Meningeal Bmps Regulate Cortical Layer Formation
Abstract
Neuronal connectivity in the cortex is determined by the laminar positioning of neurons. An important determinant of laminar positioning is likely to be the control of leading process behavior during migration, maintaining their tips directed toward the pia. In this study, we provide evidence that pial bone morphogenetic protein (Bmp) signaling regulates cortical neuronal migration during cortical layer formation. Specific disruption of pial Bmp ligands impaired the positioning of early-born neurons in the deep layer; further, cell-autonomous inhibition of Smad4, a core nuclear factor mediating Bmp signaling, in the cortical radial glial cells or postmitotic cortical neurons also produced neuronal migration defects that blurred the cortical layers. We found that leading processes were abnormal and that this was accompanied by excess dephosphorylated cofilin-1, an actin-severing protein, in Smad4 mutant neurons. This suggested that regulation of cofilin-1 might transduce Bmp signaling in the migrating neurons. Ectopic expression of a phosphorylation-defective form of cofilin-1 in the late-born wild-type neurons led them to stall in the deep layer, similar to the Smad4 mutant neurons. Expression of a phosphomimetic variant of cofilin-1 in the Smad4 mutant neurons rescued the migration defects. This suggests that cofilin-1 activity underlies Bmp-mediated cortical neuronal migration. This study shows that cofilin-1 mediates pial Bmp signaling during the positioning of cortical neurons and the formation of cortical layers.
INTRODUCTION
Cortical projection neurons extend axons to other cortical neurons or to subcortical areas such as the thalamus, the pons and the spinal cord. These cells are organized in part based on their laminar locations [1, 2]. Subcortical projection neurons mostly localize in deeper layers and are generated before more superficially located neurons. During radial migration, superficial neurons migrate toward the pia, bypassing earlier-generated neurons, to occupy locations below the marginal zone, where the early-generated Cajal-Retzius neurons form close contacts with the pial extracellular matrix [3]. The early-generated deep-layer cortical neurons also have processes that reach the pia at embryonic day (E)14.5, before late-generated neurons arrive and occupy locations closer to the pia [4]. The later-generated superficial neurons are guided by radial glial cells during migration toward the pia [5, 6].
Cortical projection neurons migrate away from the neurogenic ventricular zone and to the cortical plate, which forms below the marginal zone and the pial meninges. We previously showed that Bmp signaling from the mesenchymal cells covering the cortex, including the meninges, regulates cingulate cortical axon growth [7] and dentate gyrus neurogenesis [8]. From these previous studies, we observed that dentate granule neurons were dispersed and failed to form condensed dentate blades in the conditional Smad4 mutants, suggesting that Smad4-mediated Bmp signaling might regulate neuronal migration and layer formation as well as dentate neurogenesis. Bmp signaling induces phosphorylation of Smad1/5/8 and the formation of heterodimers of Smad1/5/8 with Smad4 before initiating Bmp target gene expression in the nucleus [9, 10], and we found that not only neurogenic progenitors but also postmitotic neurons showed phosphorylation of Smad1/5/8 [7, 8]. However, the dentate gyrus lacks clear layer boundaries as are found in the cortex; thus, it was difficult to delineate the role of Bmp signaling in neuronal migration. The cortex is covered by three layered meninges, which secrete diverse signaling molecules including Bmps during neurogenesis [7, 8]. We hypothesized that the morphogenic gradient consisting of meningeally released Bmps might direct radial migration of cortical neurons. This is perhaps consistent with a previous report showing that injection of Bmp7 in the embryonic ventricle affects neurogenesis as well as neuronal migration [11]. Here, we focused on the mesenchymal or basal Bmps, not ventricular or apical Bmps, and examined the interaction of mesenchymal cells and neuronal cells during embryonic cortical development. We present evidence for the first time supporting the idea that the mesenchymal-cortical neuronal interaction regulates radial neuronal migration in the developing cortex. Pial Bmp signaling appears to provide a positional cue that helps to determine the boundary between upper-layer and deep-layer neurons. Postmitotic neurons in the cortical plates were found to be Bmp-responsive during embryonic cortical layer formation. Inhibition of Smad4-mediated Bmp signaling in the postmitotic cortical neurons resulted in the over-migration of deep-layer neurons and the delayed migration of late-generated neurons, thus disrupting the precise boundary between deep-layer and upper-layer neurons. Reduced expression of pial Bmps produced similar over-migration of deep-layer neurons, implying that the non-neural Bmps function as signals for segregating the upper layer and the deep layer. Smad4-mediated Bmp signaling was further revealed to regulate cofilin-1 activities of cortical neurons, and introduction of a phosphorylation-mimicking variant of cofilin-1 rescued the neuronal migration phenotype in Smad4 mutant cortical neurons. This study suggests that the mesenchymal Bmp signaling controls cortical neuronal migration and laminar layer formation by regulating cofilin-1-mediated cytoskeletal changes in migrating cortical neurons.
EXPERIMENTAL PROCEDURES
Animals
Mice used in this study have been described previously (Pdgfrβ-Cre [12], hGFAP-Cre [13], Nex-Cre [14], Wnt1-Cre [15], Foxc1flx [16], Smad4flx [17], Bmp4flx [18], and Bmp7flx [19]). Experimental mice were obtained by crossing male mice carrying an allele of a Cre recombinase and a heterozygous allele of floxed gene to female mice carrying homozygous floxed genes or floxed/null alleles. The day of vaginal plug was considered to be embryonic day 0.5 (E0.5). Mouse colonies were housed at the University of California, San Francisco, in accordance with UCSF IACUC guidelines.
In utero electroporation
Timed pregnant CD1 mice were purchased from Charles River, and surgery was performed according to IACUC approved protocols at UCSF. Smad4flx homozygous mice were bred on a mixed background for the surgery. Briefly, the pregnant females at E13.5 were anaesthetized with Nembutal. The uterine horns were exposed, and embryos were injected with 2 mg/ml DNA in TE into the lateral ventricle. Electroporation was conducted at 33V, 50 ms, 950 ms with five pulses as previously described [8]. All DNA constructs were cloned into the pCIG2-IRES-EGFP vector from Dr. Franck Polleux [8], except for the cofilin-1 variants, which were cloned in the Z/EG vector [20] by replacing the Gfp with the cofilin-1 variants. Cre-inducible cofilin-1 variants were generated by PCRs using specific primers containing an S3A or S3D mutation of mouse cofilin-1 cDNA. A 1 kb region of the tubulin alpha 1 (T1α) promoter was cloned upstream of a Cre recombinase cDNA or Gfp cDNA to make T1α-Cre or T1α-GFP, respectively [21]. All template full-length cDNAs were obtained by reverse transcription PCR amplification. All experiments were repeated to yield three electroporated embryos from more than three independent experiments.
Immunostaining
Embryos were collected at noon on the indicated embryonic days. Collected brains were fixed in 4% paraformaldehyde (PFA)/PBS for four hours and cryoprotected in 15% sucrose/PBS for an additional day. OCT-embedded tissues were processed in a cryostat to produce 12 μm sections. We compared control and mutant sections on the same slide to minimize variation between groups. Primary antibodies used for the immunostaining were as follows: chicken anti-GFP (Aves Labs (GFP-1020), 1 : 1000), rabbit anti-pSmad1/5/8 (Cell Signaling (9511), 1 : 200), Ctip2 (Abcam (AB18465), 1 : 1000), mouse anti-Satb2 (Abcam (AB51502), 1 : 200), rabbit anti-cofilin-1 (Cell Signaling (5175), 1 : 300), rabbit anti-phospho-cofilin-1 (Cell Signaling (3313), 1 : 300), rabbit anti-pLimk1/2 (Santa Cruz Biotech. (SC28409), 1 : 100), mouse anti-Tuj1 (Millipore (MAB1637), 1 : 1000), rabbit anti-Cre (Covance (PRB-106P), 1 : 300), rabbit anti-Cux1 (Santa Cruz Biotech. (SC-13024), 1 : 100), rabbit anti-Map2 (Millipore (AB5622), 1 : 500), and rabbit-alpha tubulin (Abcam (AB18251), 1 : 1000). Alexa Fluor 568 Phalloidin was purchased from Invitrogen (1 : 1000). Fluorescence images were acquired at the Nikon Imaging Center, UCSF, using an upright Nikon C1 spectral confocal microscope or a fluorescence microscope equipped with a QImaging Retiga CCD-cooled camera using QCapture Pro software (QImaging). To measure the fluorescence intensity through the cortical sections, the histogram analysis function of ImageJ software was used drawing rectangular boxes between the pia and the ventricular surface.
Statistics
Values are presented as the mean±SEM. For the statistical analysis of samples, we used Student’s t-test using SigmaPlot (Systat Software Inc.) and GraphPad software (GraphPad Software) unless otherwise stated in the figure legend.
RESULTS
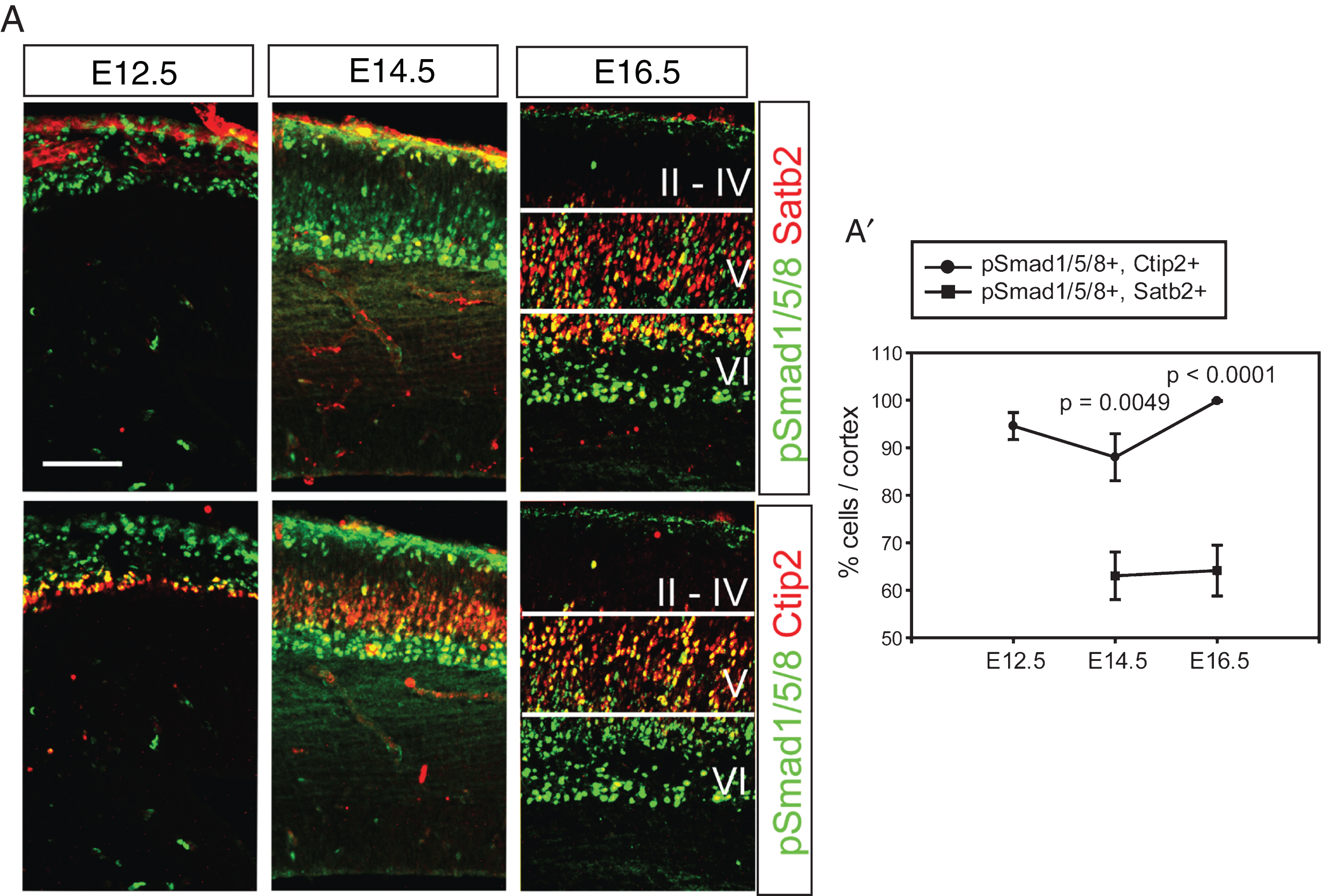
Expression of Bmps was initially detected in the dorsal midline and restricted to the non-neural tissues such as the meninges and the choroid plexus during cortical neurogenesis [7]. The meningeal Bmps and the choroidal Bmps are associated with the basal and apical aspects of cortical neurogenesis, respectively. Activation of Bmp signaling induces phosphorylation of Smad transcription factors such as Smad1, Smad5, and Smad8 and their heterodimerization with Smad4 in the responding cells [9, 10]. To visualize activation of the Bmp signaling pathway in the early- (Ctip2+) and late-born (Satb2+) neurons, triple staining for phospho-Smad1/5/8 (pSmad1/5/8), Ctip2 and Satb2 was conducted using the same brain sections. Separate images of pSmad1/5/8 (green) and Ctip2 (red) or pSmad1/5/8 (green) and Satb2 (red) at E12.5 and E14.5 clearly show that post-migratory neurons were activated by Bmp signaling and that migratory neurons in the intermediate zone are weakly positive for pSmad1/5/8 (Fig. 1A). The radial glial cells facing the lateral ventricle are also clearly stained for pSmad1/5/8 during mitosis, as shown by co-staining of phospho-Histone H3 (pH3), suggesting that Bmp signaling was active in the lateral ventricle (∼90%, Supplementary Figure 1). At E16.5, Bmp signaling was highly active in the layer V/VI and reduced in the layer II/IV and barely detected in the marginal zone and the intermediate zone (Fig. 1A). This result clearly shows that most of the Ctip2+ deep-layer neurons responded to Bmp signaling and that 60% of Satb2+ neurons were pSmad1/5/8+ during embryonic cortical development, implying that Bmp signaling may function during the formation of cortical layers (Fig. 1A and A’, p = 0.0049 (E14.5), p < 0.0001 (E16.5) comparison of pSmad1/5/8+, Ctip2+ vs. pSmad1/5/8+, Satb2+ cells (n = 3)).
Fig.1
Activation of Bmp signaling in the postmitotic cortical neurons. A) Double staining of pSmad1/5/8 and Satb2 or Ctip2 during cortical neurogenesis. The same section was used to triple label pSmad1/5/8, Satb2 and Ctip2, and separate images are presented to show Satb2- and Ctip2-expressing neurons (red) in the presence of the same pSmad1/5/8 signals (green) in a tissue. A’) Double-stained cells were plotted. Error bars represent the SEM. Student’s t-test was conducted to determine the statistical significance of the difference between pSmad1/5/8+;Ctip2+ and pSmad1/5/8+; Satb2+ n = 3). Scale bars = 100 μm.
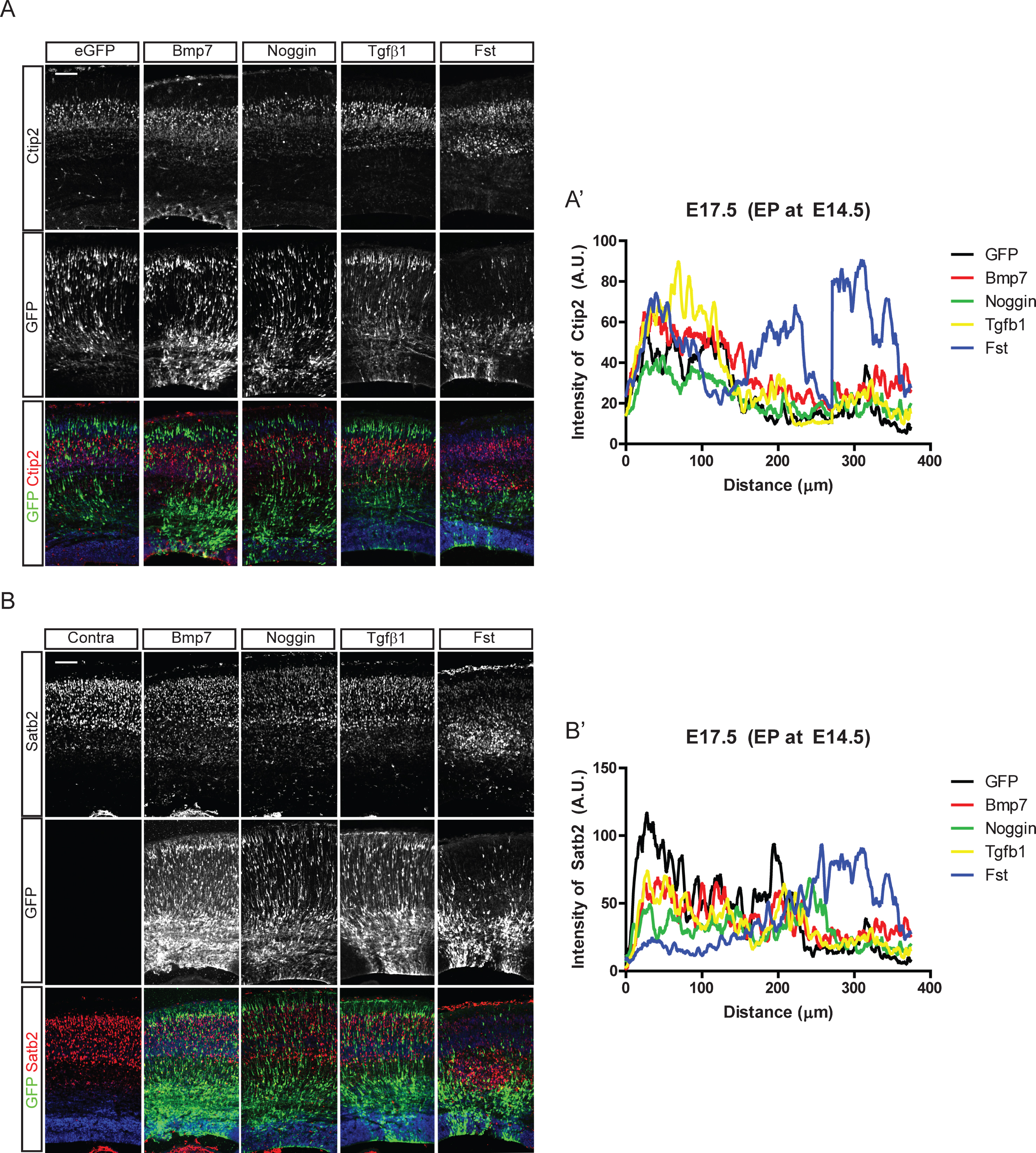
It is not known whether Bmp signaling is involved in cortical neuronal migration, the cellular basis for cortical layer formation. Thus, we first examined whether regulating Bmp activities in the embryonic cortex affects the migration of cortical neurons. We introduced Bmp7, Tgfβ1, Noggin, and Follistatin (Fst) into E14.5 cortical ventricular cells by in utero electroporation and stained cortical neurons for Ctip2, labeling early-born neurons, and Satb2, labeling late-born neurons. As shown in Fig. 2, the early-born Ctip2+ neurons did not show drastic migratory defects compared to the late-born Satb2+ neurons. Fst affected migration of Satb2+ neurons that left the ventricular zone at the time of electroporation, stalling Satb2+ upper-layer neuron migration in deeper layers. Fst and Noggin, inhibitors of Bmp signaling, were previously shown to have different developmental functions because of their different Bmp binding potency [22]. Interestingly, Noggin, another Bmp inhibitor, did not show the dramatic effect shown by Fst. However, ectopic expression of Bmp7 produced mild migratory defects represented by migrating GFP-expressing neurons in the deep layer, including the ventricular zone (Fig. 2A, A’, B, B’). Ectopic expression of Bmp4 also showed a similarly mild effect on layer formation (data not shown), but Tgfβ1 did not affect the formation of the Ctip2+ layer V. These in utero electroporation results suggest that inhibition and activation of Bmp signaling may regulate radial migration of cortical neurons and that the effect is dependent on the stage of the neuronal migration.
Fig.2
Effect of Bmp signaling on the cortical neuronal distribution. In utero electroporation with Bmp7, Noggin, Tgfβ1, and Follistatin (Fst) and IRES-driven eGFP constructs was conducted at E14.5 in CD1 mice. Three days after the electroporation, eGFP-expressing neurons were co-stained for Ctip2 (A) or Satb2 (B). A’, B’) Intensities of Ctip2+ and Satb2+ neurons in the electroporated field were plotted using the plot profile function in ImageJ software, and results from one representative experiment are shown (n = 3). For a control image of Satb2, the contralateral side of the electroporated brain (A) is used. Scale bars = 100 μm.
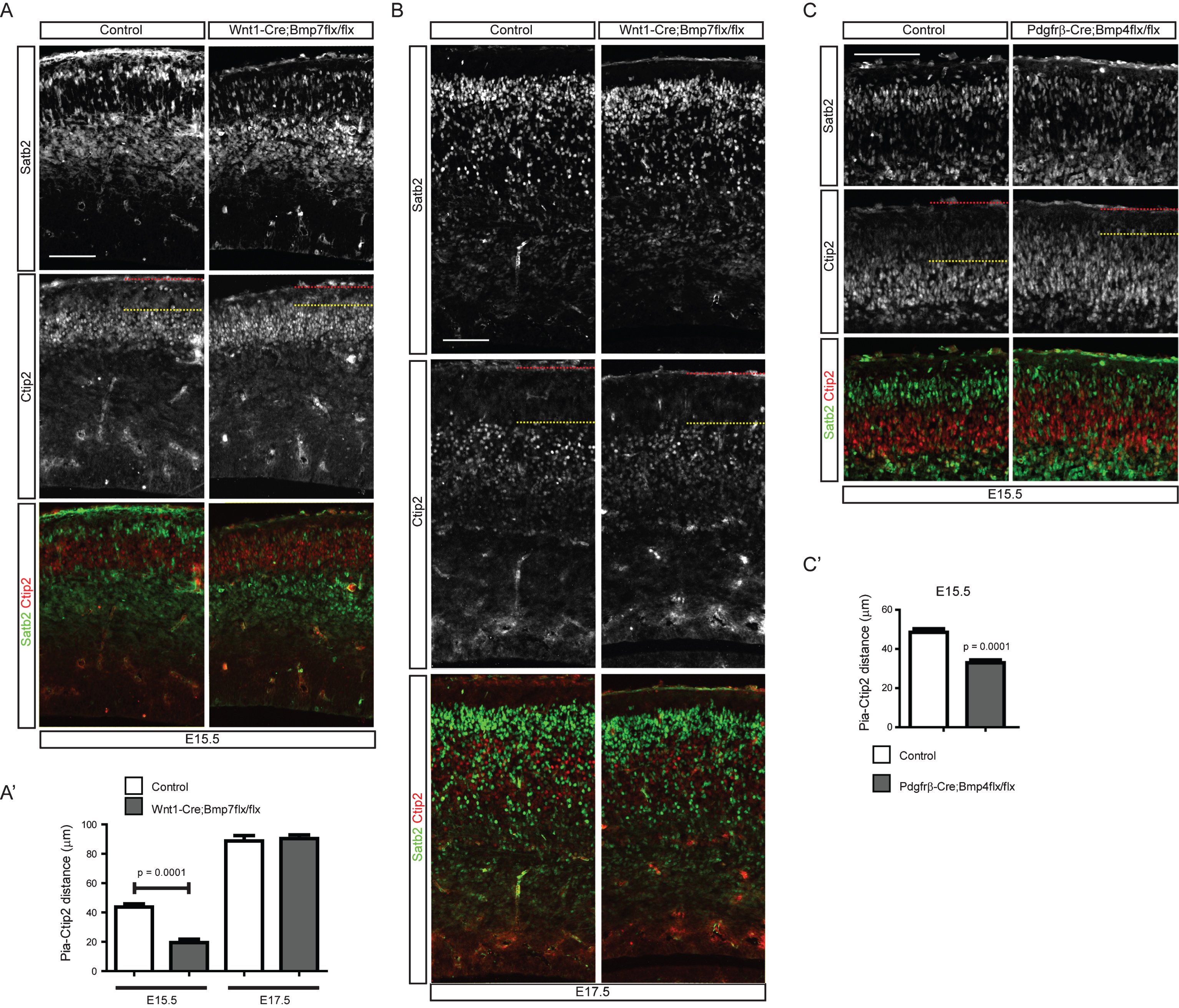
In the previous experiments, we showed that activation of Bmp signaling was detected in the postmitotic neurons close to the meninges, a potential source of Bmps. We have also reported that dentate granule neurons are abnormally scattered in meningeal mutants, implying that Bmp signaling may regulate neuronal positioning and migration [8]. To examine the effect of meningeal Bmp ligands on the cortical neuronal migration, we used two different approaches to inhibit meningeal Bmp expression. Firstly, we employed an indirect method to reduce Bmp expression by inhibiting meningeal development using Foxc1 conditional mice. Foxc1 is a critical transcription factor that regulates differentiation of the meninges from the cranial neural crest cells [23]. To reduce the formation of meningeal tissues, we used Pdgfrβ-Cre, which is expressed in the neural crest-derived mesenchymal cells, including the meninges, to conditionally inactivate Foxc1; the meninges specificity of these mutant mice was reported previously [7]. In the Pdgfrβ-Cre;Foxc1flx/flx conditional mutant mice, neurogenesis was not significantly affected for Ctip2+ and Satb2+ neurons at E15.5 and E17.5 (Fig. 3A, A’, B, and B’). However, Ctip2+ and Satb2+ neurons were broadly scattered in the cortical plate and intermediate zone with loss of clear boundaries between the cortical layers. Ctip2+ neurons migrated closer to the pia, and some of the Satb2+ neurons did not pass the Ctip2+ neurons at E15.5. At E17.5, the cortical plate of the conditional mutant mice was thicker at the level of the hippocampal commissure (Fig. 3B). In the mutants, Satb2+ neurons were observed in the intermediate zone, and Ctip2+ neurons migrated toward the pia. To quantitatively measure the neuronal scattering, we measured the distance from the front line of neurons (the line was drawn between the fifth and sixth neurons closest to the pia) to the pia, and the distance is illustrated in Fig. 3C. Ctip2+ neurons also failed to organize into the deep layer and Satb2+ neurons into the upper layer at E17.5 (Fig. 3A, A”, B, B” (n = 4, sections at the level of the hippocampal fissure were used)). Since the meninges express diverse morphogenic molecules including Bmps, we also employed more direct approaches to examine the effects of meningeal Bmp ligands on cortical neuronal migration. Bmp4 and Bmp7 are the major Bmps expressed by the meninges. Thus, we tried to directly decrease Bmp4 and Bmp7 expression using two different Cre lines [8]. Wnt1-Cre is expressed in the neural crest cells, and Pdgfrβ-Cre is expressed in mesenchymal cells, including the meninges [12, 15]. We conditionally deleted Bmp7 using Wnt1-Cre and Bmp4 using Pdgfrβ-Cre as we previously reported [8]. Interestingly, at E15.5, both mutants showed aberrant migratory behavior of cortical neurons, a failure of Ctip2+ neurons to relocate into the deep layer and a failure of Satb2+ neurons to pass the Ctip2+ neurons, similar to the Pdgfrβ-Cre;Foxc1flx/flx conditional mutants, but the defect was not detected at E17.5 (Fig. 4A, A’ (p < 0.0001 (n = 3), B, C, C’ (p = 0.0001 (n = 3)). The lack of a migration phenotype at E17.5 might be caused by compensation by the accumulation of other Bmps expressed in the mutant meninges, leading to incomplete inhibition of Bmp signaling in the cortical neurons at E17.5. We could not delete all Bmps in the developing meninges, and this will be crucial for explaining the compensatory effects. Nonetheless, these conditional mutants of meningeal Bmp ligands directly show for the first time that mesenchymal tissues are involved in the regulation of the cortical neuronal migration and the formation of the cortical layers.
Fig.3
Distribution of cortical neurons in the Pdgfrβ-Cre;Foxc1flx/flx meningeal mutant. Embryonic brains were stained for Satb2 or Ctip2 at E15.5 (A) and E17.5 (B). The numbers of Ctip2+ and Satb2+ neurons were plotted (n = 4, A’, B’). The distances between the pia and the Ctip2+ or Satb2+ neurons closest to the pia were plotted (n = 4, A”, B”) (refer to the drawing in C). Error bars represent the SEM. Student’s t-test was conducted to determine the statistical significance of the difference between the control and mutant embryos. The red dotted lines and the yellow dotted lines represent the pia and the sixth-closest Ctip2+ neurons to the pia, respectively. Scale bars = 100 μm.
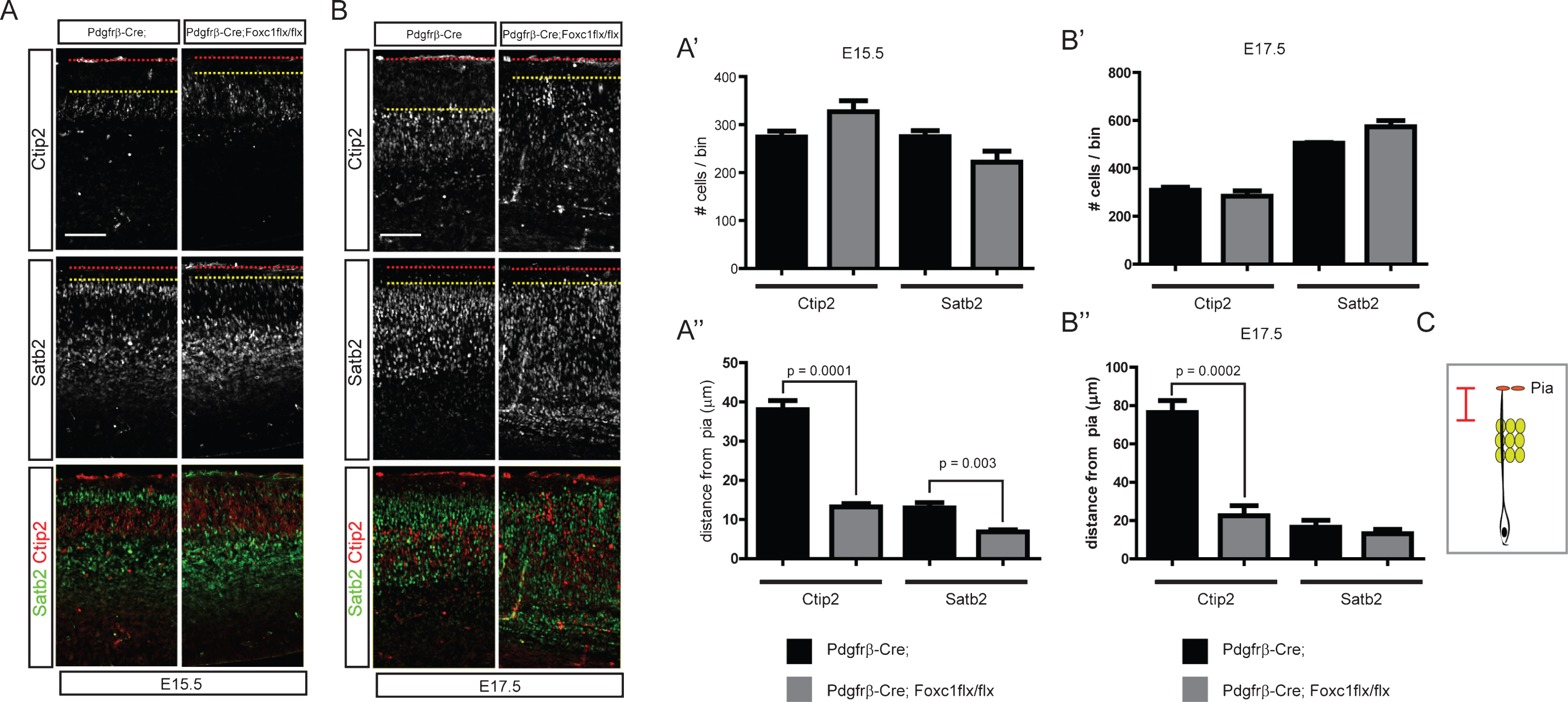
Fig.4
Distribution of cortical neurons in the meninges-specific Bmp mutants. A) E15.5 embryonic brains of Wnt1-Cre;Bmp7flx/flx mutants and their control littermates were stained for Ctip2 or Satb2. A’) The distance of the pia and the Ctip2+ neurons were plotted (n = 3). B) E17.5 embryonic brains of Wnt1-Cre;Bmp7flx/flx mutants and their control littermates were stained for Ctip2 or Satb2 (n = 3). C) E15.5 embryonic brains of Pdgfrβ-Cre;Bmp4flx/flx mutants and their control littermates were stained for Satb2 or Ctip2. C’) The distances between the pia and the Ctip2+ neurons were plotted (n = 3). The red and yellow dotted lines represent the pia and the Ctip2+ neurons closest to the pia, respectively. Student’s t-test was conducted to determine the statistical significance of the difference between the control and mutant embryos. Scale bars = 100 μm.
Bmp signaling affects proliferation of neuronal progenitor cells (Supplementary Figure 1 and unpublished results) and thus the affected cell cycle of the neuronal progenitor cells should lead to the malformation of the cortical layers. To separate the effect of Bmps on cell proliferation, we utilized two different approaches using postmitotic Cre drivers and doing in utero electroporation with a postmitotic neuronal promoter. Migration of cortical neurons is orchestrated to produce the final layered cortical structure, and this coordinated cortical development is controlled by transcription factors in the migrating neurons. The Bmp signaling pathway activates downstream targets of heterodimeric Smad transcription factors that contain Smad4, a critical heterodimeric partner [9, 10]. To examine whether the effect of meningeal Bmps on cortical neuronal migration requires Smad4-regulated gene expression, we used Smad4 conditional mice. For the conditional inactivation of Smad4 alleles, we used two different Cre lines, hGFAP-Cre and Nex-Cre, to express Cre recombinases in the cortical radial glial cells and postmitotic cortical neurons, respectively. We used hGFAP-Cre;Smad4flx/- pups instead of Smad4flx/flx mice since the double floxed mice showed low penetrance (Data not shown). Co-staining of Ctip2 and Cux1, an upper layer neuronal marker, showed the overlapping area of Cux1+ neurons and Ctip2+ neurons in the hGFAP-Cre;Smad4flx/- mutant pups at postnatal day 1 (P1). Ctip2+ neurons were localized near the pia, which may have occurred because the upper layer neurons did not successfully pass the Ctip2+ neurons, similar to the meninges mutants. This resulted in the failure of Ctip2+ neuronal localization in deeper layers and instead led to the localization of Cux1+ neurons in deep layers (Fig. 5A, A’, and A”, p = 0.0012 (n = 6) for Ctip2+ neurons). Neurogenesis was not severely affected, as shown by the numbers of Ctip2+ and Cux1+ neurons in the cortex (Fig. 5A’), but it is possible that the migration phenotype in hGFAP-Cre mutants may be due to a role for Smad4 in radial glial cells rather than in migrating neurons. Thus, to exclude the possible effect of Smad4 deletion in the radial glial cells, we examined Nex-Cre-mediated inactivation of Smad4. Nex-Cre is selective for postmitotic migrating neurons and some intermediate progenitor cells [14, 24], and the number of Ctip2+ neurons was not changed in the Nex-Cre;Smad4flx/flx mutants at E17.5 (Data not shown). Similar to the hGFAP-Cre-mediated Smad4 mutants, Nex-Cre;Smad4flx/flx mutants also showed failure of Ctip2+ neurons to localize in layer V at E17.5 (Fig. 5B, and B’, p < 0.0001 (n = 6)). Cell-autonomous inhibition of Bmp signaling in the migrating cortical neurons showed drastic migration defects of the Cux1+ neurons, which is comparable to the Satb2+ neurons in the developing meningeal mutants. This difference probably implies pleiotropic roles of Bmp family molecules in the formation of cortical layers, affecting stop/go signals in the different types of cortical neurons. At E16.5, Ctip2+ neurons were mostly activated by Bmps, but the trespassing Satb2 neurons were partially activated (Fig. 1). Considering the region of active Bmp signaling, we reasoned that this migration phenotype might be caused by the defective final localization of neurons in relation to the pial surface. The leading processes of the migrating neurons are important for interaction with the pial layer [25]. We used Nex-Cre;Smad4flx/flx mutants to stain Map2, which marks the leading process (which will later become the primary dendrite) of migrating cortical neurons at E17.5. This result showed aberrant staining of Ctip2+ neurons expressing Map2 in the marginal zone, likely resulting from the abnormal projection of the leading processes (Fig. 5B). It has been shown that Bmp regulates dendritic development of in vitro cultured neurons [26–28], but the present result shows for the first time that Bmp controls the leading processes, future dendrites, of migrating neurons in vivo. To examine the cytoskeletal changes in cortical neurons in response to Bmp signaling, we treated cortical neurons with Bmp7 (20 ng/ml) or Noggin (40 ng/ml) after 5 days of in vitro culture. Examining cell morphology after treatment showed that the tips of neurites were severely affected. We counted the numbers of cells containing “collapsed” or “motile” neurites (containing filopodia or lamellipodia) from more than 100 neurons per experiment, as shown in Fig. 6A”. We found that Bmp7 induced collapse of neurites, while Noggin showed the opposite effect (Fig. 6A, A’, B, and B’, vehicle vs. Bmp7; p < 0.001 (n = 3), vehicle vs. Noggin; p < 0.05 (n = 3)). Recent studies showed that Cajal-Retzius cells regulate cortical neuronal migration through regulating cofilin-1, an actin-severing protein [25, 29]. Reelin (Cajal-Retzius cells) and Bmps (meningeal cells) are both basally localized signaling molecules; thus, it may be possible that they interact to regulate neuronal migration by controlling the same cellular machinery. To test this idea, we incubated cortical neurons cultured in vitro for 5 days with Bmp7 (20 ng/ml) for various lengths of time and stained them for phosphorylated Limk1/2, kinases that phosphorylate cofilin-1, and phosphorylated cofilin-1. Phosphorylation of both Limk1/2 and cofilin-1 was increased at 8 hr after treatment and decreased at 24 hr (Fig. 6C). This result suggests that the cytoskeletal changes observed in cortical neurons treated with Bmp7 or Noggin might be affected by the regulation of cofilin-1 by Bmp signaling.
Fig.5
Distribution of cortical neurons in the Smad4 conditional mutant mice. A) Brain sections of postnatal day 1 (P1) hGFAP-Cre;Smad4flx/- mutants and their control littermates were stained for Cux1, an upper layer neuronal marker, and Ctip2. Representative images obtained from the level of the corpus callosum (anterior) and the hippocampal commissure (posterior) are presented. The drawing in the upper panel shows the loss of the boundary between the Cux1 (red) and the Ctip2 (green) layers in mutants. A’) The numbers of Ctip2+ and Cux1+ neurons were plotted (n = 6). A”) The distances between the pia and the Ctip2+ and Cux1+ neurons are shown (n = 6). B) E17.5 Nex-Cre;Smad4flx/flx mutants were stained for Ctip2 and Map2. B’) The distances between the pia and the Ctip2+ neurons were plotted (n = 6). The red and yellow dotted lines represent the pia and the Ctip2+ neurons closest to the pia, respectively. Student’s t-test was conducted to determine the statistical significance of the difference between the control and mutant embryos. Scale bars = 100 μm.
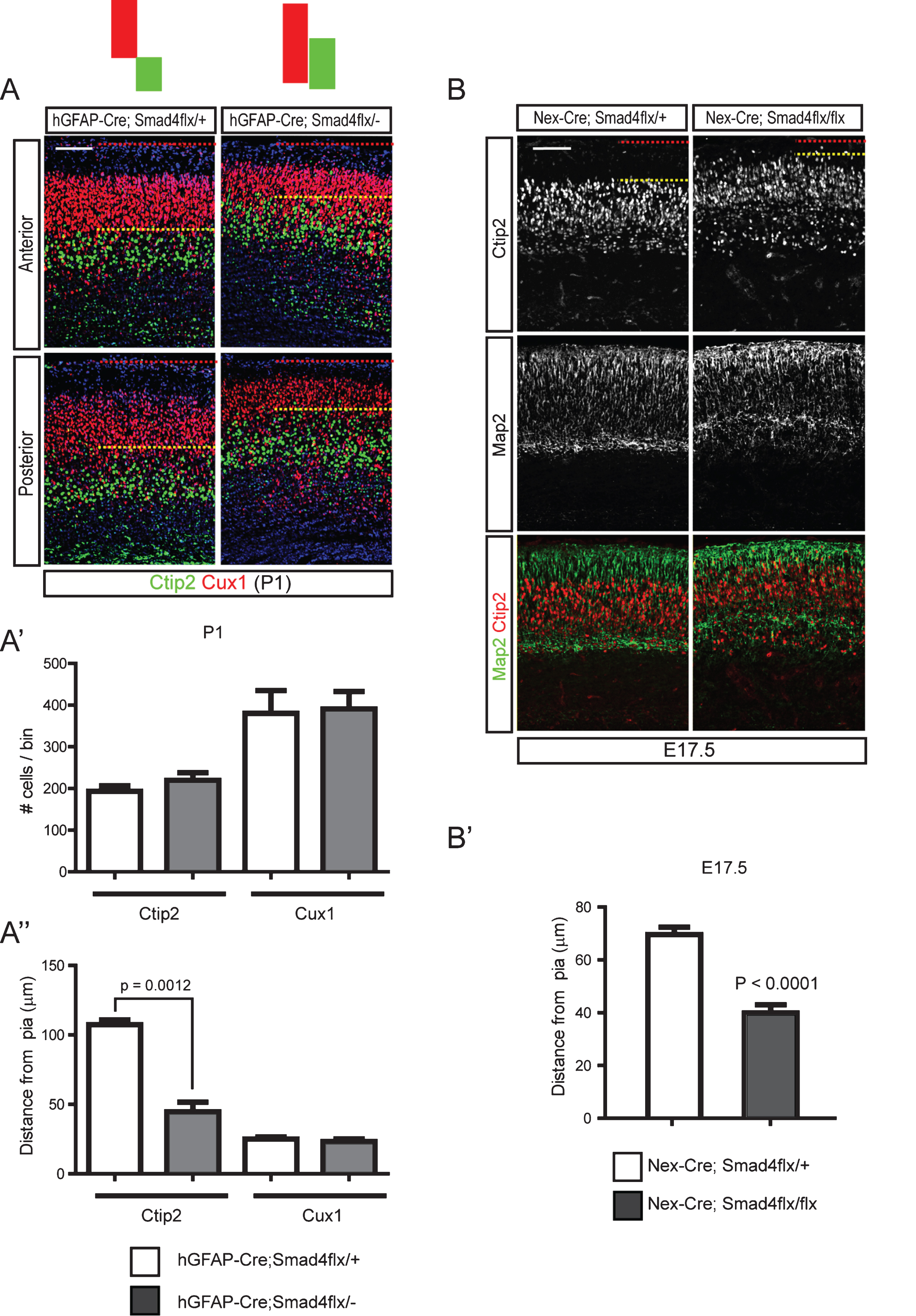
Fig.6
Cytoskeletal changes in cortical neurons induced by Bmp signaling through regulation of cofilin-1 activity. A) E15.5 cortices were used to establish cortical neuronal culture for 5 days in vitro, when more than 50% of the neurons were positive for Ctip2+ (data not shown). Cells were treated with Bmp7 (20 ng/ml) or Noggin (40 ng/ml) for 2 hr on day 6 or 24 hr on day 5 (A) and collected at the same time on day 6 (n = 3). B) High-magnification images of representative cells are presented. The numbers of cells having motile or collapsed neurites (A”) were plotted for the 2 hr and 24 hr treatments (A’, B’). We defined a neuron as having collapsed neurites when it had more than two collapsed neurites, and we counted more than 100 neurons with fewer than 3 neurites. C) Cortical neurons (Tuj1+, red) cultured for 5 days in vitro were stained for phospho-Limk1/2 (pLimk1/2, green) or phospho-cofilin-1 (pcofilin-1, green) after treatment with Bmp7 (20 ng/ml) (n = 3). Tuj1 signal is not presented separately. D) P1 hGFAP-Cre;Smad4flx/- mutants and their control littermates were stained for cofilin-1, Ctip2 (top panels) and pcofilin-1 and Ctip2 (middle panels). High-magnification images from the boxed areas in the middle panels are presented in the bottom panels (n = 3). Student’s t-test was conducted to determine the statistical significance of the difference between the groups (n = 3). Scale bars = 10 μm (A’, B), 50 μm (A, C), 100 μm (D).
To examine the regulation of cofilin-1 activity in vivo, we examined the phosphorylation of cofilin-1 using hGFAP-Cre;Smad4flx/- mutant pups at P1. Cofilin was expressed abundantly in the marginal zone and in the Ctip2+ neurons and at slightly lower levels in the Smad4 mutant. Phosphorylation of cofilin-1 in cortical Ctip2+ neurons and leading processes was also reduced in the mutant. This suggests that there was increased actin-severing activity of Cofilin proteins in the hGFAP-Cre;Smad4flx/- mutant neurons, indicating that Bmp-Cofilin signaling may be involved in regulating the final localization of the migrating neurons (Fig. 6D). The marginal zone of the Smad4 mutants was infiltrated by cells showing cytoplasmic phospho-cofilin-1 (Fig. 6D). Reduced phosphorylation of cofilin-1 in hGFAP-Cre;Smad4flx/- mutants suggests that Bmp signaling regulates cofilin-1 activity in a gene expression-dependent manner.
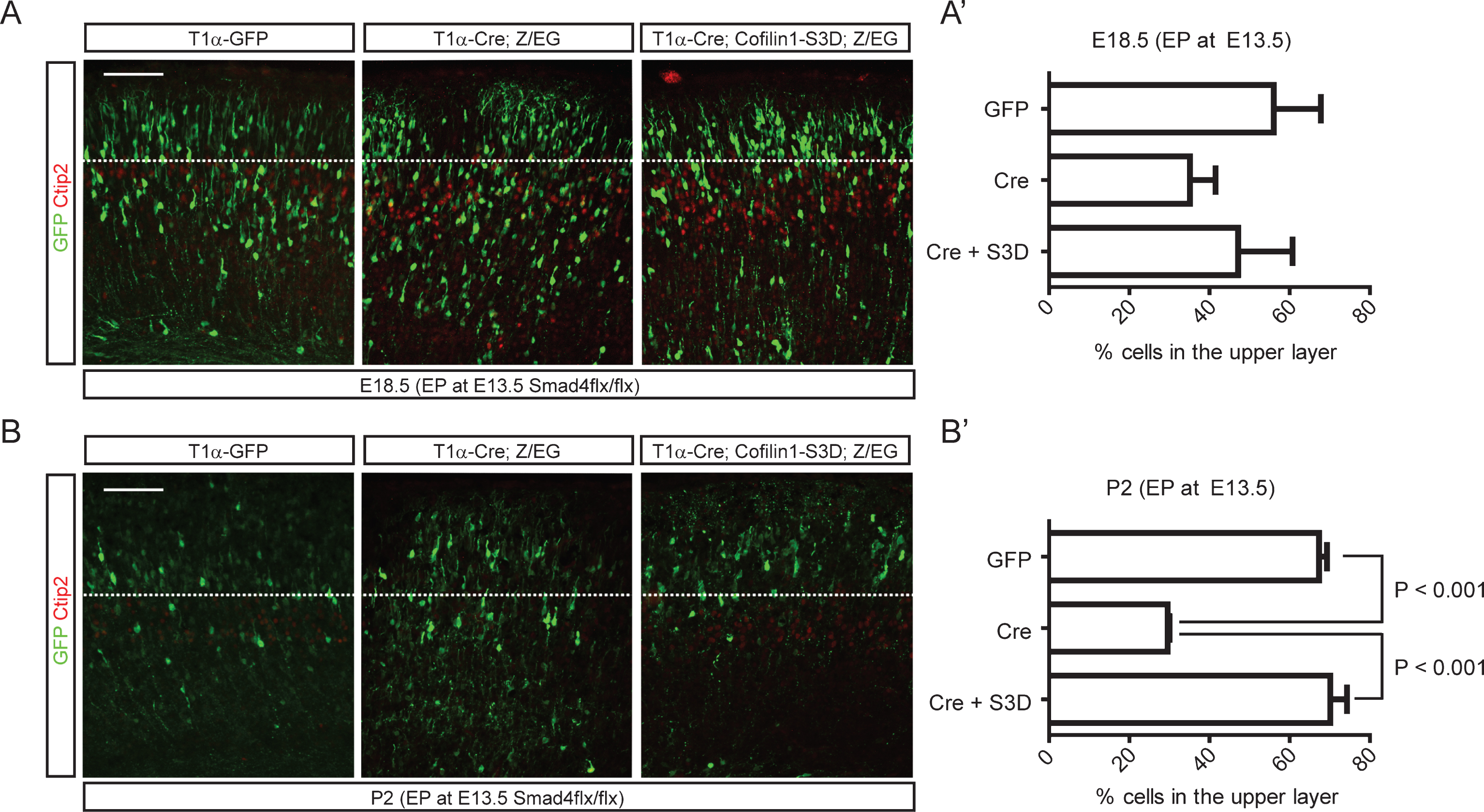
Because regulation of cofilin-1 activity through upstream kinases or phosphatases is well known, we next examined whether ectopic inhibition of cofilin-1 activity in Smad4 mutant embryos could rescue the migration phenotypes of Smad4 mutant neurons in vivo. We electroporated neurons migrating away from the intermediate zone with wild-type Cofilin-1, a phosphomimetic mutant (cofilin-1-S3D), or a cofilin-1 mutant that is resistant to phosphorylation at a critical serine residue (cofilin-1-S3A), which has been reported to rescue apical neurogenesis defects by Rnd3 inhibition [30]. To circumvent the effects of cofilin-1 during apical neurogenesis, we designed an in utero electroporation experiment utilizing Cre-inducible cofilin-1 expression in postmitotic neurons. We co-electroporated a tubulin alpha 1 (T1α) promoter-driven Cre expression construct that drives expression in postmitotic neurons with Cre-inducible cofilin-1 variants such as cofilin-1-S3A or cofilin-1-S3D. Cre-inducible cofilin-1 variants were cloned under stop sequences that can be removed by Cre recombination (Fig. 7A). To visualize electroporated neurons, we performed immunostaining for Cre. To test whether the inhibition of serine phosphorylation of cofilin-1 alone could phenocopy neuronal migration defects observed in Smad4 mutants, we co-electroporated T1α-Cre with cofilin-1 variants into E13.5 CD1 embryonic cortices. We found that neurons electroporated with cofilin-1-S3A showed delayed migration, leaving electroporated neurons in the deeper layers (Fig. 7B). Staining the electroporated tissues for Satb2 clearly showed that more Cre+ neurons had not even entered the upper layer 4 days after electroporation. To confirm the idea that Cofilin is regulated by Bmp signaling, we tested the ability of phosphomimetic cofilin-1 (cofilin-1-S3D) to rescue the neuronal migration phenotype in the Smad4 mutant neurons in vivo. We used E13.5 Smad4flx/flx conditional mutant embryos for the electroporation of T1α-Cre, cofilin-1-S3D and a Z/EG reporter. We used the Z/EG Cre reporter to visualize the electroporated cortices and label the Cre expressing neurons without staining for Cre protein. Smad4 mutant neurons electroporated with only T1α-Cre showed delayed migration compared to T1α-GFP electroporated control neurons at 5 days after electroporation, supporting the deletion of Smad4 in the Cre-expressing neurons (Fig. 8A, and A’). At P2, more than 67% of the T1α -GFP electroporated neurons reached the upper layer, whereas 71% of the Smad4 mutant neurons electroporated with T1α-Cre failed to reach the upper layer. As we predicted, 70% of the T1α-Cre-expressing Smad4 mutant neurons were observed in the upper layer after co-expression of cofilin-1-S3D. This supports the idea that Bmp signaling regulates cortical neuronal migration through the regulation of cofilin-1 activity (Fig. 8B, and B’, p < 0.001 (n = 3)).
Fig.7
A phosphorylation-defective form of cofilin-1 reduced migration of late-born neurons. A) A schematic drawing of Cre-inducible expression constructs of cofilin-1 variants. The tubulin alpha 1 (T1α) promoter was used to express Cre. Cre expression removes a loxP-flanked beta-galactosidase (β-gal) and induces expression of an S3A (phosphorylation defective) or S3D (phosphomimetic) form of cofilin-1. B) E13.5 CD1 embryos were electroporated with T1α-Cre, T1α-Cre;cofilin-1-S3A, or T1α-Cre;cofilin-1-S3D. Representative images of electroporated brains at E17.5 stained for Cre and Satb2 are presented (n = 3). Scale bars = 100 μm.
Fig.8
Rescue of neuronal migration defects of Smad4 mutant neurons by cofilin-1-S3D expression. E13.5 Smad4flx/flx embryos were electroporated with 1) T1α-GFP, 2) T1α-Cre;Z/EG, or 3) T1α-Cre;cofilin-1-S3D;Z/EG, a Cre reporter expressing GFP after recombination, and analyzed at E18.5 (A, A’) and P2 (B, B’) (n = 3). Electroporated brains were stained for GFP and Ctip2, and GFP+ neurons in the upper layer above the Ctip2+ layer were counted. Student’s t-test was conducted to determine the statistical significance of the difference between the groups. Scale bars = 100 μm.
DISCUSSION
The laminar positioning of cortical neurons that is established during embryonic development is determined by intrinsic and extrinsic factors influencing the migrating cortical neurons. Cortical pyramidal neurons are born from E11 to E18 and migrate through the subventricular zone with a multipolar morphology, passing the intermediate zone and adopting their polarized bipolar morphology with a leading process oriented toward the pia and a trailing process, which will become the apical dendrite and the axon, respectively [31–33]. The reestablishment of bipolar morphology from the multipolar morphology is important for the radial migration of cortical neurons, and the retraction of multiple processes involves many molecules associated with cell polarization [31, 33]. The radially migrating bipolar neurons are passed by late-born neurons, and neurons born even later will pass these by, eventually generating an inside-out pattern of cortical layer formation [2]. The interactions of migrating neurons with the molecular signals that regulate their radial migration are finely regulated, and the impairment of these interactions is associated with disturbances of neuronal function, such as cortical heterotopia, mental retardation and other neurodevelopmental disorders including seizure and schizophrenia [34]. Our study provides evidence that pial Bmp ligands function as key regulatory cues for the radially migrating cortical neurons during embryonic development and that Smad transcription factors mediate the Bmp signaling in the migrating neurons.
The leading processes are critical cellular compartments of migrating neurons that interact with environmental guidance cues including Netrins, Semaphorins, Ephrins, and Slits [35]. Guidance molecules such as Sema3A and Slits function as attractants or repellants for cortical neurons during migration by regulating growth cone collapse, which implies the involvement of the leading processes in this process [36–38]. The mobilization of actin-based cytoskeletal structures in the leading process is thus important for the guidance of the migrating centrosomes and cell body, and members of the cofilin family of actin filament depolymerizing proteins such as cofilin-1, m-cofilin, and actin depolymerizing factor (ADF) are known to control actin filament length and cell migration [39–42]. The complete loss of non-muscle cofilin (cofilin-1) affects the migration of neural crest cells and the formation of the neural tube, and the conditional loss of cofilin-1 in the Nestin-Cre+ neural progenitors impairs radial migration of cortical neurons [39, 43]. Reelin, secreted by the Cajal-Retzius neurons in the marginal zone, is one of the environmental cues that regulate cofilin-1 in radially migrating neurons [25, 29]. Reelin drives phosphorylation of cofilin-1 in the leading processes of migrating neurons, sending a stop signal to the neurons. Specifically, mutant upper-layer neurons expressing Apoer2, one of the Reelin receptors, are incapable of passing early-generated neurons and remain in the deep layer, producing two bands of Cux2+ neurons in the cortex; this suggests that the Reelin-cofilin-1 feedback influences cytoskeletal changes during neuronal migration [44]. The Smad4 mutant neurons also failed to pass by the early-born neurons residing in the deep layer, and this phenotype was rescued by the ectopic expression of phosphomimetic cofilin-1. This suggests that Reelin is not the only environmental factor translating cofilin-1 activity into neuronal migration. Since cell-autonomous inactivation of Bmp signaling in the migrating neurons produces radial migration impairments in late-born neurons similar to the migration defects observed in Apoer2 mutant neurons, signaling downstream of Reelin and Bmps may converge onto the regulation of cofilin-1 activity through Smad4-dependent gene transcription. However, the molecular interaction between Bmp signaling in the nucleus and Reelin signaling through Apoer2 but not Vldlr remains to be further studied. Inactivation of Smad4 signaling in the early-born neurons also impaired the localization of those neurons in the deep layer, which was not observed in the Apoer2 mutant. We could not test whether cofilin-1 was also involved in this process because of the technical difficulty of in utero electroporation at E11, when early-born neurons are exposed to the lateral ventricle [45]. Cofilin-1 may be involved in Bmp-mediated migration of early-born neurons since cofilin-1 is expressed as early as E8 in the developing neuroectoderm [43], and cofilin-1 is enriched in the leading processes of late-born neurons and in the cytoplasm of early-born neurons [25]. Since early-born neurons migrate via glia-independent somal translocation, unlike the glia-guided locomotion of the upper-layer cortical neurons [5], it is also possible that cofilin-1-independent molecular interactions may mediate the Bmp-mediated regulation of the early born neuronal migration. For example, region-specific proneural factors such as Ascl1 and Neurog2 modulate RhoA-Limk-cofilin-1-dependent and -independent signaling through the atypical Rho GTPases Rnd3 and Rnd2, respectively, in different modes of cell migration [30, 46]. Future studies are needed to uncover the differential roles of the leading processes in the neuronal migration that occurs at different developmental stages.
Defects in the Smad4 gene cause Myhre syndrome, which is characterized by growth deficiency, mental retardation, midface hypoplasia, and microcephaly [47]. The present study provides evidence that Bmp-mediated regulation of Smad4-dependent gene expression may involve cytoskeletal changes and cellular motility, and this may be the molecular mechanism underlying some of the phenotypes associated with Myhre syndrome.
FUNDING
This work was supported by KBRI-2231-415 and NRF-2015M3C7A1030964 (Y.C.).
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ACKNOWLEDGMENTS
We thank Kurt Thorn and the Nikon Imaging Center at University of California, San Francisco, for use of the confocal microscope.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/BPL-170048.
REFERENCES
[1] | Douglas RJ , Martin KA . Neuronal circuits of the neocortex. Annu Rev Neurosci (2004) ;27: :419–51. |
[2] | Nadarajah B , Parnavelas JG . Modes of neuronal migration in the developing cerebral cortex. Nat Rev Neurosci (2002) ;3: (6):423–32. |
[3] | Trommsdorff M , Gotthardt M , Hiesberger T , Shelton J , Stockinger W , Nimpf J , et al. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell (1999) ;97: (6):689–701. |
[4] | Arlotta P , Molyneaux BJ , Chen J , Inoue J , Kominami R , Macklis JD . Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron (2005) ;45: (2):207–21. |
[5] | Nadarajah B , Brunstrom JE , Grutzendler J , Wong RO , Pearlman AL . Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci (2001) ;4: (2):143–50. |
[6] | Tarabykin V , Stoykova A , Usman N , Gruss P . Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development (2001) ;128: (11):1983–93. |
[7] | Choe Y , Siegenthaler JA , Pleasure SJ . A cascade of morphogenic signaling initiated by the meninges controls corpus callosum formation. Neuron (2012) ;73: (4):698–712. |
[8] | Choe Y , Kozlova A , Graf D , Pleasure SJ . Bone morphogenic protein signaling is a major determinant of dentate development. J Neurosci (2013) ;33: (16):6766–75. |
[9] | Colak D , Mori T , Brill MS , Pfeifer A , Falk S , Deng C , et al. Adult neurogenesis requires Smad4-mediated bone morphogenic protein signaling in stem cells. J Neurosci (2008) ;28: (2):434–46. |
[10] | Yang X , Li C , Herrera PL , Deng CX . Generation of Smad4/Dpc4 conditional knockout mice. Genesis (2002) ;32: (2):80–1. |
[11] | Ortega JA , Alcantara S . BDNF/MAPK/ERK-induced BMP7 expression in the developing cerebral cortex induces premature radial glia differentiation and impairs neuronal migration. Cereb Cortex (2010) ;20: (9):2132–44. |
[12] | Foo SS , Turner CJ , Adams S , Compagni A , Aubyn D , Kogata N , et al. Ephrin-B2 controls cell motility and adhesion during blood-vessel-wall assembly. Cell (2006) ;124: (1):161–73. |
[13] | Zhuo L , Theis M , Alvarez-Maya I , Brenner M , Willecke K , Messing A . hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis (2001) ;31: (2):85–94. |
[14] | Goebbels S , Bormuth I , Bode U , Hermanson O , Schwab MH , Nave KA . Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis (2006) ;44: (12):611–21. |
[15] | Danielian PS , Muccino D , Rowitch DH , Michael SK , McMahon AP . Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol (1998) ;8: (24):1323–6. |
[16] | Hayashi H , Kume T . Forkhead transcription factors regulate expression of the chemokine receptor CXCR4 in endothelial cells and CXCL12-induced cell migration. Biochem Biophys Res Commun (2008) ;367: (3):584–9. |
[17] | Bardeesy N , Cheng KH , Berger JH , Chu GC , Pahler J , Olson P , et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev (2006) ;20: (22):3130–46. |
[18] | Chang W , Lin Z , Kulessa H , Hebert J , Hogan BL , Wu DK . Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements. PLoS Genet (2008) ;4: (4):e1000050. |
[19] | Zouvelou V , Passa O , Segklia K , Tsalavos S , Valenzuela DM , Economides AN , et al. Generation and functional characterization of mice with a conditional BMP7 allele. Int J Dev Biol (2009) ;53: (4):597–603. |
[20] | Novak A , Guo C , Yang W , Nagy A , Lobe CG . Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis (2000) ;28: (3-4):147–55. |
[21] | Wang S , Roy NS , Benraiss A , Goldman SA . Promoter-based isolation and fluorescence-activated sorting of mitotic neuronal progenitor cells from the adult mammalian ependymal/subependymal zone. Dev Neurosci (2000) ;22: (1-2):167–76. |
[22] | Massague J , Chen YG . Controlling TGF-beta signaling. Genes Dev (2000) ;14: (6):627–44. |
[23] | Siegenthaler JA , Ashique AM , Zarbalis K , Patterson KP , Hecht JH , Kane MA , et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell (2009) ;139: (3):597–609. |
[24] | Belvindrah R , Graus-Porta D , Goebbels S , Nave KA , Muller U . Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. J Neurosci (2007) ;27: (50):13854–65. |
[25] | Chai X , Forster E , Zhao S , Bock HH , Frotscher M . Reelin stabilizes the actin cytoskeleton of neuronal processes by inducing n-cofilin phosphorylation at serine3. J Neurosci (2009) ;29: (1):288–99. |
[26] | Guo X , Lin Y , Horbinski C , Drahushuk KM , Kim IJ , Kaplan PL , et al. Dendritic growth induced by BMP-7 requires Smad1 and proteasome activity. J Neurobiol (2001) ;48: (2):120–30. |
[27] | Lee-Hoeflich ST , Causing CG , Podkowa M , Zhao X , Wrana JL , Attisano L . Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. EMBO J (2004) ;23: (24):4792–801. |
[28] | Lein PJ , Beck HN , Chandrasekaran V , Gallagher PJ , Chen HL , Lin Y , et al. Glia induce dendritic growth in cultured sympathetic neurons by modulating the balance between bone morphogenetic proteins (BMPs) and BMP antagonists. J Neurosci (2002) ;22: (23):10377–87. |
[29] | Chai X , Zhao S , Fan L , Zhang W , Lu X , Shao H , et al. Reelin and cofilin cooperate during the migration of cortical neurons: A quantitative morphological analysis. Development (2016) ;143: (6):1029–40. |
[30] | Pacary E , Heng J , Azzarelli R , Riou P , Castro D , Lebel-Potter M , et al. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron (2011) ;69: (6):1069–84. |
[31] | LoTurco JJ , Bai J . The multipolar stage and disruptions in neuronal migration. Trends Neurosci (2006) ;29: (7):407–13. |
[32] | Noctor SC , Martinez-Cerdeno V , Ivic L , Kriegstein AR . Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci (2004) ;7: (2):136–44. |
[33] | Tabata H , Nakajima K . Multipolar migration: The third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci (2003) ;23: (31):9996–10001. |
[34] | Fox JW , Walsh CA . Periventricular heterotopia and the genetics of neuronal migration in the cerebral cortex. Am J Hum Genet (1999) ;65: (1):19–24. |
[35] | Guan KL , Rao Y . Signalling mechanisms mediating neuronal responses to guidance cues. Nat Rev Neurosci (2003) ;4: (12):941–56. |
[36] | Chen G , Sima J , Jin M , Wang KY , Xue XJ , Zheng W , et al. Semaphorin-3A guides radial migration of cortical neurons during development. Nat Neurosci (2008) ;11: (1):36–44. |
[37] | Gonda Y , Andrews WD , Tabata H , Namba T , Parnavelas JG , Nakajima K , et al. Robo1 regulates the migration and laminar distribution of upper-layer pyramidal neurons of the cerebral cortex. Cereb Cortex (2013) ;23: (6):1495–508. |
[38] | Zheng W , Geng AQ , Li PF , Wang Y , Yuan XB . Robo4 regulates the radial migration of newborn neurons in developing neocortex. Cereb Cortex (2012) ;22: (11):2587–601. |
[39] | Bellenchi GC , Gurniak CB , Perlas E , Middei S , Ammassari-Teule M , Witke W . N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev (2007) ;21: (18):2347–57. |
[40] | Ghosh M , Song X , Mouneimne G , Sidani M , Lawrence DS , Condeelis JS . Cofilin promotes actin polymerization and defines the direction of cell motility. Science (2004) ;304: (5671):743–6. |
[41] | Gungabissoon RA , Bamburg JR . Regulation of growth cone actin dynamics by ADF/cofilin. J Histochem Cytochem (2003) ;51: (4):411–20. |
[42] | Kuhn TB , Meberg PJ , Brown MD , Bernstein BW , Minamide LS , Jensen JR , et al. Regulating actin dynamics in neuronal growth cones by ADF/cofilin and rho family GTPases. J Neurobiol (2000) ;44: (2):126–44. |
[43] | Gurniak CB , Perlas E , Witke W . The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Dev Biol (2005) ;278: (1):231–41. |
[44] | Hack I , Hellwig S , Junghans D , Brunne B , Bock HH , Zhao S , et al. Divergent roles of ApoER2 and Vldlr in the migration of cortical neurons. Development (2007) ;134: (21):3883–91. |
[45] | McKenna WL , Betancourt J , Larkin KA , Abrams B , Guo C , Rubenstein JL , et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci (2011) ;31: (2):549–64. |
[46] | Maekawa M , Ishizaki T , Boku S , Watanabe N , Fujita A , Iwamatsu A , et al. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science (1999) ;285: (5429):895–8. |
[47] | Le Goff C , Mahaut C , Abhyankar A , Le Goff W , Serre V , Afenjar A , et al. Mutations at a single codon in Mad homology 2 domain of SMAD4 cause Myhre syndrome. Nat Genet (2011) ;44: (1):85–8. |



