Significance and Mechanisms Analyses of RB1 Mutation in Bladder Cancer Disease Progression and Drug Selection by Bioinformatics Analysis
Abstract
BACKGROUND:
Bladder cancer is still a disease of significant morbidity and mortality. In bladder cancer, RB1 is one of the most common mutant genes.
METHODS:
In this study, we explored the Genomics of Drug Sensitivity in Cancer (GDSC) database for drug sensitivity. The latest TCGA data were downloaded for analysis. To deal with functional enrichment analysis, GSEA, KEGG and GO were used. Prognostic analyses have been carried out using the GEPIA online tool.
RESULTS:
Results from the GDSC database showed that bladder cancer cells with RB1 mutation are more resistant to Dactolisib, MK-2206 and GNE-317. RB1 mutation was found in 25%bladder cancer patients. Patients with RB1 mutation often had lower RB1 mRNA expression level and higher histologic grade. In addition, we identified 999 differentially expressed genes in both groups. Functional enrichment analysis suggested that DEGs were primarily enriched in multiple metabolic progressions, cell proliferation and cancer related pathways. There were strong correlations between WT1, GPR37, CHRM2 and EZH2 expression levels and the prognosis.
CONCLUSIONS:
In all, the significance of RB1 mutation in disease progression and drug selection in bladder cancer was suggested by our results, and multiple genes and pathways related to such a program were identified.
INTRODUCTION
Bladder cancer is one of the most common malignancies worldwide and causes about 150,000 deaths per year [1]. Bladder cancer is characterized by easy recurrence and strong invasiveness. Transurethral resection of the bladder (TURB) is currently the main treatment for non-muscle invasive bladder cancer (NMIBC). After that, intravesical chemotherapy is the key therapy to prevent relapse. As for high-risk NMIBC, intravesical Bacillus Calmette-Guérin (BCG) is one of the most successful immunotherapies as the standard of treatment [2]. For muscle invasive bladder cancer (MIBC), surgery and chemotherapy are currently the main treatments. Nowadays, individualized treatment can help clinicians select target chemotherapy drugs according to the characteristics of tumor cells. This can achieve better treatment results, help patients delay the progression of the disease, and improve the prognosis. Therefore, an in-depth study of the molecular characteristics of bl-adder cancer can provide candidate targets and strategies for individualized treatment.
Retinoblastoma 1, also called RB Transcriptional Corepressor 1 (RB1), is one of the key regulators of cell division and acts as a tumor suppressor [3, 4]. The active, hypophosphorylated form of RB1 binds to E2F family members to exert the regulatory roles. A previous study has shown that RB1 could bind to the transcription factor E2F1 to negatively regulate cell cycle and stabilize constitutive heterochromatin to maintain the overall chromatin structure [5]. It has determined that defects in RB1 could cause various types of cancer, including bladder cancer [6–8]. Because the RB1 mutation is one of the most commonly detected mutations in patients with bladder cancer [8], we tried to further analyze the molecular characteristics of RB1 mutant bladder tumors to help identify some novel treatment methods.
In the present study, we analyzed the genetic al-terations of RB1 in bladder cancer, which include mutation and copy number alterations. Then we fo-cused on the characteristics associated with RB1 mutated bladder cancer. Results from the GDSC database showed the significance of RB1 mutation in bladder cancer drug selection. We also analyzed the RNA sequencing (RNA-Seq) dataset of MIBC to find out the association of RB1 mutation with disease progression. Meanwhile, the identification of the critical pathways associated with RB1 mutation could contribute to uncovering the potential mechanisms and therapeutic targets.
MATERIALS AND METHODS
Analysis of the GDSC database
The establishment of the GDSC (Genomics of Cancer Drug Sensitivity) project is mainly for cancer molecular therapy and mutation exploration [9]. In this article, we explored compounds that are selective for RB1 mutation in bladder cancer cells. Related volcano plots, scatter plots, and the statistical analysis was downloaded directly from the GDSC online platform.
Cell culture and Dactolisib treatment
The human bladder carcinoma cell lines J82, 5637, RT4 and TCCSUP cells were obtained from the American Type Culture Collection (ATCC). All cell lines were maintained in RPMI-1640 supplemented with 10%FBS and an antibiotic solution (100 units/ml penicillin and 0.1 mg/ml streptomycin), and growing at 37 in standard cell culture conditions (5%CO2, 95%humidity). After treatment with 0.1μM Dactolisib (NVP-BEZ235), cell viability was assessed using MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (Promega) at indicated time points.
RNA-Seq data
The RNA-Seq data of bladder cancer gene expression were downloaded through the TCGA database. Corresponding clinical information was obtained through the cBioPortal for Cancer Genomics website [10]. TCGA: https://portal.gdc.cancer.gov/.; cBioPortal: http://www.cbioportal.org/index.do
Identification of differentially expressed genes (DEGs)
In order to find the DEGs between RB1 mutation and wild-type bladder cancer patients, we conducted the study using the EdgeR method [11, 12]. DEGs were identified with the following criterion: fold change (FC)≥2 or ≤0.5; the P-value <0.05.
Gene set enrichment analysis (GSEA)
In order to further discover the differences in gene expression levels of biological annotations and pathways, we analyzed using GSEA software to help understand the effect of RB1 mutations on the biological functional genome of tumor tissue in patients with bladder cancer. Enrichment results satisfying P-value <0.05 with a false discovery rate (FDR) <0.25 were considered statistically significant.
Functional annotation and pathway enrichment analysis of DEGs
In this study, we used DAVID online tools to complete gene function and related enrichment analysis [13]. GO and KEGG analysis was uploaded to DAVID website for enrichment analysis, while specifying a P-value <0.05 for statistical significance. DAVID: https://david.ncifcrf.gov/.
Statistical analysis
The prognostic analyses of hub genes were downloaded directly from the GEPIA website. We compared the expression levels of RB1 mRNA in RB1 wild-type and mutant bladder cancer tissues using t-test. The χ2 test was used to assess whether the RB1 mutation affected the patient’s clinical progression data. The clinical prognostic analysis was mainly carried out by Kaplan-Meier method with log rank test through Graphpad. All statistical analyses were performed using R 3.3. 0 and Graphpad. P < 0.05 was considered statistically significant.
RESULTS
Bladder cancer cells with RB1 mutation are more resistant to Dactolisib, MK-2206 and GNE-317
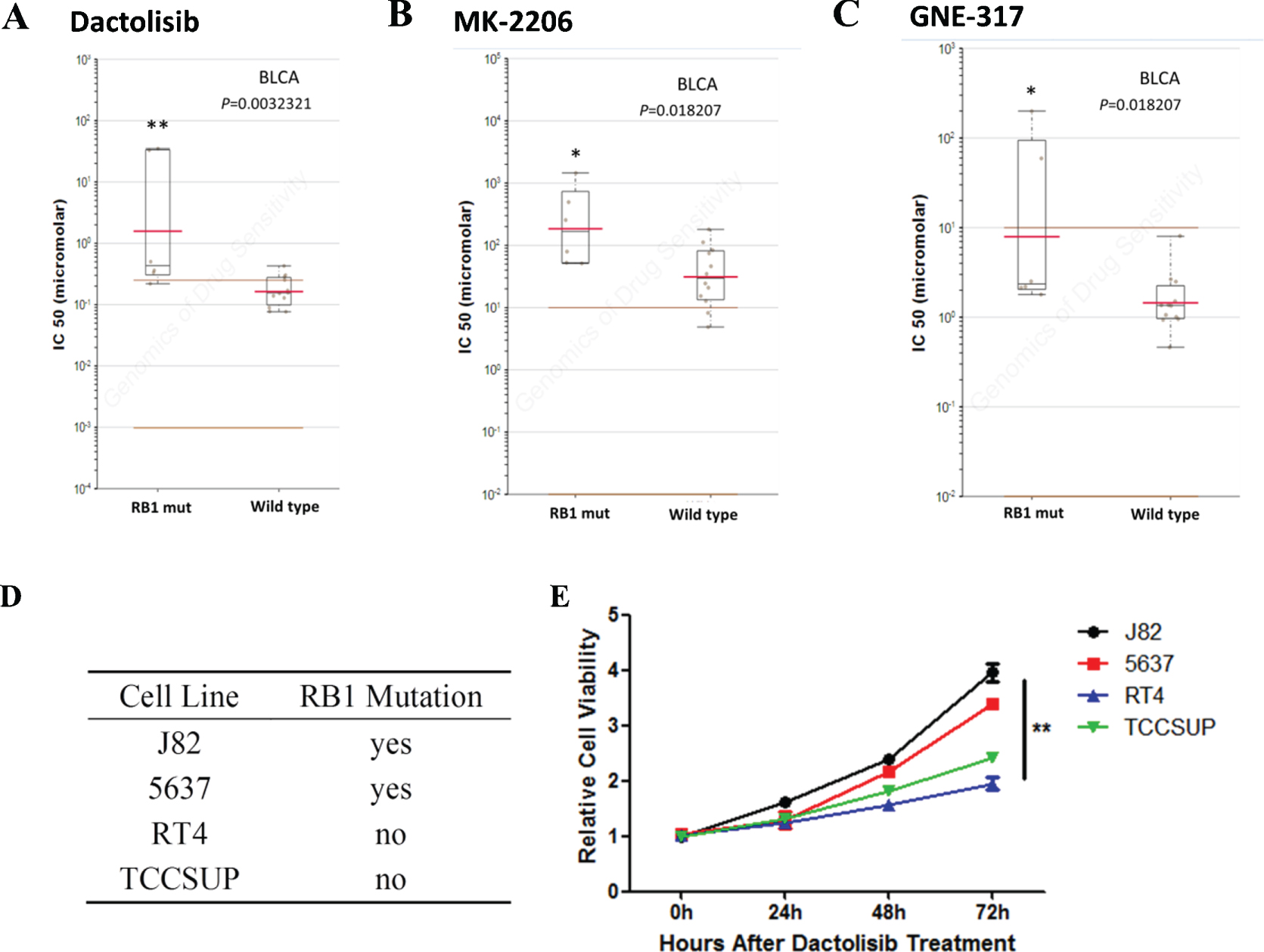
The sensitivity to chemotherapy drugs is important to the therapeutic effect. In order to find out some details in drug selection, we explored the GDSC database, a public online platform that has been developed to help researchers to analyze the astronomical data matrix and download data [14]. Results indicated that bladder cancer cells that harbor RB1 mutation, not other types of tumors, were significantly resistant to Dactolisib, MK-2206 and GNE-317 (Fig. 1). To confirm the results, we then did a cell viability assay using bladder cancer cell lines with different RB1 status. We found that bladder cancer cells with RB1 mutations were selectively resistant to Dactolisib (Figs. 1D and E). As a result, Dactolisib, MK-2206 and GNE-317 might not be suitable for bladder cancer patients with RB1 mutation.
Fig. 1
Mutation of RB1 influences drug selection of bladder cancer. A- C. Reproduction of GDSC database showed that bladder cancer cells with RB1 mutation, but not cancer of other types, was significantly resistant to Dactolisib, MK-2206 and GNE-317. D. RB1 mutation status in different bladder cancer cell lines. E. Cell viability assay showed that bladder cancer cells with RB1 mutations were selectively resistant to Dactolisib.
RB1 mutation in bladder cancer
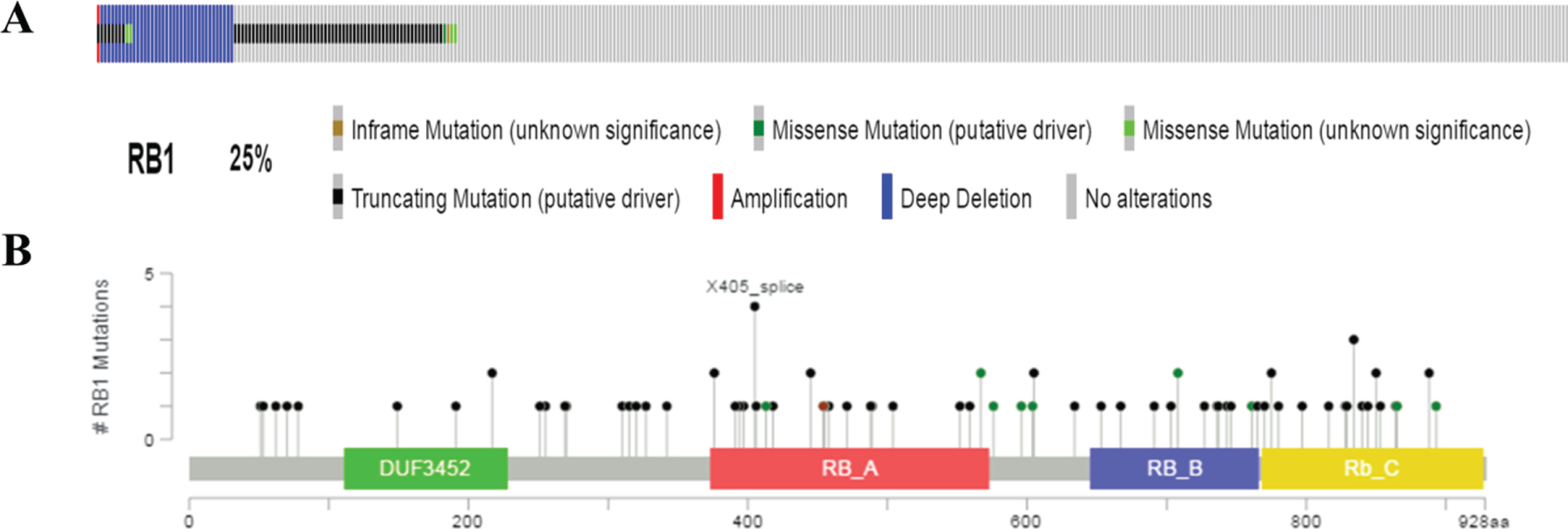
To further investigate the characteristics associated with RB1 mutated bladder cancer, we then downloaded related clinical information of 408 bladder cancer patients through the cBioPortal for Cancer Genomics website, and their cancer tissue express-ion data from TCGA database. There were 100 pat-ients (25%) who had genetic alterations in RB1 gene (Fig. 2A). As shown in Fig. 2B, the main alteration types included mutation (inframe mutation, trunc-ating and missense mutations spanning over entire gene) and copy number alterations (amplification and deep deletion). In the following researches, we mainly focused on the characteristics of RB1 mutated bladder cancer in molecular features, disease progression and prognosis.
Fig. 2
Mutation frequency and types of RB1 in bladder cancer from the cancer Genome Atlas (TCGA) database. A. Mutation frequency of RB1 in bladder cancer. B. Mutation types of RB1 in bladder cancer.
RB1 mutation in disease progress and prognosis
In clinical affairs, we explored the characteristics of RB1 mutated bladder cancer in disease development and prognosis. We compared the clinical in-formation in both groups. Results indicated that patients with RB1 mutation have higher neoplasm histologic grade, indicating that RB1 mutation might contribute to the disease progression (Table 1).
Table 1
Clinical Characteristics of BLCA Patients and RB1 status in TCGA
| Characteristics | RB1 status | P value | |
| wild type | mutated | ||
| Age, years | 67.64 | 70.4 | |
| range | 34–90 | 54–89 | |
| Gender | P = 0.1772 | ||
| Female | 92 | 14 | |
| Male | 243 | 57 | |
| Tumor T stage | P = 0.2138 | ||
| T1 | 2 | 1 | |
| T2 | 35 | 2 | |
| T2a | 22 | 4 | |
| T2b | 46 | 10 | |
| T3 | 37 | 5 | |
| T3a | 53 | 18 | |
| T3b | 63 | 18 | |
| T4 | 8 | 3 | |
| T4a | 37 | 4 | |
| T4b | 3 | 1 | |
| N stage | P = 0.0912 | ||
| N0 | 197 | 39 | |
| N1 | 40 | 6 | |
| N2 | 54 | 21 | |
| N3 | 8 | 0 | |
| NX | 30 | 6 | |
| M stage | P = 0.0590 | ||
| M0 | 166 | 25 | |
| M1 | 9 | 2 | |
| MX | 157 | 45 | |
| AJCC Neoplasm Disease Stage | P = 0.7044 | ||
| Stage I | 2 | 0 | |
| Stage II | 109 | 20 | |
| Stage III | 115 | 25 | |
| Stage IV | 107 | 27 | |
| Neoplasm Histologic Grade | P = 0.0284 | ||
| high | 311 | 72 | |
| low | 21 | 0 |
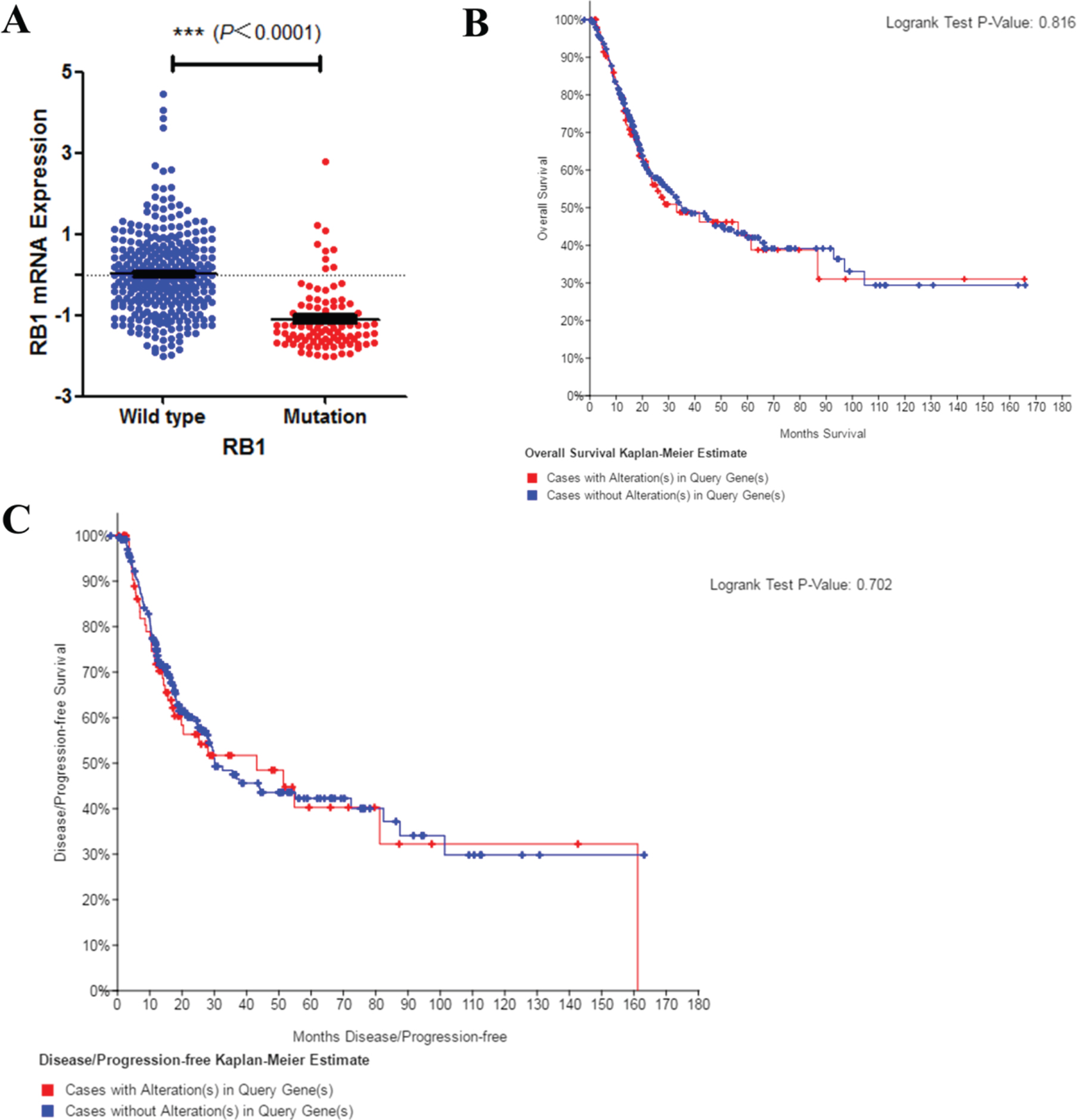
Next, lower RB1 expression level was found in mutated bladder cancer patients’ tissues (Fig. 3A). However, RB1 mutation did not affect disease prognosis of overall survival (Fig. 3B) and recurrence (Fig. 3C). Results above indicated that RB1 mutation may be associated with bladder cancer disease progression, but not the prognosis. Early intervention may be benefit to such patients.
Fig. 3
Mutation of RB1 and bladder cancer prognosis. A. Correlation between RB1 mutation and mRNA expression. B. C. Kaplan–Meier survival and disease recurrence curves for bladder cancer patients stratified by RB1 mutation.
Results of gene set enrichment analysis
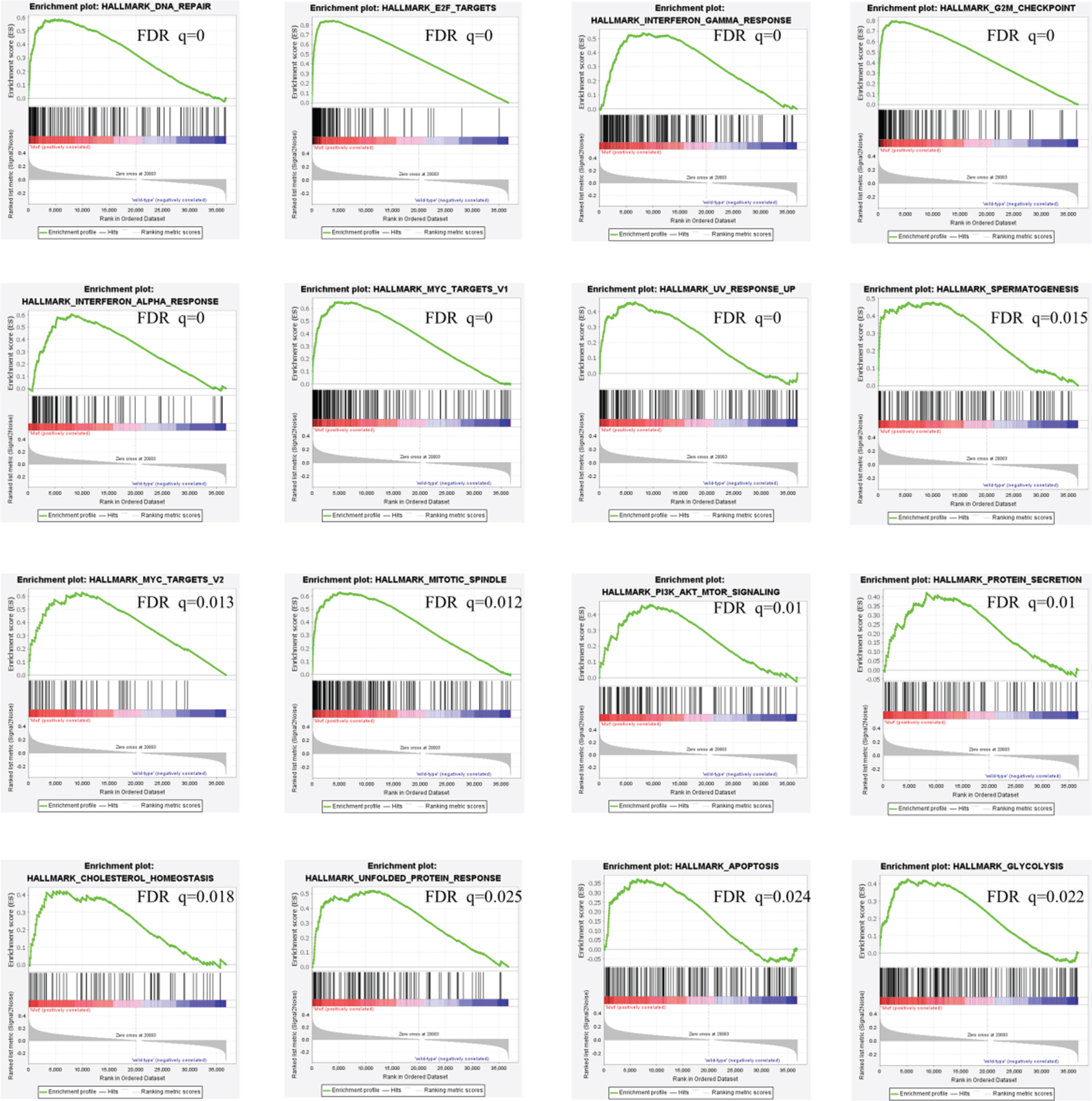
With the scope of exploiting the underlying mechanisms, we analyzed the characteristics of RB1 mutated bladder cancer in molecular features and cellular processes. We first used GSEA approach to analyze biological functional gene sets. Results in Fig. 4 indicated that DNA repair, E2F targets, interf-eron gamma response, G2M checkpoint, interferon alpha response, MYC targets v1, UV response up, spe-rmatogenesis, MYC targets v2, mitotic spindle, PI3K-AKT-mTOR signaling, protein secretion, cholesterol homeostasis, unfolded protein response, apoptosis, glycolysis and fatty acid metabolism were signific-antly enriched, which suggests that pathways in cancer, metabolism and DNA repair may be associated with disease progression in RB1 mutated bladder cancer.
Fig. 4
GSEA results of RB1 mutation in bladder cancer patients.
Identification of DEGs
We then identified the DEGs to deal with further analysis. A total of 999 DEGs were identified, among which 476 genes were up-regulated and 523 genes were down-regulated in RB1 mutated bladder cancer. The volcano diagram of DEGs was shown in Fig. 5A.
Fig. 5
DAVID enrichment results of differentially expressed genes. A. Volcano plot for differentially expressed genes. B. The GO enrichment terms of differentially expressed genes. C. The KEGG pathway analysis of differentially expressed genes. D-F. The correlationship of expression levels of WT1 (D), GPR37 (E) and EZH2 (F) with disease-free survival. Log rank Test was used to analysis and a P-value <0.05 for statistical significance. G-I. The correlationship of CHRM2 (G), WT1 (H) and GPR37 (I) expression levels with overall survival. Log rank Test was used to analysis and a P-value <0.05 for statistical significance.
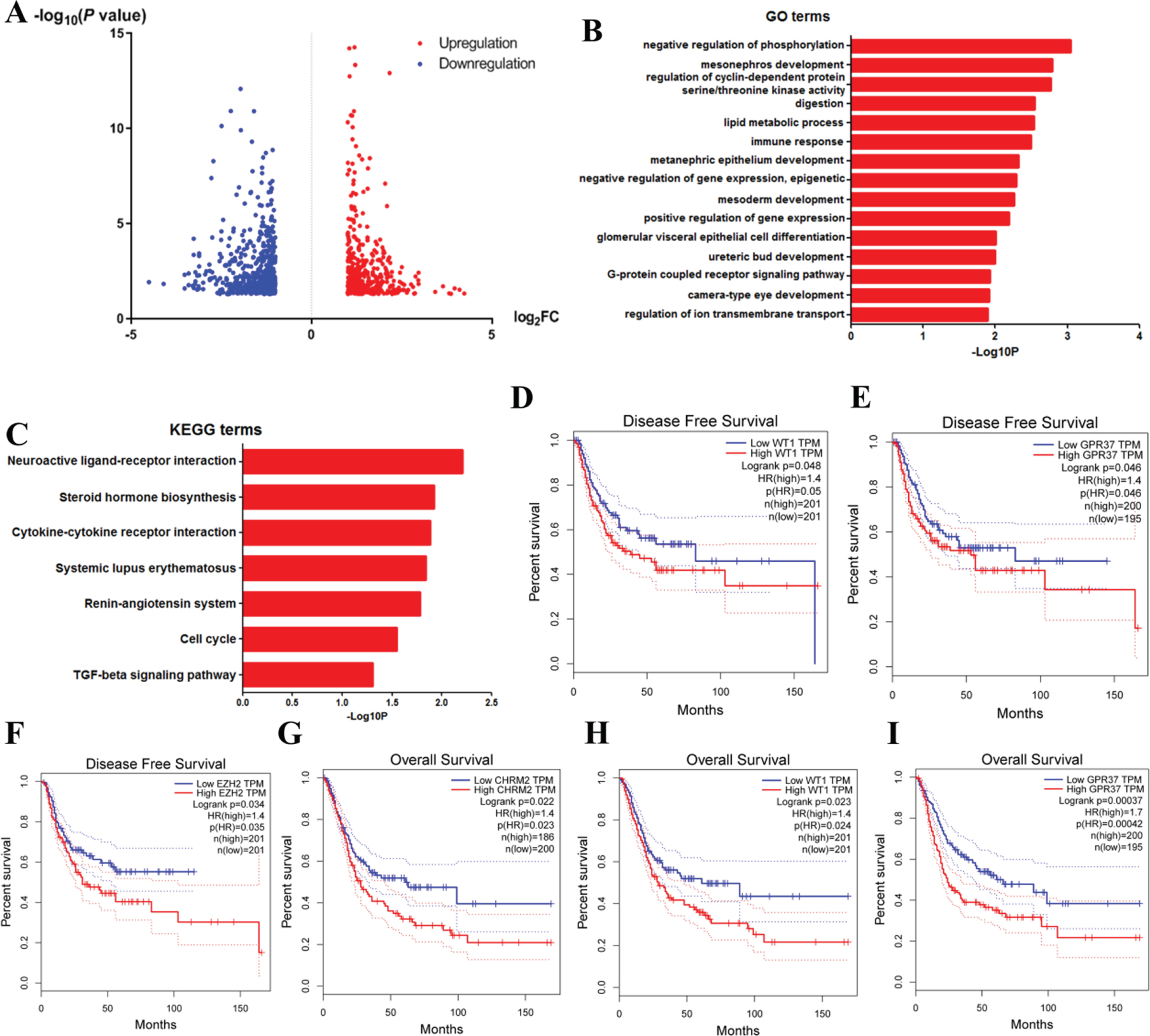
GO and KEGG analysis of DEGs
Then, we conducted functional analyses using the obtained DEGs. We uploaded 999 DEGs to the DAVID online tool. Results of GO analysis (Fig. 5B) suggested significant enrichment in positive regulation of negative regulation of phosphorylat-ion, mesonephros development, regulation of cyclin-dependent protein serine/threonine kinase activity, digestion, lipid metabolic process, immune response, metanephric epithelium development, negative regulation of gene expression, epigenetic, mesoderm development, positive regulation of gene expression, glomerular visceral epithelial cell differentiation, ureteric bud development, G-protein coupled receptor signaling pathway, camera-type eye development and regulation of ion transmembrane transport.
What’s more, KEGG pathway analysis showed enrichment mainly in neuroactive ligand-recep-tor interaction, steroid hormone biosynthesis, cyto-kine-cytokine receptor interaction, systemic lupus erythematosus, renin-angiotensin system, cell cycle and TGF-beta signaling pathway (Fig. 5C).
Identification of the hub gene and prognostic analysis
To find out the hub genes that might contribute to disease progression and drug selection, we then uploaded all the DEGs above to screen the informat-ion in the STRING database. We treated the top 15 genes ranked by degree as hub genes, which included EZH2, CDKN2A, SHH, GNG4, IGF2, CXCL9, POU5F1, CSF2, IFNG, WT1, CDKN1C, GPR37, MELK, CHRM2 and ITGA2B (Table 1). EZH2, which has the highest degree of 28, is the top nodule. To further investigate the roles of such hub genes in disease clinical outcome, we explored the GEPIA website [15]. Figure 5D-F showed that the expression levels of WT1, GPR37 and EZH2 strongly associated with disease-free survival, while Fig. 5G-I showed that CHRM2, WT1 and GPR37 correlated with overall survival. These results suggested that these 4 genes might be the most critical genes for further research. To confirm our results, we verified the prognostic-related hub genes above using the data of dataset GSE48075. We found that only WT1 of these genes was significantly related to the prognosis of bladder cancer patients (Log-rank P = 0.0053, data not shown). However, because the sample size is quite small, we will conduct further verification in subsequent researches.
DISCUSSION
Bladder cancer is still one of the most common malignancies all around the world [1]. The characte-ristics of bladder tumors determine that it is more prone to recurrence, invasion and metastasis. Chemo-therapy, including intravesical chemotherapy, and intravesical BCG are the critical therapies dealing with bladder cancer cells and preventing recurrence [16, 17]. Nowadays, individualized treatment can help clinicians select target chemotherapy drugs acc-ording to the characteristics of tumor cells. In our study, we explored the characteristics associated with RB1 mutated bladder cancer in drug selection, clinical affairs and molecular features. Results from the GDSC dataset showed preliminary evidence that Dactolisib, MK-2206 and GNE-317 exhibit less sensitivity for bladder cancer cells that harbor RB1 mutation. Our in vitro assays also confirmed the resistance of bladder cancer cells with RB1 mutation to Dactolisib. The results above provide potential evidence for not to use such drugs to patients above in clinical practice.
RB1 acts as a negative regulator of the cell cy-cle and stabilizes constitutive heterochromatin to maintain the overall chromatin structure. RB1 promotes G0-G1 transition when phosphorylated by CDK3/cyclin-C, while hypophosphorylated RB1 bi-nds transcription factor E2F family members as a transcription repressor [5, 18, 19]. CDK 4/6 and cyclin D complex could also phosphorylate RB1 to promote G1 to S phase in many cancer cells [19]. RB1 is a tumor suppressor and critical mutation could influence its function [4]. Studies have shown that RB1 mutation is frequently found in bladder cancer.
In clinical terms, we found about 25%of patients with RB1 mutation among bladder cancer patients. Further analysis showed that RB1 mutation has no significant influence on prognosis. The clinical characteristic analysis showed higher neoplasm histologic grade in bladder cancer patients with RB1 mutation, suggesting the role of RB1 mutation as an indicator to advanced tumor. Our results showed that bladder cancer patients with RB1 mutation are more likely to progress to the advanced stage. Early and comprehensive intervention might be important for such patients to live longer.
Nowadays, molecular pathological diagnosis and individualized medicine testing can help doctors select target drugs according to the characteristics of the patient’s tumor cells, including bladder cancer. Different patients may have tumor cells with different pathways activated in their bodies, which results in different molecular and pathological characteristics. These are the main mechanisms for disease progression and drug resistance in many tumors, which is also the basis of individualized treatment. Analyzing the activation of different pathways can help us understand the characteristics of different subtypes of tumors, and might help us explore potential personalized treatment at the same time. If a certain pathway is specifically activated in certain subtype patients, the drugs targeted this pathway might be more effective in such patients. Through these analyses, we can find out the potential mechanisms of drug resistance and clinical phenotypes that are associated with RB1 mutation, and provide some theoretical basis and directions for further research and verification. This might be quite preliminary and only have some theoretical functions. To explore these issues deeply, the gene expression data was analyzed in depth to find the key pathways and core genes associated with RB1 mutation. Results of GSEA analysis suggested that RB1 mutation were mainly associated with DNA repair, multiple cancer related pathways, cell proliferation and division, and metabolism. Previous researches have shown that defects in DNA repair contributed the disease progression of bladder cancer and influenced the treatment, especially in MIBC. Genomic alterations in the DNA repair-associated genes could render tumors sensitive to cisplatin-based chemotherapy for MIBC [20]. RB1 regulates the cell cycle. In RB1 mutated tumors, our findings showed that many processes involved in cell proliferation were enriched. RB1 mutation could result in abnormal regulation of cell cycle and cell division proliferation, which lead to continuous cell growth and tumor progression. Therefore, besides the three drugs above, other drugs, such as Docetaxel (Taxotere) which effectively induces G2M arrest and apoptosis might be more sensitive in patients with RB1 mutation [21].
Next, we searched for DEGs and performed functional enrichment analysis on them. The results show a total of 999 DEGs. Enrichment analysis suggested that DEGs in RB1 mutated bladder cancer patients implicated with multiple cellular programs. Among all programs, those related to metabolism were the most important terms. As we all know, adaptations across multiple metabolic processes are necessary to satisfy the energy required for an increased rate of proliferation in the malignant transformation progression. In bladder cancer, dysregulation of cell metabolism has been a hallmark of the malignant phenotype [22, 23]. Our results indicated the possibility that RB1 mutation might induce more active cell metabolism in tumor cells. Drugs targeting this might have better therapeutic effects in such patients.
In the hub gene and prognostic analyses, we found that the enhancer of zeste homolog 2 (EZH2) is the top hub gene with the highest degree. EZH2 is a critical component of the polycomb repressive complex 2 (PRC2). EZH2 could induce gene silencing mainly through catalyzing histone H3K27me3 and is always overexpressed in various cancers [24]. Our results indicate the possibility that EZH2 might play a critical role in drug selection and disease progression of RB1 mutated bladder cancer. Moreover, prognostic analysis indicated that 4 genes, WT1, CHRM2, GPR37 and EZH2, are strongly associated with disease prognosis. These results suggested that these 4 genes might be the potential target genes for further research. To confirm the roles of these 4 genes in bladder cancer prognosis, we used the prognostic information from another two published researches called MSK-IMPACT and Nature 2014. Results in Sup Figure s1 showed that GPR37 and CHRM2 are strongly associated with overall survival, but no gene was found to be associated with disease-free survival.
Our study contained one limitation. In our resea-rch, our results mostly base on online datasets and bi-oinformatics analysis. Although we confirmed some of our results using another dataset GSE48075, the sample size of this cohort with prognostic information was still too small. In the future, we still need further researches in clinical and molecular biology experiments to confirm our results and investigate the roles of hub genes that were identified by us.
CONCLUSIONS
In conclusion, this study found out the clinical significance of RB1 mutation in bladder cancer. What’s more, we identified the main pathways and genes associated with RB1 mutation. All results above could facilitate developing early intervention and providing better therapeutic strategies against such a special su-btype of bladder cancer. Furthermore, the mechanism and validation of RB1 mutation in bladder cancer still need further research in clinical and molecular biology experiments.
ACKNOWLEDGMENTS
The authors have no acknowledgments.
FUNDING
The authors report no funding.
AUTHOR CONTRIBUTIONS
DGZ and BG: conception and design, and drafting the manuscript; JJT and QEX: acquisition and analysis of the data and drafting the manuscript; ZYY and BG: statistical analysis and technical support. All authors have read and approved the manuscript, and ensure that this is the case.
ETHICAL CONSIDERATIONS
This work is exempt from any requirement for Institutional Review Board approval since it is a database study and no human or animal research was involved in the elaboration of this manuscript.
CONFLICT OF INTEREST
DGZ, JJT, QEX, ZYY and BG have no conflicts of interest to report.
REFERENCES
[1] | Siegel RL , Miller KD , Jemal A . Cancer statistics. CA Cancer J Clin (2016) ;66: (1):7–30. |
[2] | Nicholas M , Donin Andrew T , Lenis, et al. Immunotherapy for the Treatment of Urothelial Carcinoma. J Urol. 2017. |
[3] | Dyson NJ . RB a prototype tumor suppressor and an enigma. Genes Dev (2016) ;30: (13):1492–502. |
[4] | Corradini P , Inghirami G , Astolfi M , Ladetto M , Voena C , Ballerini P , et al. Inactivation of tumor suppressor genes, p53 and Rb1, in plasma cell dyscrasias. Leukemia (1994) ;8: (5):758–67. |
[5] | Morris EJ , Ji JY , Yang F , Di Stefano L , Herr A , Moon NS , et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature (2008) ;455: (7512):552–6. |
[6] | Babenko OV , Zemliakova VV , Saakian SV , Brovkina AF , Strel’nikov VV , Zaletaev DV , et al. [RB1 and CDKN2A functional defects resulting in retinoblastoma]. Mol Biol (Mosk) (2002) ;36: (5):777–83. |
[7] | Hansen MF , Cavenee WK . Retinoblastoma and the progression of tumor genetics. Trends Genet (1988) ;4: (5):125–8. |
[8] | Gui Y , Guo G , Huang Y , Hu X , Tang A , Gao S , et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet (2011) ;43: (9):875–8. |
[9] | Vanden Heuvel JP , Maddox E , Maalouf SW , Iorns E , Tsui R , Denis A , et al. Replication Study: Systematic identification of genomic markers of drug sensitivity in cancer cells. eLife 2018;7. |
[10] | Cerami E , Gao J , Dogrusoz U , Gross BE , Sumer SO , Aksoy BA , et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery (2012) ;2: (5):401–4. |
[11] | Robinson MD , McCarthy DJ , Smyth GK . edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) ;26: (1):139–40. |
[12] | McCarthy DJ , Chen Y , Smyth GK . Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res (2012) ;40: (10):4288–97. |
[13] | Dennis G Jr , Sherman BT , Hosack DA , Yang J , Gao W , Lane HC , et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome BiolP (2003) ;4: (5):3. |
[14] | Yang W , Soares J , Greninger P , Edelman EJ , Lightfoot H , Forbes S , et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res (2013) ;41: (Database issue):D955–61. |
[15] | Tang Z , Li C , Kang B , Gao G , Zhang Z . GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) ;45: (W1):W98–W102. |
[16] | Kaufman DS , Shipley WU , Feldman AS . Bladder cancer. Lancet (2009) ;374: (9685)239–49. |
[17] | Sternberg CN , Donat SM , Bellmunt J , Millikan RE , Stadler W , De Mulder P , et al. Chemotherapy for bladder cancer: treatment guidelines for neoadjuvant chemotherapy, bladder preservation, adjuvant chemotherapy, and metastatic cancer. Urology (2007) ;69: (1 Suppl):62–79. |
[18] | Peyressatre M , Prevel C , Pellerano M , Morris MC . Targeting cyclin-dependent kinases in human cancers: from small molecules to Peptide inhibitors. Cancers (2015) ;7: (1):179–237. |
[19] | O’leary B , Finn RS , Turner NC . Treating cancer with selective CDK4/6 inhibitors. Nature reviews Clinical Oncology (2016) ;13: (7):417–30. |
[20] | Plimack ER , Dunbrack RL , Brennan TA , Andrake MD , Zhou Y , Serebriiskii IG , et al. Defects in DNA Repair Genes Predict Response to Neoadjuvant Cisplatin-based Chemotherapy in Muscle-invasive Bladder Cancer. Eur Urol (2015) ;68: (6):959–67. |
[21] | Martin SK , Kyprianou N . Exploitation of the Androgen Receptor to Overcome Taxane Resistance in Advanced Prostate Cancer. Adv Cancer Res (2015) ;127: :123–58. |
[22] | Boyland E , Williams DC . The metabolism of tryptophan. 2. The metabolism of tryptophan in patients suffering from cancer of the bladder. Biochem J (1956) ;64: (3):578–82. |
[23] | Ritterson Lew C , Guin S , Theodorescu D . Targeting glycogen metabolism in bladder cancer. Nature reviews Urology (2015) ;12: (7):383–91. |
[24] | Margueron R , Reinberg D . The Polycomb complex PRC2 and its mark in life. Nature (2011) ;469: (7330):343–9. |




