Non-canonical post-transcriptional RNA regulation of neural stem cell potential
Abstract
Adult brain structures and complexity emerge from a single layer of neuroepithelial cells that early during the development give rise to neural stem cells (NSCs). NSCs persist in restricted regions of the postnatal brain where they support neurogenesis throughout life thus allowing brain plasticity and adaptation. NSC regulation involves a precise coordination of intrinsic and extrinsic mechanisms that finely modulate the neurogenic process. Here we will discuss new mechanisms of post-transcriptional gene regulation that act in the embryonic and adult brain to regulate NSC maintenance and differentiation. In our recent work we found that the RNAaseIII Drosha not only regulates microRNA production, but also directly affects the stability of mRNAs and thereby controls proteome composition. This non-canonical (miRNA-independent) function of Drosha is central in the maintenance and fate choices made by adult hippocampal NSCs in the healthy brain. We found that Drosha targets the mRNA of the gliogenic transcription factor Nuclear Factor I/B and thereby blocks its expression in the NSCs. In the absence of Drosha, NSCs aberrantly differentiate into oligodendrocytes and are lost leading to an impairment of neurogenesis. Overall these findings reveal an unprecedented Drosha-mediated post-transcriptional mechanism for the regulation of hippocampal NSC potential.
INTRODUCTION
Adult stem cells generate progenies throughout the course of life. However, their fates are usually restricted and they produce specific cell-types in their respective tissue. During brain development NSCs have the capacity to self-renew and give rise to neurons and glia until they transform into parenchymal astrocytes, ependymal cells, or remain as adult stem cells in the two adult niches, the subventricular zone (SVZ) of the wall of the lateral ventricles [1] and the subgranular zone of the hippocampal dentate gyrus (DG) [2]. Adult NSCs have characteristics of astrocytes, including marker expression, and have distinct features in the lateral ventricular and hippocampal neurogenic areas [31]. NSCs in the lateral ventricle produce immature neuroblasts that migrate in chains towards the olfactory bulb where they differentiate into local interneurons [3, 4], while in the hippocampus NSCs generate glutamatergic granule neurons and parenchymal astrocytes [5, 6]. Moreover, SVZ but not DG NSCs also generate myelinating oligodendrocytes both in physiological condition and upon a demyelinating insult [6–8].
In the adult neurogenic niches, fine regulation of the balance between stem cell preservation and production of differentiated progeny is achieved by interactions between extrinsic signals and intrinsic pathways [9]. Cell-intrinsic regulatory mechanisms can take place at the level of transcriptional activity. In addition, recent works have highlighted a crucial role of post-transcriptional control of gene expression in neural progenitors of the developing and adult brain [10–13].
MIRNA-INDEPENDENT FUNCTION OF DROSHA DURING NEUROGENESIS
Adult neurogenesis requires fine regulation of signaling pathways and gene expression. The RNaseIII Drosha and the RNA binding protein (RBP) DGCR8 (Pasha in flies and worms) constitute the Microprocessor, a large complex responsible for the biogenesis of most miRNAs. Long primary transcripts (pri-miRNAs) that contain the local stem-loop structure of the miRNA sequence are processed by the Microprocessor. Drosha crops the pri-miRNA and produces a 60–70 nucleotide (nt) stem-loop pre-miRNA [14]. This pre-miRNA is subsequently exported to the cytoplasm to be further processed by the RNaseIII Dicer, generating a ∼22 nt double-stranded RNA duplex [15–17]. The mature single-stranded lead-miRNA binds to the RNA-induced silencing complex (RISC) directing it to complementary mRNA targets resulting in transcript repression either through mRNA cleavage and degradation or inhibition of translation [18, 19].
Besides the well studied function of miRNAs in the regulation of embryonic and adult NSC maintenance and differentiation, recent work propose an alternate function for the Microprocessor as regulator of embryonic and adult neurogenesis [12, 20, 21]. The first indication that the Drosha can act independent of miRNAs arise from genome-wide studies of Drosha- and Dicer-knockdown in Drosophila Schneider S2 cells [22]. Drosha- but not Dicer-knockdown leads to accumulation not only of several miRNA precursors but surprisingly also of mRNAs. Interestingly these Drosha mRNA targets have strongly conserved structural hairpins in their sequences, which can undergo direct cleavage by Drosha [23]. In line with this, the first Microprocessor mRNA-target identified was that of DGCR8/Pasha [22, 24]. DGCR8 mRNA contains hairpins in the coding sequence and the 5’ untranslated region (UTR), which are evolutionarily conserved amongst organisms and that are targeted and processed by the Microprocessor [24]. Drosha-depletion leads to DGCR8 mRNA accumulation indicating that Drosha inhibits DGCR8 expression in an auto-regulatory mechanism to control Microprocessor levels [22, 24]. Since then, and thanks to the development of novel high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation, the non-canonical function of Drosha on regulation of mRNA has been extended to other cell types, including embryonic stem cells, thymocyte progenitors and dendritic cell progenitors highlighting its relevance in a broad range of biological processes [25–28].
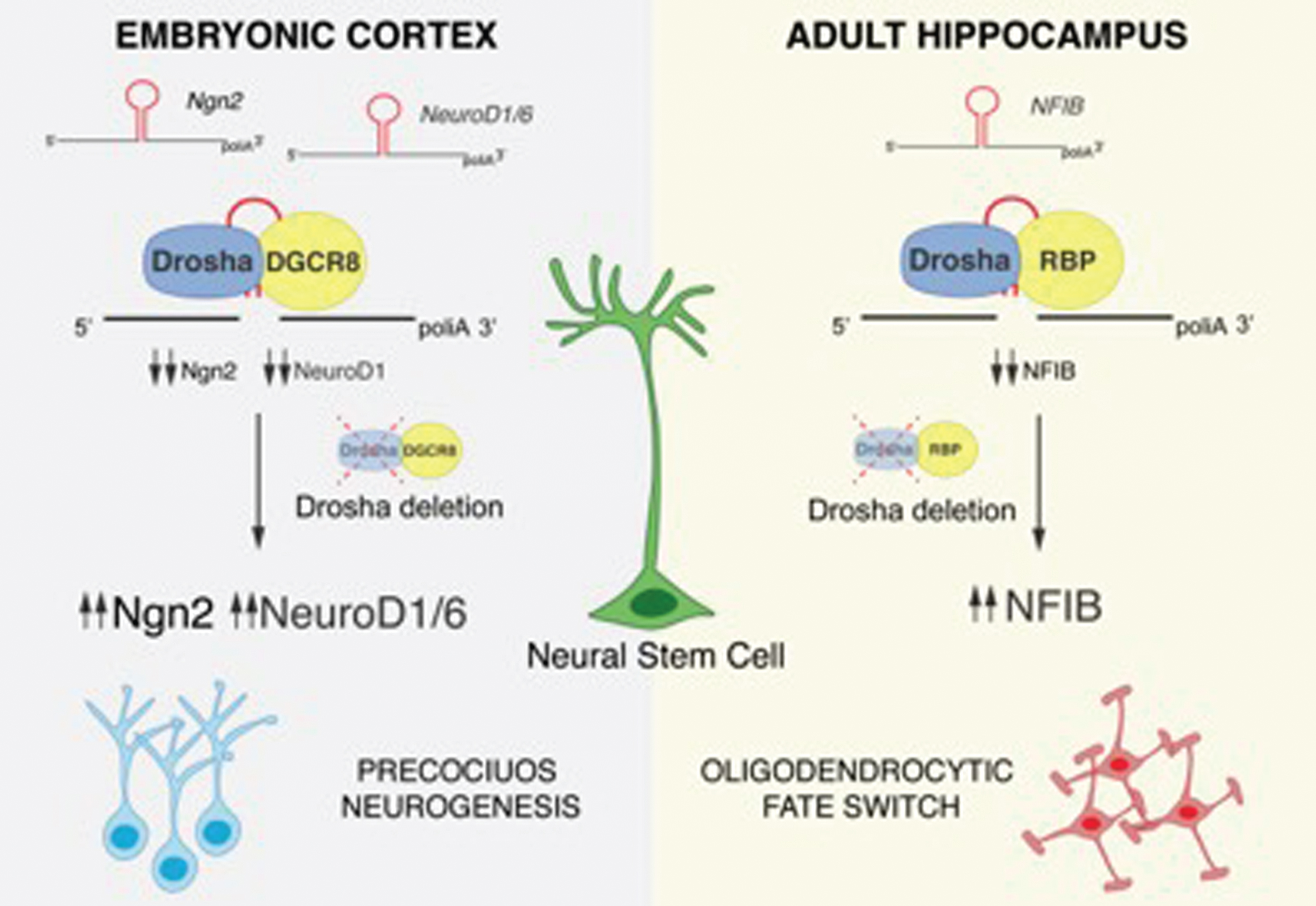
During neurogenesis, a fast regulation of the transcriptome and proteome is essential for the maintenance and differentiation of NSCs. Notch signaling in NSCs activates the expression of the transcription factors Hes1 and Hes5, which are required for NSC maintenance by inhibiting the expression of the proneural factors including Neurogenin2 (Ngn2). A sustained expression of Ngn2 induces NSC to differentiate into neurons [29, 30]. Elimination of Drosha or DGCR8 in embryonic NSCs results in a loss of the NSC pool and precocious neuronal differentiation, whereas Dicer-deficiency does not. In this context, Drosha binds to and negatively regulates the stability of the mRNA of the proneural gene Ngn2 and the neural determination factor NeuroD1 and 6, thereby maintaining the NSC pool. Ngn2, NeuroD1 and NeuroD6 mRNAs contain evolutionarily conserved hairpins, which can be bound by Drosha. These data indicate that Drosha facilitates embryonic NSC maintenance by directly blocking the accumulation of mRNAs encoding for neuronal differentiation factors (Fig. 1) [21].
Fig.1
Drosha-mediated post-transcriptional regulation of NSC fate potential. The proneural factors Ngn2, NeuroD1, NeuroD6 and the gliogenic factor NFIB contain evolutionarily conserved hairpin structures in their mRNA sequences that are targeted and cleaved by Drosha. Following Drosha deletion and accumulation of Ngn2, NeuroD1 and NeuroD6, embryonic NSCs precociously differentiate. NFIB accumulation in Drosha cKO hippocampal NSCs induces a fate conversion into the oligodendrocytic lineage.
DROSHA RESTRICTS ADULT DG NSC POTENTIAL BY TARGETING MRNA OF GLIOGENIC TRANSCRIPTION FACTOR
Adult NSCs in the postnatal brain are multipotent, are instructed by their local niche to divide, and produce neurons, astrocytes and oligodendrocytes, the myelin-producing support cells of the central nervous system [9, 31]. However, the DG NSCs are fate restricted and are able to generate neurons and astrocytes during physiological conditions but they do not generate oligodendrocytes in vivo, suggesting an intrinsic and niche-independent fate restriction that prevents their generation of specific progeny [6, 8, 32, 33]. Interestingly, oligodendrocyte differentiation can be induced by overexpression of the proneural transcription factor Ascl1 [34, 35] or by deletion of neurofibromin 1 (Nf1) [36] and also in vitro when co-cultured with neurons [37]. Adult DG NSCs are a heterogeneous population and include radial and non-radial NSCs that shuttle between active and quiescent states. Both radial and non-radial DG NSCs express Hes5 and therefore have active Notch signaling that control the balance between proliferation and differentiation [8, 38].
How lineage fate restriction is achieved and whether different NSC pool can differentially regulate their own fates was still unknown. Interestingly we found that in the adult hippocampus Drosha is expressed by most cells including radial and non-radial NSCs. We found that knockout of Drosha from Hes5 expressing NSCs negatively impacted the number of NSC/progenitors, neuroblasts and newborn neurons in the adult DG. Due to the loss of NSCs and aberrant differentiation in the DG of Drosha conditional knockout (cKO) mice, we examined cell fate in detail and observed that a significant proportion of the NSC-derived newborn cells in the granule cell layer expressed Olig2 and Sox10, markers of oligodendrocyte progenitor cells. Hence, loss of Drosha induced a surprising fate switch in DG NSCs to the oligodendrocyte lineage. Taken together the data demonstrate that Drosha is essential to maintain NSCs and neurogenesis in the adult DG and loss of Drosha forces NSCs to leave the stem cell pool and produce oligodendrocytes. Interestingly, we found that only when we targeted gene deletion to radial NSCs with adenoviruses expressing Cre-expression under the control of the gfap promoter [39] did we observe a reduction in the progenitor pool and neuroblasts. Indeed, deletion of Drosha from active proliferative NSCs/progenitors did not result in a reduction of the stem cell pool and oligodendrocytic fate commitment thus indicating that radial quiescent NSCs are preferentially affected by the fate-switching Drosha deletion. Furthermore, S100β+ parenchymal astrocytes were increased in the granule cell layer of the Drosha cKO DG at the expense of neurons. Together the data suggest that Drosha regulates neurogenesis and that Drosha-deficient NSCs may prematurely differentiate into glia at the expense of neurons, a process observed during aging and NSC exhaustion [32].
Drosha is a central component of the miRNA biogenesis pathway and Dicer regulates mature miRNA production downstream of Drosha. When we deleted Dicer (Dicer cKO) from DG NSCs, this did not affect the progenitor pool and caused a minor decrease in neuroblasts consistent with the demonstrated role of Dicer in neuronal survival and maturation [40]. Interestingly, we found that deletion of Dicer did not induce oligodendrocytic differentiation of DG NSCs suggesting that Drosha but not Dicer inhibits oligodendrocyte differentiation from adult DG NSCs in vivo. This also indicate that the mechanism of induced fate switching caused by the loss of Drosha does not involve miRNAs as Dicer is critical for miRNA maturation. Furthermore, comparison of Drosha- and Dicer-deficient NSCs did not reveal significant changes in miRNA profile, suggesting that the miRNAs are relatively stable and that Microprocessor-induced phenotypes are miRNA-independent.
We and others have shown that Drosha can bind and cleave hairpin loops in mRNAs and regulate their expression [21, 28, 41]. Nuclear factor I family transcription factors (NFIA, B, C,X) are crucial during embryonic development and in the adult brain [42]. NFIB influences stem cell maintenance and differentiation in several tissues, including in the SVZ, as part of a cross-regulatory network together with Pax6/Brg1 [43, 44]. In addition, NFIB can repress Notch signaling in embryonic hippocampal NSCs by repressing Hes1 promoter activity [45]. NFIB knockout mice exhibit severe defects in corpus callosum, the major white matter tract of the brain, consistent with defects in oligodendrocyte development [46]. In silico analysis (Evofold) [23] revealed two evolutionarily conserved hairpins in the mRNA of NFIB, a short sized of 20 base hairpin in 5’UTR (5’UTR HP) and a longer hairpin of 83 bases in the 3’UTR (3’UTR HP). We found that Drosha directly regulates the stability of NFIB mRNA in the DG NSCs. We performed cross-linked immunoprecipitation (CLIP) for endogenous Drosha protein and found that Drosha binds both the 5’ and 3’ UTR HP in NFIB mRNA in DG NSCs. Furthermore, we showed that only Drosha interaction with the 3’UTR HP and not with the 5’UTR HP in NFIB mRNAs resulted also in mRNA cleavage. Our results also demonstrated that Drosha ablation induced an accumulation of NIFB mRNA, thus indicating that Drosha down regulates the transcripts in DG NSCs. In line with this, single-cell transcriptomic data of adult DG NSCs revealed that NFIB is absent from radial NSCs and is upregulated in the late progeny, thus indicating that NFIB needs to be downregulated in NSCs to prevent aberrant differentiation and NSC exhaustion [47].
In summary, we found that Drosha regulates NFIB mRNA levels in DG NSCs and that NFIB accumulation leads to oligodendrocyte differentiation in Drosha cKO NSCs. As a result, DG NSCs retain both Drosha function and blockade of oligodendrocytic differentiation. When we ablated Drosha from DG NSCs in vitro and simultaneously prevented NFIB mRNA accumulation by a RNAi knockdown approach, we reduced oligodendrocytic differentiation of Drosha cKO NSCs and these rescued NSCs adopted neuronal and astrocytic fates. Therefore, Drosha negatively regulates DG NSC differentiation towards an oligodendrocytic fate by suppressing NFIB mRNA levels (Fig. 1).
CONCLUSION
Neurogenesis is controlled by a hypostable transcriptome [48]. Post-transcriptional gene regulatory mechanisms ensure constant and rapid adaptation of NSCs during brain development and in the adult. How hippocampal NSC lineage potential is restricted has been a fundamental question in developmental biology and adult neurogenesis. Findings over the last few years uncovered different and unexpected non-canonical roles of the RNAseIII Drosha.
We identified a novel non-canonical Drosha regulation of the adult stem fate through a niche independent intrinsic pathway. Our findings show that Drosha sustains hippocampal neurogenesis and represses oligodendrocyte production from DG NSCs. Drosha regulates NSC differentiation in the adult hippocampus at least partially through a miRNA-independent, intrinsic cell fate program. Moreover, we identified the transcription factor NFIB as a direct target of Drosha and showed that the blockade of NFIB expression is necessary for inhibiting oligodendrocyte formation in DG NSCs in vitro. Thus, hippocampal NSCs retain the potential to generate all three cell-lineages of the brain but an intrinsic Drosha-mediated regulation restricts their differentiation potential in vivo.
It still remains to be understood how Drosha-mediated post-transcriptional regulation of mRNA stability is achieved and how Drosha can specifically target mRNAs of the pro-neurogenic factor Ngn2 and NeuroD1 in embryonic NSCs and of NFIB in adult NSCs. It is possible that the activity of Drosha is regulated through interaction with RNA binding proteins that mediate the recognition specific mRNA targets in a cell-specific and developmentally regulated fashion. There is evidence that the interaction between Drosha and TDP-43 but not DCGR8 mRNA is required for Drosha-dependent cleavage of Ngn2 mRNA in vitro [49]. Future studies will elucidate how different Microprocessor complexes are assembled to obtain RNA target regulation.
Our findings could have wider implications for understanding how stem and progenitors cells regulate their own differentiation and fate at the post-transcriptional level. The identification of non-canonical functions for the Microprocessor complex opens new perspectives in the field of neural stem cell biology. Further analysis will aim to provide new insights into the complex role of the Microprocessor in controlling gene expression during neurogenesis.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors declare no competing financial interests.
ACKNOWLEDGMENTS
We thank the members of the Taylor lab for critical reading of the manuscript and for helpful discussions. The authors declare no conflict of interest. This work was supported by the Swiss National Science Foundation (310030_143767).
REFERENCES
[1] | Furutachi S , Miya H , Watanabe T , Kawai H , Yamasaki N , Harada Y , et al., Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat Neurosci. (2015) ;18: (5):657–65. |
[2] | Kriegstein A , Alvarez-Buylla A . The glial nature of embryonic andadult neural stem cells. Annual Reviews of Neuroscience. (2011) ;149–84. |
[3] | Lois C , Alvarez-Buylla A Long-distance neuronal migration in the adult mammalian brain. Science. (1994) ;264: (0036-80725 (Print)): 1145–8. |
[4] | Hack Ma , Saghatelyan A , de Chevigny A , Pfeifer A , Ashery-Padan R , Lledo P-M , et al., Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat Neurosci. (2005) ;8: (7):865–72. |
[5] | Seri B , García-Verdugo JM , McEwen BS , Alvarez-Buylla A . Astrocytes give rise to new neurons in the adult mammalianhippocampus. The Journal of Neuroscience: The Official Journal ofthe Society for Neuroscience. (2001) ;21: (18):7153–60. |
[6] | Bonaguidi MA , Wheeler MA , Shapiro JS , Stadel RP , Sun GJ , Ming GL , et al., In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. (2011) ;145: (7):1142–55. |
[7] | Menn B , Garcia-Verdugo JM , Yaschine C , Gonzalez-Perez O , Rowitch D , Alvarez-Buylla A . Origin of oligodendrocytes in the subventricular zone of the adult brain. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. (2006) ;26: (30):7907–18. |
[8] | Lugert S , Basak O , Knuckles P , Haussler U , Fabel K , Götz M , et al., Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. (2010) ;6: (5):445–56. |
[9] | Ihrie RA , Álvarez-Buylla A . Lake-front property: A unique germinal niche by the lateral ventricles of the adult brain. Neuron. (2011) ;70: (4):674–86. |
[10] | Dai W , Li W , Hoque M , Li Z , Tian B , Makeyev EV . Apost-transcriptional mechanism pacing expression of neural geneswith precursor cell differentiation status. Nat Commun. (2015) ;6: , 7576. |
[11] | Knuckles P , Vogt MA , Lugert S , Milo M , Chong MM , Hautbergue GM , et al., Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat Neurosci. (2012) ;15: (7):962–9. |
[12] | Rolando C , Erni A , Grison A , Beattie R , Engler A , Gokhale PJ , et al., Multipotency of Adult Hippocampal NSCs In Vivo Is Restricted by Drosha/NFIB. Cell Stem Cell. (2016) ;19: (5):653–62. |
[13] | Pilaz L-J , Silver DL . Post-transcriptional regulation in corticogenesis: How RNA-binding proteins help build the brain. Wiley Interdisciplinary Reviews: RNA. (2015) ;6: (5):501–15. |
[14] | Lee Y , Ahn C , Han J , Choi H , Kim J , Yim J , et al., The nuclear RNase III Drosha initiates microRNA processing. Nature. (2003) ;425: (6956):415–9. |
[15] | Bohnsack MT , Czaplinski K , Görlich D . Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. (2004) ;10: (2):185–91. |
[16] | Lund E , Güttinger S , Calado A , Dahlberg JE , Kutay U . Nuclear export of microrna precursors. Science. (2004) ;303: (5654):95–8. |
[17] | Ketting RF , Fischer SEJ , Bernstein E , Sijen T , Hannon GJ , Plasterk RHA . C elegans. Genes Dev. (2001) (516):2654–9. |
[18] | Hammond SM , Boettcher S , Caudy AA , Kobayashi R , Hannon GJ . Argonaute2, a link between genetic and biochemical analyses of RNAi. Science (New York, NY). (2001) 293: 5532;1146–50. |
[19] | Ha M , Kim VN . Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology. (2014) ;15: (8):509–24. |
[20] | Cheng LC , Pastrana E , Tavazoie M , Doetsch F . miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. (2009) ;12: (4):399–408. |
[21] | Knuckles P , Vogt Ma , Lugert S , Milo M , Chong MMW , Hautbergue GM , et al., Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nature Neuroscience. (2012) ;15: (7):962–9. |
[22] | Kadener S , Rodriguez J , Abruzzi KC , Khodor YL , Sugino KEN , Ii MTM , et al., Genome-wide identification of targets of the drosha – pasha/DGCR8 complex ((2009) ;537–45. |
[23] | Pedersen JS , Bejerano G , Siepel A , Rosenbloom K , Lindblad-Toh K , Lander ES , et al., Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. (2006) ;2: (4):33. |
[24] | Han J , Pedersen JS , Kwon SC , Belair CD , Kim Y-K , Yeom K-H , et al., Posttranscriptional crossregulation between Drosha and DGCR8. Cell. (2009) ;136: (1):75–84. |
[25] | Chong MM , Zhang G , Cheloufi S , Neubert TA , Hannon GJ , Littman DR . Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. (2010) ;24: (17):1951–60. |
[26] | Johanson TM , Keown Aa , Cmero M , Yeo JHC , Kumar A , Lew AM , et al., Drosha controls dendritic cell development by cleaving messengerRNAs encoding inhibitors of myelopoiesis. Nat Immunol. (2015) ;16: (11):1134–41. |
[27] | Karginov FV , Cheloufi S , Chong MMW , Stark A , Smith AD , Hannon GJ . Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Molecular Cell. (2010) ;38: (6):781–8. |
[28] | Macias S , Plass M , Stajuda A , Michlewski G , Eyras E , Caceres JF . DGCR8 HITS-CLIP reveals novel functions for theMicroprocessor. Nat Struct Mol Biol. (2012) ;19: (8):760–6. |
[29] | Shimojo H , Ohtsuka T , Kageyama R . Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. (2008) ;58: (1):52–64. |
[30] | Imayoshi I , Isomura A , Harima Y , Kawaguchi K , Kori H , Miyachi H , et al., Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. (2013) ;342: (6163):1203–8. |
[31] | Kriegstein A , Alvarez-Buylla A . The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. (2009) ;32: (1):149–84. |
[32] | Encinas JM , Michurina TV , Peunova N , Park JH , Tordo J , Peterson DA , et al., Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. (2011) ;8: (5):566–79. |
[33] | Steiner B , Zurborg S , Horster H , Fabel K , Kempermann G . Differential 24h responsiveness of Prox1-expressing precursor cells in adult hippocampal neurogenesis to physical activity, environmental enrichment, and kainic acid-induced seizures. Neuroscience. (2008) ;154: (2):521–9. |
[34] | Jessberger S , Toni N , Clemenson GD Jr , Ray J , Gage FH . Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat Neurosci. (2008) ;11: (8):888–93. |
[35] | Braun Simon MG , Pilz G-A , Machado Raquel AC , Moss J , Becher B , Toni N , et al., Programming hippocampal neural stem/progenitor cells into oligodendrocytes enhances remyelination in the adult brain after injury. Cell Reports. (2015) ;11: (11):1679–85. |
[36] | Sun GJ , Zhou Y , Ito S , Bonaguidi MA , Stein-O’Brien G , Kawasaki NK , et al., Latent tri-lineage potential of adult hippocampal neural stem cells revealed by Nf1 inactivation. Nat Neurosci. (2015) ;18: (12):1722–4. |
[37] | Suh H , Consiglio A , Ray J , Sawai T , D’Amour KA , Gage FH . In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. (2007) ;1: (5):515–28. |
[38] | Lugert S , Vogt M , Tchorz JS , Muller M , Giachino C , Taylor V . Homeostatic neurogenesis in the adult hippocampus does not involve amplification of Ascl1(high) intermediate progenitors. Nat Commun. (2012) ;3: , 670. |
[39] | Merkle FT , Mirzadeh Z , Alvarez-Buylla A . Mosaic organization of neural stem cells in the adult brain. Science. (2012) ;317: (5836):381–4. |
[40] | Davis TH , Cuellar TL , Koch SM , Barker AJ , Harfe BD , McManus MT , et al., Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. (2008) ;28: (17):4322–30. |
[41] | Heo I , Joo C , Kim YK , Ha M , Yoon MJ , Cho J , et al., TUT4 inConcert with Lin28 Suppresses MicroRNA Biogenesis throughPre-MicroRNA Uridylation. Cell. (2009) ;138: (4):696–708. |
[42] | Harris L , Genovesi LA , Gronostajski RM , Wainwright BJ , Piper M . Nuclear factor one transcription factors: Divergent functions in developmental versus adult stem cell populations. Developmental Dynamics. (2015) ;244: (3):227–38. |
[43] | Chang CY , Pasolli HA , Giannopoulou EG , Guasch G , Gronostajski RM , Elemento O , et al., NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature. (2013) ;495: (7439):98–102. |
[44] | Ninkovic J , Steiner-Mezzadri A , Jawerka M , Akinci U , Masserdotti G , Petricca S , et al., The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. (2013) ;13: (4):403–18. |
[45] | Piper M , Barry G , Hawkins J , Mason S , Lindwall C , Little E , et al., NFIA controls telencephalic progenitor cell differentiation through repression of the notch effector hes1. The Journal of Neuroscience. (2010) ;30: (27):9127–39. |
[46] | Piper M , Moldrich RX , Lindwall C , Little E , Barry G , Mason S , et al., Multiple non-cell-autonomous defects underlie neocortical callosal dysgenesis in Nfib-deficient mice. Neural Dev. (2009) ;4: , 43. |
[47] | Shin J , Berg Daniel A , Zhu Y , Shin Joseph Y , Song J , Bonaguidi Michael A , et al., Single-cell RNA-Seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. (2015) ;17: (3):360–72. |
[48] | Hsieh J . Orchestrating transcriptional control of adult neurogenesis. Genes Dev. (2012) ;26: (10):1010–21. |
[49] | Di Carlo V , Grossi E , Laneve P , Morlando M , Dini Modigliani S , Ballarino M , et al., TDP-43 regulates the microprocessor complex activity during in vitro neuronal differentiation. Mol Neurobiol. (2013) ;48: (3):952–63. |



