | 50 | 64 |
| Reverse | tcattttctcagttatcttttcagttcaat | 26.7 | 64.7 |
2.5.High resolution melting
Aliquot of 20 ng of DNA were amplified in a final volume of 10 μL containing 5 μL of Master Mix EvaGreen (Bio-Rad), 10 pmol of primers (Fermentas). PCR for PIK3CA exons 9 and 20 was performed in a real-time PCR thermal cycler (Precision Melt Analysis, Bio-Rad) using 1 cycle at 98 °C for 2 min followed by 35 cycles of 30 s at 95 °C, 1 min at 60 °C, and a final extension at 60 °C for 5 min. A melting profile was performed from 68 °C to 90 °C with 0.1 °C increments. Samples and standards were analyzed in duplicate.
2.6.COLD - PCR
COLD-PCR is a developed PCR approach to enrich minor allele variants and low-abundance mutations [23]. Reaction was performed in a 25 μL final volume, using 10 ng DNA 20 pmol of each primers, 1× PCR buffer included 20 mM MgCl2, 200 mM dNTP and 1 unit of Taq DNA polymerase (Fermentas). The COLD-PCR cycling protocol was assessed by varying the Tc in a range close to the amplicon Tm, as evaluated by HRM. The Tc values for exons 9 and 20 are 76.9 °C and 77.6 °C respectively. COLD-PCR conditions were presented in Table 3.
Table 3
Cycling conditions for COLD-PCR
| Step | Temperature (°C) | Time | Number of cycle |
| Initial denaturation | 94 | 5 min | 1 |
| Denaturation | 94 | 30 s | 10 |
| Primer hybridization | 60 | 30 s | |
| Extension | 72 | 30 s | |
| Final extension | 72 | 5 min | 1 |
| Initial denaturation | 94 | 2 min | 1 |
| Denaturation | 94 | 20 s | 40 |
| Denaturation intermediary | 70 | 1 min | |
| Denaturation of hetero-duplex | Tc | 5 s | |
| exon 9: 76.9 | |||
| exon 20: 77 | |||
| Primer hybridization | 60 | 30 s | |
| Extension | 72 | 30 s | |
| Final extension | 72 | 5 min | 1 |
2.7.Sequencing analysis
Before sequencing, tumor samples amplicons were purified by the kit “Spinklean Gel Extraction”. The sequencing of product of COLD-PCR and conventional PCR was performed on the (ABI 3100, Applied Biosystem) using the primers described in Table 4.
Table 4
Correlation Between PIK3CA hotspot mutations, and Clinicopathological features using multivariate analysis
| Covariates | P | HRa | 95% CIb | |
| Lower | Upper | |||
| Age | 0.459 | 0.347 | 0.021 | 5.697 |
| Tumor size | 0.133 | 9.543 | 0.502 | 181.539 |
| T-stage | 0.188 | 4.45 | 0.481 | 41.188 |
| Grade | 0.059 | 0.018 | 0.021 | 1.171 |
| ER | 0.057 | 32.404 | 0.906 | 2.112 |
| PR | 0.141 | 0.105 | 0.05 | 6.893 |
| Her2 | 0.179 | 0.128 | 0.06 | 2.56 |
[i] HR: hazard ratio; CI: confidence interval.
2.8.Statistical analysis
Statistical analyses were performed using the SPSS 17 statistical software for Windows. The two sided 𝜒2 test was used to determine associations between the mutation al status of PIK3CA gene and various clinico-pathological features. The correlation with overall survival was performed using Kaplan–Meier survival plots, and the significance was tested using the logrank test.
3.Results
3.1.Sequencing of standards DNA and HRM analysis
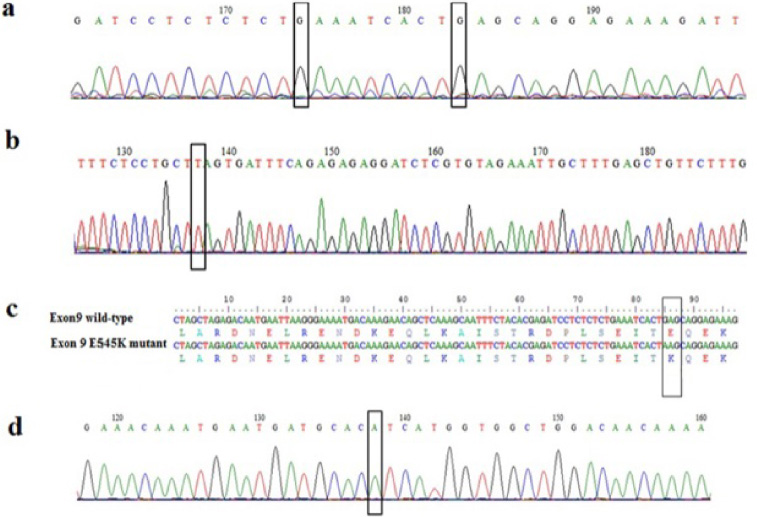
Firstly, we constructed standards using DNA extracted from breast cancer cell lines that contained the wild type PIK3CA gene or exon 9 mutated. PCR fragments were cloned and 3 recombinant vectors containing the wild type exon 9 (9 WT), exon 20 (20WT) and the E545K mutation were obtained and sequenced (Fig. 1).
Fig. 1.
Chromatograms showing DNA sequences of exon 9 (a) wild-type, and (b) mutant. Alignment of the nucleotide and protein sequences of wild-type, and E545K mutant (c). Chromatogram showing DNA sequence of wild-type exon 20 (d). Nucleotides corresponding to the hotspot mutations E542K and E545K in exon 9 and H1048Y in exon 20 are boxed.
The sensitivity and reliability of the melting profile are obtained by optimizing different parameters such as: the serial dilutions of the mutated and wild-type DNA, derived from standards, the amount of DNA in order to obtain similar values of Ct with the aim of minimizing the end point dispersion of the curves, the HRM conditions specifically the degree of temperature increase and the time interval (0.1 °C increments with 5 s) in order to optimize the discrimination between the different standard curves, and the choice of the normalization interval of the melting curve separating the region of interest from the “pre-melting” and “post-melting” regions.
3.2.Mutated allele enrichment by “COLD-PCR”
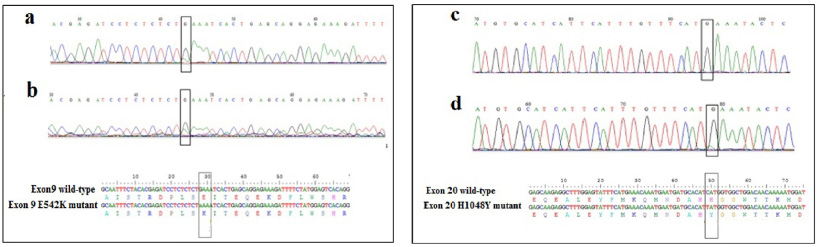
DNA sequencing of samples displaying distinct HRM profiles compared to the wild type, did not reveal any mutation. This is probably due to the low proportion of mutated DNA in tissues sample. Therefore, we carried out the COLD-PCR, which allows the enrichment of the mutated allele to identify the variant. After the optimization of some parameters such as the Tc for exon 9 and exon 20, the cycling conditions, and the DNA quantity, we showed that the amplification was obtained using with 10 ng of DNA (Fig. 2). DNA sequencing of sample with different HRM profiles compared to those of WT exon 9, showed a low fluorescence peak slightly higher than the background when the amplicon was obtained by conventional PCR (Fig. 3a). However, sequencing of the same DNA after amplification by COLD-PCR showed more clearly the presence of the variant at the nucleotide G1624 resulting in the substitution of E542K (Fig.3a). In addition, we were also able to show the presence of the H1048Y mutation using the COLD-PCR, as presented in the Fig. 3b.
Fig. 2.
Analysis of exon 9 amplicons obtained by COLD-PCR. L: 100 bp ladder, Lanes 1, 2, and 3 correspond to the COLD-PCR results performed with 20 ng, 10 ng and 3 ng of DNA respectively. 4: negative control.
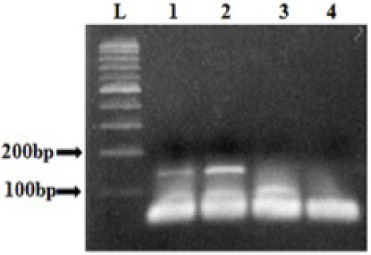
Fig. 3.
Chromatograms and sequence alignment showing the E542K mutation in exon 9 using the amplicons obtained by conventional PCR (a) and by COLD PCR (b). Chromatograms and sequence alignment showing the H1048Y mutation in exon 20 using the amplicons obtained by conventional PCR (c) and by COLD PCR (d).
3.3.Identification of PIK3CA hotspot mutations in breast cancer
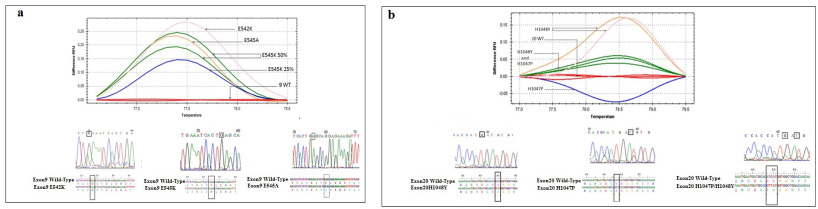
In the group of patients with sporadic BC, we found that among 42 cases, 28 (66.7%) carried hotspot mutations. The E545K, and the H1048Y were the most prevalent mutations identified in 7 (16.7%) and 9 patients (21.4%) respectively. In patients with hereditary BC, the frequency of PIK3CA mutation was low since only 5 cases out of 21 (23.8%) were carriers. Similarly, the E545K and the H1048Y were also the most identified mutations in this group. Interestingly, 8 cases (12.7%) harbored double mutations namely the H1047P and the H1048Y. Figure 4 illustrated the HRM curves for the exon 9 (Fig. 4a), and exon 20 (Fig. 4b) hotspot mutations as well as the alignment of the mutated and the wild type sequences.
Fig. 4.
HRM curves, chromatograms, and sequence alignment for (a) exon 9 wild-type, mutant E542K, E545k, and E545A, and (b) exon 20 wild-type, mutant H1047P and H1048Y.
3.4.Correlation between PIK3CA mutations and clinico-pathological parameters
In sporadic BC patients, the mutated status of PIK3CA associated with tumor size (p = 0.036). Furthermore, if we consider only PIK3CA mutations at exon 9, significant associations were observed with tumor grade (p = 0.028) and expression of Her2-neu (p = 0.022). High grade tumors (GII + GIII) are more frequently mutated in exon 9 than low grade tumors (GI). Regarding mutations in exon 20, we noted significant association with age at diagnosis (p = 0.023).
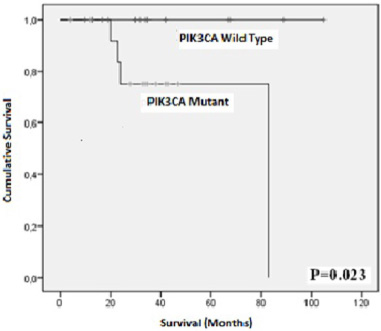
In hereditary cases, significant correlations were observed between the exon 9 mutations and the age at diagnosis (p = 0.023), tumor size (p = 0.001), and tumor stage (p = 0.008). Mutation in exon 20 correlated only with age at diagnosis (p = 0.023). Regarding, the overall survival, we showed that patients carrying PIK3CA mutations had shorter survival rate compared to those without PIK3CA mutations (p log rank = 0.023, Fig. 5).
Fig. 5.
Kaplan Meier plots showing the association of PIK3Ca hotspot mutations with overall survival.
4.Discussion
PIK3CA is the catalytic subunit alpha of the phosphatidylinositol-4,5-bisphosphate 3-kinase, which is involved in crucial biological process such as cell proliferation and survival, differentiation, motility, and intracellular trafficking [5,7]. Previous studies has reported that PIK3CA mutations were detected in 20 to 40% of breast cancer [14,24–26]. The recent study of Ren et al., using the TCGA dataset, showed that 34.49% of samples harbored PIK3CA mutations, and the H1047R, H1047L, E542K, and E545K accounted for 61.95% of the identified mutations [27]. Among the identified mutations, the H1047R is the most common as shown by previous reports [14,24,28].
In our study, we used the PCR-HRM to screen for PIK3CA hotspot mutations in 63 Tunisian patients with sporadic (42) or hereditary (21) breast cancer. Our results showed that 66.7% (28 out of 42) of sporadic BC cases, and 14.3% (3 out of 21) of hereditary BC carried PIK3CA hotspot mutations. In sporadic BC group, the mutation frequency was high compared to other studies reporting that the mutation rate varied between 8 and 40% [24,29]. Furthermore, the E545K and H1048Y were the most prevalent mutations identified in patients with sporadic or hereditary BC, whereas the known hotspot mutation H1047R was not found in our patients. This finding suggests that the H1048Y is likely to be specific to Tunisian BC patients, nevertheless this should be confirmed on larger series.
On the other hand, it was well demonstrated that double PIK3CA mutations in cis activate PI3K pathway and considerably promote tumor growth more than single mutation [30]. In sporadic BC patients, we identified 8 patients among 42 (19%) carrying double PIK3CA mutations while only 2 out of 21 (9.5%) hereditary BC patients harbored double mutations. In a large study including data from 6338 BC patients, 35% of patients carried mutations, the H1047R was identified in 35% of patients [14].
The clinical relevance of PIK3CA mutations has been evaluated in BC. These studies suggested that PIK3CA mutations are associated with a good outcome in patients with ER+/PR+/Her2- BC [31–33]. High frequencies (>30%) of PIK3CA gene mutations were detected in tumors with more favorable features (early tumor grade, ER-positive, PR-positive, and HER2-negative) [29]. In Contrast, the rate of PIK3CA mutations was lower in tumors with aggressive behavior such as the Triple Negative subtype (ER/PR/Her2 negative) [14,24,34]. In patients with metastatic ER+/PR+/Her2- BC, PIK3CA mutations associated with a poor outcome and resistance to chemotherapy [35].
Regarding the prognosis, conflicting results on the association of PIK3CA-mutations were reported. Indeed, some studies found an association to better survival [36,37] whereas other studies found that PIK3CA mutations correlated with short survival [34,38]. The recent study of Reinhardt et al. did not find any association neither with recurrence free interval (RFI) nor with overall survival (OS) within a large cohort of 1123 patients [24]. In our study, we found that BC patients with mutated PIK3CA had a significant overall shorter survival rate compared to those with wild-type PIK3CA tumors, however this finding need to be confirmed on a larger series. These divergent results concerning PIK3CA mutation and prognosis might probably result from the patient origin, sample size, detection method, subgroups, etc.
In conclusions, our results showed a novel PIK3CA hotspot mutation (H1048Y) in Tunisian breast cancer patients detected by HRM-COLD-PCR. The absence of PIK3CA hotspot mutation associated with good prognosis.
Acknowledgements
The authors thank the patients involved in the study.
Ethical approval
This work is approved by Ethic committee of Centre Hospitalo-Universitaire Habib Bourguiba. The study was conducted according to the declaration of Helsinki and written informed consent was obtained from all the patients prior to their participation in the study.
Consent for publication
All authors agree to publish.
Conflict of interest
All authors declare no conflict of interest.
Data availability
The data sets used or analyzed during the study are included in the manuscript.
Funding
None.
Declarations
None.
Author contributions
SDJ designed and performed the experiments, the statistical analysis and wrote the manuscript. FT contributed to the preparation of the samples. WA supervised the HRM experiments. AK gave permission to collect the samples, WBKR, TSB, JD and AK discussed the results and revised the manuscript, and RMG supervised and revised it. All authors approved the final version of the manuscript.
References
[1] | Perou CM, , Sorlie T, , Eisen MB, , van de Rijn M, , Jeffrey SS, , Rees CA , Molecular portraits of human breast tumours, Nature, 406: : 747–752, (2000) . |
[2] | Sung H, , Ferlay J, , Siegel RL, , Laversanne M, , Soerjomataram I, , Jemal A , Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J Clin, 71: : 209–249, (2021) . |
[3] | Missaoui N, , Jaidene L, , Abdelkrim SB, , Ben Abdelkader A, , Beizig N, , Ben Yaacoub L , Breast cancer in tunisia: Clinical and pathological findings, Asian Pac J Cancer Prev, 121: : 69–72, (2011) . |
[4] | Parkin DM, , Ferlay J, , Hamdi-Cherif M, , Sitas F, , Thomas J, , Wabinga H , Breast cancer in Africa: Epidemiology and prevention, IARC Sci Publ Lyon Fr, 153: : 262–267, (2003) . |
[5] | Goncalves MD, , Hopkins BD, , Cantley LC, Phosphatidylinositol 3-kinase, growth disorders, and cancer, N Engl J Med, 21: : 2052–2062, (2018) . |
[6] | Samuels Y, , Wang Z, , Bardelli A, , Silliman N, , Ptak J, , Szabo S , High frequency of mutations of the PIK3CA gene in human cancers, Science, 304: : 554, (2004) . |
[7] | Phillips J, , Domingo E, PIK3CA (phosphoinositide-3-kinase, catalytic, alpha polypeptide), Atlas Genet Cytogenet Oncol Haematol, (2019) . doi:10.4267/2042/70181. |
[8] | Vivanco I, , Sawyers CL, The phosphatidylinositol 3-Kinase AKT pathway in human cancer, Nat Rev Cancer, 2: : 489–501, (2002) . |
[9] | Katso R, , Okkenhaug K, , Ahmadi K, , White S, , Timms J, , Waterfield MD, Cellular function of phosphoinositide 3-kinases: Implications for development, homeostasis, and cancer, Annu Rev Cell Dev Biol, 17: : 615–675, (2001) . |
[10] | Samuels Y, , Velculescu VE, Oncogenic mutations of PIK3CA in human cancers, Cell Cycle, 3: : 1221–1224, (2004) . |
[11] | Karakas B, , Bachman KE, , Park BH, Mutation of the PIK3CA oncogene in human cancers, Br J Cancer, 94: : 455–459, (2006) . |
[12] | Fendri A, , Khabir A, , Mnejja W, , Sellami-Boudawara T, , Daoud J, , Frikha M , PIK3CA amplification is predictive of poor prognosis in Tunisian patients with nasopharyngeal carcinoma, Cancer Science, 11: : 2034–2039, (2009) . |
[13] | Arafeh R, , Samuels Y, PIK3CA in cancer: The past 30 years, Semin Cancer Biol, 59: : 36–49, (2019) . |
[14] | Martínez-Sáez O, , Chic N, , Pascual T, , Adamo B, , Vidal M, , González-Farré B , Frequency and spectrum of PIK3CA somatic mutations in breast cancer, Breast Cancer Res, 22: (1): 45, (2020) . |
[15] | Jin J, , Shi Y, , Zhang S, , Yang S, PIK3CA mutation and clinicopathological features of colorectal cancer: A systematic review and Meta-Analysis, Acta Oncol, 59: (1): 66–74, (2020) . |
[16] | Campbell IG, , Russell SE, , Phillips WA, PIK3CA mutations in ovarian cancer, Clin Cancer Res, 1: : 7042–7043, (2005) . |
[17] | Ligresti G, , Militello L, , Steelman LS, , Cavallaro A, , Basile F, , Nicoletti F , PIK3CA mutations in human solid tumors: Role in sensitivity to various therapeutic approaches, Cell Cycle, 8: : 1352–1358, (2009) . |
[18] | Taylor CF, Mutation scanning using high-resolution melting, Biochem Soc Trans, 37: 433–437, 2009. |
[19] | Graham R, , Liew M, , Meadows C, , Lyon E, , Wittwer CT, Distinguishing different DNA heterozygotes by high-resolution melting, Clin Chem, 51: : 1295–1298, (2005) . |
[20] | Margraf RL, , Mao R, , Highsmith WE, , Holtegaard LM, , Wittwer CT, RET proto-oncogene genotyping using unlabeled probes, the masking technique, and amplicon high-resolution melting analysis, J Mol Diag, 9: : 184–196, (2007) . |
[21] | Krypuy M, , Ahmed AA, , Etemadmoghadam D, , Hyland SJ, , deFazio A, , Fox SB , High resolution melting for mutation scanning of TP53 exons 5-8, BMC Cancer, 7: : 168, (2007) . |
[22] | Takano T, , Ohe Y, , Tsuta K, , Takano T, , Ohe Y, , Tsuta K , Epidermal growth factor receptor mutation detection using high-resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib, Clin Cancer Res, 13: : 5385–5390, (2007) . |
[23] | Pritchard CC, , Akagi L, , Reddy PL, , Jonathan LJ, COLD-PCR enhanced melting curve analysis improves diagnostic accuracy for KRAS mutations in colorectal carcinoma, BMC Clinical Pathology, 10: : 6, (2010) . |
[24] | Reinhardt K, , Stückrath K, , Hartung C, , Kaufhold S, , Uleer C, , Hanf V , PIK3CA-mutations in breast cancer, Breast Cancer Res Treat, 196: (3): 483–493, (2022) . |
[25] | Pevzner AM, , Gaptulbarova KA, , Tsyganov MM, , Ibragimova KM, , Vvedensky VA, , Zhusina GY , Investigation of somatic PIK3CA gene mutations in breast cancer patients, J BUON, 26: (3): 747–752, (2021) . |
[26] | Kim JW, , Lim AR, , You JY, , Lee JH, , Song SE, , Lee NK , PIK3CA mutation is associated with poor response to HER2-targeted therapy in breast cancer patients, Cancer Res Treat, 55: (2): 531–541, (2023) . |
[27] | Ren X, , Cui H, , Dai L, , Chang L, , Liu D, , Yan W , PIK3CA mutation-driven immune signature as a prognostic marker for evaluating the tumor immune microenvironment and therapeutic response in breast cancer, J Cancer Res & Clin Oncol, 150: : 119, (2024) . |
[28] | Chang DY, , Ma WL, , Lu YS, Role of alpelisib in the treatment of PIK3CA-mutated breast cancer: Patient selection and clinical perspectives, Ther Clin Risk Manag, 17: : 193–207, (2021) . |
[29] | Forbes SA, , Bindal N, , Bamford S, , Cole C, , Kok CY, , Beare D , COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer, Nucleic Acids Res, 39: : 945–950, (2011) . |
[30] | Vasan N, , Razavi P, , Johnson JL, , Shao H, , Shah H, , Antoine A , Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors, Science, 366: (6466): 714–723, (2019) . |
[31] | Kalinsky K, , Jacks LM, , Heguy A, , Patil S, , Drobnjak M, , Bhanot UK , PIK3CA mutation associates with improved outcome in breast cancer, Clin Cancer Res, 15: (16): 5049–5059, (2009) . |
[32] | Zardavas D, , Te Marvelde L, , Milne RL, , Fumagalli D, , Fountzilas G, , Kotoula V , Tumor PIK3CA genotype and prognosis in early-stage breast cancer: A pooled analysis of individual patient data, J Clin Oncol, 36: : 981–990, (2018) . |
[33] | Cizkova M, , Susini A, , Vacher S, , Magdalena C, , Susini A, , Vacher S , PIK3CA mutation impact on survival in breast cancer patients and in ERa, PR and ERBB2-based subgroups, Breast Cancer Res, 14: : 28, (2012) . |
[34] | Sobhani N, , Roviello G, , Corona SP, , Scaltriti M, , Ianza A, , Bortul M , The prognostic value of PI3K mutational status in breast cancer: A meta-analysis, J Cell Biochem, 119: : 4287–4292, (2018) . |
[35] | Mosele F, , Stefanovska B, , Lusque A, , Tran Dien A, , Garberis I, , Droin N , Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer, Ann Oncol, 31: (3): 377–386, (2020) . |
[36] | Dumont AG, , Dumont SN, , Trent JC, The favorable impact of PIK3CA mutations on survival: An analysis of 2587 patients with breast cancer, Chin J Cancer, 31: : 327–334, (2012) . |
[37] | Bo P, , Shi C, , Shi-Peng S, , An C, , Liu ZY, , Feng X , Prognostic role of PIK3CA mutations and their association with hormone receptor expression in breast cancer: A meta-analysis, Sci Rep, 4: : 1–9, (2015) . |
[38] | Fan H, , Li C, , Xiang Q, , Xu L, , Zhang Z, , Liu Q , PIK3CA mutations and their response to neoadjuvant treatment in early breast cancer: A systematic review and meta-analysis, Thorac Cancer, 9: : 571–579, (2018) . |




