Ten Years of Proteomics in Bladder Cancer: Progress and Future Directions
Abstract
Research efforts targeting the identification of bladder cancer biomarkers have been extensive during the past decade. Investigations have been performed at the genome, transcriptome, proteome, and metabolome levels and outputs have started appearing including the sketching of disease molecular subtypes. Proteins are directly linked to cell phenotype hence they accumulate special interest as both biomarkers and therapeutic targets. Multiple technical challenges exist, of the main, being the protein concentration vast dynamic range and presence of proteins in modified forms. The scope of this review is to summarize the contribution of proteomics research in this quest of bladder cancer biomarkers. To obtain an unbiased and comprehensive overview, the scientific literature was searched for manuscripts describing proteomic studies on urothelial cancer from the last ten years and those including independent verification studies in urine, tissue and blood are briefly presented. General observations include: a) in most cases, suboptimal experimental design including healthy controls in biomarker discovery and frequently biomarker verification, is followed; b) variability in protein findings between studies can be observed, to some extent reflecting complexity of experimental approaches and proteome itself; c) consistently reported biomarkers include mainly plasma proteins and d) compilation of protein markers into diagnostic panels appears the most promising way forward. Two main avenues of research can now be foreseen: targeting integration of the existing disparate data with proteomic findings being placed in the context of existing knowledge on bladder cancer subtypes and in parallel, accumulation of clinical samples to support proper validation studies of promising marker combinations.
Abbreviations/Glossary
2DE | Two-dimensional gel electrophoresis |
A1AT | Alpha-1-antitrypsin |
ADAM28 | Disintegrin and metalloproteinase domain-containing protein 28 |
ApoA-I | Apolipoprotein A1 |
BLCAP | Bladder cancer-associated protein |
CE | Capillary Electrophoresis |
ELISA | Enzyme Linked Immunosorbent Assay |
FDA | US Food and Drug Administration |
FGB | Fibrinogen beta chain precursor |
IHC | Immunohistochemistry |
iTRAQ | Isobaric Tags for Relative and Absolute Quantitation |
LC | Liquid Chromatography |
MIBC | Muscle Invasive Bladder Cancer |
MRM | Multiple Reaction Monitoring |
MS | Mass Spectrometry |
MS/MS | Tandem mass spectrometry |
NIF-1 | Zinc finger protein 335 |
NMIBC | Non Muscle Invasive Bladder Cancer |
UROM | Uromodulin |
WB | Western Blot |
INTRODUCTION
Given the existing shortcomings in bladder cancer management (invasiveness and suboptimal sensitivity of cystoscopy, high cost of a surveillance program, unmet need to tailor treatment selection to cancer molecular profile, [reviewed in 1–4]) research on establishing new, ideally non-invasive biomarkers of added diagnostic and prognostic/predictive value has been extensive: as a rough reflection of this activity, the number of Medline hits using keywords “biomarker” and “bladder cancer” between 2005–2016 is 3842 (searched on 15/08/2016). Main targeted contexts of use include screening of high risk populations, monitoring for cancer recurrence and progression and prediction of treatment effectiveness [1, 4]. Several molecular markers have been approved by the Food and Drug Administration (FDA) and extensively tested for use in bladder cancer diagnosis and/or prognosis [reviewed in 1, 4, 5], nevertheless, these have not been introduced into the clinical guidelines and the main goal of achieving a tangible improvement in disease management has not been achieved yet [1].
The scope of this review is to summarize and perform an initial evaluation of the contribution of proteomics research in this quest of bladder cancer biomarkers and/or understanding disease pathophysiology. To obtain an unbiased and comprehensive overview, the Web-of-Science was searched for manuscripts from the last ten years, containing the key words “bladder cancer” OR “urothelial cancer” and proteom*, limiting to original manuscripts. A total of 247 records were retrieved (search performed 06/2016). These were inspected for relevance and validity (as defined by confirmation of findings in an independent sample set) by both co-authors, and 52 manuscripts [listed as references 11–44, 47–64] were found to be actual reports on application of proteomics in the context of bladder cancer, forming the basis of this review. Two topics appear to be in the center of interest (Fig. 1): 1) assessing the urine proteome for diagnosis of the disease, and 2) investigating the tissue proteome aiming at prognosis; structuring respectively the main text below. It should be noted that while submitting this manuscript, a review on bladder cancer protein markers was published [6], focusing however solely on soluble urinary proteins, applying inclusion criteria based on reported biomarker performance and clinical characteristics of the case-control groups, and without special emphasis on proteomics (i.e. large scale protein analysis). Based on these differences, an overlap in the retrieved references of approximately 25% (13/52) between the presented and previous review [6] may be observed which suggests that collectively the two manuscripts provide an in-depth coverage of the field.
URINE PROTEOMICS
As outlined in several recent publications, urine has the major advantage of being easily accessible, and, in the case of bladder cancer, being in direct contact with the tumor [5, 7]. In addition, the urine proteome is fairly well characterized and reference standards are available [7, 8]. The clinical focus in this case has been on the discovery of bladder cancer diagnostic markers. The workflow typically involved a biomarker discovery phase, with application of high resolution techniques (such as various types of liquid chromatography in combination to tandem mass spectrometry (Liquid Chromatography Mass Spectrometry-LC-MS/MS) followed by validation of selected findings in a new set of samples, using targeted proteomic techniques: antibody-based (Enzyme Linked Immunosorbent Assay -ELISA) or Mass spectrometry-based (Multiple reaction Monitoring-MRM/ Capillary electrophoresis mass spectrometry CE-MS). From a methodological point of view, investigations at the proteome level grossly may be divided in those focusing on the characterization of the low molecular weight proteome (native peptidome, protein mass <15 kD) and those targeting the analysis of the larger proteins and modified forms thereof, requiring different sample preparation techniques in each case. A presentation of the individual techniques would be out of the realm of this review but the interested reader may visit several recent reviews on the topic [7, 9, 10].
In the case of the urine peptidome analysis, capillary electrophoresis coupled to mass spectrometry (CE-MS) has been extensively applied. In a first publication in 2006 [11], the authors demonstrated superior performance of a classifier based on 22 urinary peptides in the training set (100% sensitivity and 100% specificity). Upon independent validation including 31 samples from bladder cancer patients and a total of 366 controls (samples from renal diseases, urinary tract infection, prostate hyperplasia and prostate cancer), the 22- peptide biomarker panel classified correctly all bladder cancer patients (100% sensitivity), with a 73% specificity. However, only one peptide, fibrinopeptide A, out of the 22 of the classifier, could be identified in this study [11]. In a later study [12], it became evident that this classifier, while differentiating between advanced cancer and controls, required further adjustment to allow correct classification of less advanced tumors. In this study, the authors could demonstrate relevance of urinary peptides also in the differentiation between non-invasive and invasive bladder cancer (with 92% sensitivity and 58% specificity [12]. In a very recent study [13], the same group has further refined this approach and presented two distinct panels, one targeting diagnosis of bladder cancer at first presentation (primary disease) and the second biomarker panel to be potentially employed for surveillance (detection of recurrences) and demonstrated their value in a multi-center study, encompassing a total of 1357 patients. Interestingly, several peptides included in the panels, such as fibrinogen chains, apolipoprotein A1 (ApoA-1), beta-2-macroglobulin and basement membrane-specific heparan sulfate proteoglycan are also described in the form of larger proteins in other bladder cancer proteomic studies (Tables 1 and 2, summarized below) [13].
Multiple studies focusing on larger mass proteins and modifications thereof have been reported: Kreunin et al. [14] focused on urinary glycoproteins and described several potential biomarkers for bladder cancer, including alpha-1B-glycoprotein. In a subsequent report, the same group reported the identification of additional bladder cancer glycoprotein biomarkers, with the most prominent being alpha-1-antitrypsin (A1AT) [15]. A1AT was similarly highlighted in the study by Linden et al. who also suggested among others, fibrinogen and apolipoprotein E as biomarkers of bladder cancer [16]. Additional forms of apoliprotein proteins as markers for bladder cancer have been highlighted in multiple studies: Lei et al. reported on the identification of multiple biomarkers distinguishing bladder cancer from controls using two dimensional electrophoresis, of which upregulation of ApoA-1 in urine of bladder cancer patients was confirmed by Western Blot (WB) [17]. Along the same lines, Li et al. [18, 19], found increased abundance of urinary fibrinogen, lactate dehydrogenase B, Apo-A1, clusterin and haptoglobin associated with bladder cancer, and further confirmed Apo-A1 using ELISA in two studies [18, 20] supporting overall sensitivities ranging from 89–92% and specificities (for healthy controls) of 85–86% in a total number of 535 included subjects. Increased urinary levels of ApoA1 were also observed in a study involving application of LC-MS in combination to peptide labeling (Isobaric Tags For Relative And Absolute Quantitation -iTRAQ), where pooled samples from 37 urine samples from bladder cancer patients or those with hernia were analyzed [21]. The result was confirmed by ELISA in a total of 126 urine samples supporting overall sensitivity in bladder cancer detection of 94.6% with a specificity of 92% (for discrimination between bladder cancer and hernia [21]). Additional findings from the same study were confirmed in a subsequent analysis using MRM techniques and samples from bladder cancer (n = 76), hernia (n = 57) and urinary tract infections/hematuria (n = 23), where a six-peptide panel including afamin, adiponectin, complement C4 gamma chain, apolipoprotein A-II precursor, ceruloplasmin, and prothrombin was suggested as having diagnostic value (AUC of 0.814) [22].
Aiming at identifying urinary biomarkers associated with bladder cancer aggressiveness, Zoidakis et al. investigated pools of urine samples from patients with non-invasive or invasive bladder cancer [23]. Differential abundance of aminopeptidase N (APN), myeloblastin and profilin-1 was confirmed in independent validation by WB (APN, myeloblastin) or ELISA (profilin-1) in a total of 108 samples from bladder cancer patients and 97 samples from patients with benign diseases. Profilin-1 was further investigated at the tissue level using tissue microarrays suggesting a prognostic potential for the molecule and the association of its levels with stage (p = 0.002), overall survival (p = 0.0005) and disease specific survival (p = 0.0005). Functional relevance of the protein was further suggested by in vitro and in vivo profilin-1 blocking experiments resulting in decreased cell motility and tumor growth [23 and Frantzi et al, Oncotarget, accepted for publication). Profilin-1 was also detected as being differentially expressed in bladder cancer by Linden et al 16]) who also reported fibrinogen B, apolipoprotein E, alpha-2-macroglobulin, alpha-1-antitrypsin, carbonic Anhydrase 1 as potential bladder cancer biomarkers. Confirmation of these findings was performed for the case of fibrinogen B and alpha-1-antitrypsin, using urine samples from bladder cancer patients and healthy controls and providing sensitivities of 77% and 66% and specificities of 77% and 85% respectively.
Based on the hypothesis that metal-binding proteins may be of substantial relevance in bladder cancer, Frantzi et al. [24] employed immobilized metal affinity chromatography in combination with high-resolution LC-MS/MS for the discovery of urinary peptides associated with bladder cancer in urine samples from patients with muscle invasive bladder cancer (MIBC), non-muscle invasive bladder cancer (NMIBC), and benign controls. The differential expression of two candidate biomarkers, histone H2B and Nuclear interacting factor 1/Zinc finger 335 (NIF-1) were verified by ELISA in urine and immunohistochemistry (IHC) in tissue, in a total of 166 urine samples from bladder cancer patients and benign urological diseases and 32 bladder cancer tissue specimens respectively, collectively suggesting an association of the proteins with tumor stage.
Tyan et al. [25] investigated urine samples from bladder cancer patients and identified several abundant proteins, including human Disintegrin and metalloproteinase domain-containing protein 28 (ADAM28). Further analysis using WB and ELISA assays indicated that ADAM 28 was indeed increased in urine of bladder cancer patients, in comparison to controls [25].
Based on the assumption that bladder cancer-specific biomarkers should also be reflected by a change in gene expression, Shimwell et al. [26] combined urinary proteomics with transcriptome analysis. This elegant approach resulted in the identification and subsequent validation of Midkine and Hepatocyte growth factor activator inhibitor type 1 (HAI-1) by ELISA assays in 642 bladder cancer patients. The performance in discriminating between bladder cancer and healthy controls, increased as bladder cancer progresses to muscle invasive disease, with AUC values for Midkine of: 0.574 for Ta vs controls, 0.708 for T1 vs controls and 0.851 for T2+ vs controls. The same observation was made for HAI-1 with AUC values of 0.566, 0.748 and 0.886, respectively.
Similarly to Zoidakis et al. [23], Taylor et al. [27], investigated the association of urinary aminopeptidases with bladder cancer using activity assays. In a small set of 16 bladder cancer patients, the authors could identify significant change in the activities of five aminopeptidases: aminopeptidase N (ANPEP), glutamyl aminopeptidase (ENPEP), dipeptidyl peptidase 4 (DPP4), dipeptidyl peptidase 7 (DPP7), and dipeptidyl peptidase 1 (CTSC), in comparison to healthy controls.
Several additional reports have been published on single biomarkers, awaiting further larger scale independent validation. Orenes-Pinero reported the identification of Reg1-1 as biomarker for bladder cancer [28]. Kawanishi and colleagues identified growth-regulated alpha protein (CXCL1) as being upregulated in the T24 cell line [29]. Investigating further the distribution of CXCL1 in urine from bladder cancer patients and controls, the authors could confirm the finding, and suggested CXCL1 as potential biomarker for the disease [29]. Employing immunodepletion, Chen et al. [30] identified several apolipoproteins as positively associated with bladder cancer, confirming previous results (summarized in Table 2). The authors further identified serum amyloid A-4 protein (SAA4) as significantly increased in urine of bladder cancer patients, while pro-epidermal growth factor (ProEGF) was significantly reduced. The same group also analysed proteins of urinary microparticles from patients with bladder cancer and patients with hernia as controls by LC-MS/MS from which they highlighted calcium-signal transducer 2 (TACSTD2) as potential bladder cancer diagnostic marker, following confirmation of the LC-MS results by ELISA in a total of 221 urine samples [31]. Tan et al, identified serine/threonine-protein kinase PLK2 (PLK2) as upregulated and its differential expression was verified in a small cohort [32]. Tsui et al. [33] demonstrated significant reduction of bikunin, and pro-urolinase-plasminogen activator associated with bladder cancer. Urinary vitamin D-binding protein (also termed gc-globulin) was identified among other proteins as being significantly increased in bladder cancer, and confirmed in a small validation cohort [19]. Lu et al. [34] investigated urine of patients with upper urinary tract bladder cancer and controls and identified several proteins as significantly changing in abundance; among those, the authors chose zinc-alpha-2-glycoprotein, calreticulin, annexin A2, annexin A3 and haptoglobin and verified their increased abundance in independent urine samples as well as tissue [34]. Urquidi et al [35] evaluated the performance of interleukin 8 (IL-8), matrix metallopeptidase 9 (MMP-9) and syndecan-1 (SDC1) in a set of 127 urine samples (corresponding to 64 cancer cases versus 63 benign controls) by ELISA supporting increased levels of all proteins in the former with a slightly better diagnostic performance for IL-8 with reported AUC value of 0.790) [35].
Based on previous results in plasma, Ebbing et al. [36] developed the hypothesis that calprotectin, the heterodimer of S100A8/S100A9, may be upregulated in bladder cancer, and constitute a potential urinary biomarker. The authors investigated urinary calprotectin in 181 samples from patients with bladder cancer, renal and prostate cancers, and healthy controls, using ELISA. A significant increase of calprotectin in bladder cancer could be detected, presenting with an AUC value of 0.880 (sensitivity of 81%, specificity of 93%). Recently, Kumar et al. [37] reported the identification of 5 urinary proteins, coronin-1A, apolipoprotein A4, semenogelin-2, gamma synuclein and protein deglycase DJ-1 (DJ-1/PARK7) (), all significantly upregulated in the urine of bladder cancer patients. An assay combining these five biomarkers showed an AUC of 0.920 nevertheless its independent validation is pending.
Collectively, in the vast majority of the aforementioned proteomic discovery studies an over-representation of bladder cancer and use of healthy or benign controls can be generally observed. A main consensus appears to be that combination of markers to multi-parametric classifiers in general increases accuracy in disease detection. Besides the aforementioned peptide classifiers [11–13], multiple additional groups have combined findings into panels (summarized in Table 1): Goodison et al. [38] in 2012 reported a combination of 8 proteins: IL-8, MMP-9, plasminogen activator inhibitor 1 (PAI-1), vascular endothelial growth factor A (VEGF), angiogenin (ANG), carbonic anydrase-9 (CA-9), apolipoprotein E (APOE), matrix metallopeptidase 10 (MMP-10) measured by ELISA, in a case-control study involving 127 urine samples, and comparing bladder cancer and healthy controls, showing an AUC of 0.980 (sensitivity of 92% and specificity of 97%); these markers were subsequently also tested in another set of 102 samples from patients with bladder cancer and 206 with various urological disorders, providing BC detection with 74% sensitivity and 90% specificity [39]. The same group had also tested combinations of C-C motif chemokine 18 (CCL18), plasminogen activator inhibitor 1 (PAI-1) and CD44 antigen [40], and more recently [41] combinations of ten proteins (IL8, MMP9, MMP10, serpin family A member 1 (SERPINA1)), VEGFA, ANG, CA9, APOE, Serpin Family E Member 1 (SERPINE1) and SDC1, to classify patients under surveillance prior to cystoscopy [41]. The latter study involving the analysis of 125 subjects (53 recurrent cases and 72 negative for tumor relapse) supported that the panel could detect recurrent disease with a sensitivity of 79% and specificity of 88% [41]. Similar analysis of a similar panel (except for including A1AT and PAI1 instead of SERPINA1 and SERPINE1) in the form of a multiplex assay on 200 samples collected prior to cystoscopy, supported an overall diagnostic sensitivity of 85%, with a specificity of 81% [42]. Recently, the panel was evaluated in combination to demographic features in a set of 686 subjects (including 394 with bladder cancer) corresponding to an AUC of 0.891 (95% CI, 0.86–0.92) for the combined (biomarker-demographic) model [43].
In a parallel approach, Soukap et al. [44] recently assessed a panel of 27 biomarkers, neuron specific enolase (NSE), attractin (ATT), adipocyte fatty acid-binding protein (AFABP), resistin, midkine, clusterin, uromodulin, zinc-alpha 2 glycoprotein (ZAG2), heat shock protein beta-1 (HSP27), 60 kDa heat shock protein, mitochondrial (HSP60), neural cell adhesion molecule 1 (NCAM1/CD56), (angiogenin, calreticulin, chromogranin A, carcinoembryonic antigen related cell adhesion molecule 1 (CEACAM1), chemokine (C-X-C Motif) ligand 1 (CXCL1), interleukin-13 receptor subunit alpha-2 (IL13Ra2), progranulin, vascular endothelial growth factor A (VEGFA), carbonic anhydrase IX, annexin-V, T cell immunoglobulin mucin 4 (TIM4), galectin-1, cystatin B, synuclein G, ApoA1 and ApoA2) by ELISA or by electrochemiluminiscence immunoassay in 70 patients with bladder cancer and 49 healthy control subjects. Cytology combined with midkine and synuclein G showed superior performance in this cohort providing sensitivity of 91.8% and specificity 97.5% in bladder cancer detection. The authors also investigated the value of biomarkers when monitoring for recurrence and found that a combination of cytology and erythrocytes count in urine sediment with midkine, ZAG2, CEACAM1, and synuclein G results in bladder cancer detection with 92.7%, sensitivity and 90.2% specificity in a cohort of 44 cases and 61 controls [44].
Collectively these studies advocate the diagnostic added value of using marker combinations (in comparison to individual markers) nevertheless, still in their vast majority their clinical value is compromised by an over-representation of bladder cancer cases and use of healthy or benign urological diseases as controls (instead of clinically relevant patients with hematuria and/or patients under surveillance), likely resulting in over-estimates of accuracy rates. The next logical step is the assessment of ideally all potential biomarkers in a properly powered prospective study in a representative cohort, as advocated in several articles by now [1, 4, 45, 46].
SECRETED PROTEINS
Based on the hypothesis that urinary biomarkers for bladder cancer would be secreted from the tumor, the secretome of tumor cells has also been investigated using proteomics approaches. One of the first efforts in this direction was reported by Lin et al. [47]. The authors investigated the secretome of malignant U1 and pre-malignant U4 cells, and identified multiple differences, including Laminin alpha-5 chain, ADP-ribosylation factor guanine nucleotide-exchange factor 2 and urokinase-type plasminogen activator (u-PA). The latter (u-PA) was further investigated and data were presented indicating that loss of u-PA is associated with malignant transformation [47].
Comparing the secretome from T24 and the aggressive T24M variant, Makridakis et al. [48] identified several proteins apparently associated with the transformation towards metastatic tumor, including, secreted protein acidic and cysteine rich (SPARC), tissue-type plasminogen activator (tPA), and clusterin, all of which were further validated by WB [48]. The increased levels of SPARC in urine of bladder cancer patients in association to tumor stage were further recently shown (Critselis et al, submitted). In addition, a biological relevance of SPARC in bladder cancer was suggested as blocking SPARC with specific antibodies resulted in decreased cell motility in vitro.
Besides these in vitro studies, secreted proteins in the form of exosomes have also been studied in vivo. Welton et al. [49] described isolation of exosomes from the bladder cancer HT1376 cells, and demonstrated feasibility of isolating exosomes from urine from bladder cancer patients. This concept was further developed by Chen et al. [31] who analyzed urinary microparticles from bladder cancer and hernia patients highlighting calcium-signal transducer 2 as a potential marker (described above). While exosomes may hold information on bladder cancer, they do not appear to be amendable for routine diagnosis, a result of the quite tedious isolation procedure which may also be reflected in the overall lack of overlap between findings of existing studies.
TISSUE PROTEOMICS
Several studies have been reported, where initial findings from urine analysis using proteomics were further verified at the tissue level. These include the aforementioned studies highlighting associations of profilin-1 [23], NIF-1 and H2B [24] as well as studies on S100 proteins found upregulated in urine of bladder cancer patients by several groups, but also in tissue [50].
Using a more bona-fide tissue proteomics approach, Peng et al. [51] investigated bladder cancer and adjacent tissue by two-dimensional electrophoresis (2DE) and identified phosphoglycerate mutase 1 (PGAM1), significantly up-regulated in cancer cells. Using IHC in 60 bladder cancer (30 high and 30 low grade) and 30 adjacent normal urothelium tissue specimens, the authors demonstrated a significant correlation of PGAM1 expression with the severity of histological grade. Knockdown of PGAM1 expression by RNAi suggested antitumor activity in vivo, which led to the hypothesis that PGAM1 might serve as a promising therapeutic target for bladder cancer [51]. To obtain insight into molecular pathophysiology, differential proteome analysis between grade II and III bladder cancer was also carried out in [52]. Several proteins, including galectin-1, annexin 1, annexin V were found at increased abundance in grade III cells, while fatty acid binding proteins, heat-shock protein 27, 15-hydroxyprostaglandin dehydrogenase, lysophospholipase, mitochondrial short-chain enoyl-coenzyme A hydratase 1 precursor and L-lactate dehydrogenase (LDH) were identified as being downregulated in the advanced grade III samples. Independent validation of these results is pending.
In a parallel approach, the prognostic potential of cystatin B at the tissue level has been suggested, as increased expression levels of the protein were correlated with stage (p = 0.0047) and grade (p = 0.062), time to disease recurrence (p = 0.0047) and progression (p = 0.007) [53]. These results are supported by the finding of increased cystatin secretion by the metastatic T24M cells in comparison to non-metastatic variants (48), and by an observed increase of cystatin in urine of bladder cancer patients in comparison to controls (53).
Barboro et al. [54] investigated invasive bladder cancer tissue, aiming at identifying prognostic biomarkers. By comparing muscle invasive bladder cancer tissue specimens (n = 21) with normal and non-tumor sections (n = 3 and n = 9, respectively) by 2DE, the authors could demonstrate significant upregulation of lamin B1 and fibrinogen beta chain in the former, and reduction of actin, desmin and vimentin. Further, the authors found a significant correlation of the protein p54 (nrb) with vascular invasion (0.645, pearson correlation; p = 0.002) and survival.
Moreira et al. [55] investigated the bladder cancer prognostic value of bladder cancer-associated protein (BLCAP), earlier described by the group, in 2.108 tissue specimens, retrieved from archival datasets. Based on the protein expression levels and cell localization, cancer cells could be grouped into 4 categories with BLCAP decreased staining intensity correlating with tumor grade (p < 0.0001) and stage (p < 0.0001). Orenes-Pinero et al. employed protein arrays to identify serum proteins associated with BC [28]. Two of the identified candidate biomarkers, dynamin and clusterin, were further evaluated using IHC in tissue arrays. Reduced clusterin was found to be associated with muscle invasive bladder cancer whereas reduced dynamin was in addition associated with adverse outcome [28].
Chung et al. [56] identified 12 proteins differentially expressed in muscle versus non-muscle invasive bladder cancer. Of these, the upregulation of cofilin was further validated and its potential biological function was investigated in T24 cells. The results suggested a role of the protein in epidermal growth factor (EGF)-induced migration, involving cofilin phosphorylation following epidermal growth factor administration [56].
Using Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) labelling, Grau et al. [57] performed comparative proteomics on the T24 bladder cancer cell line and its aggressive derivate, T24T. The authors identified cullin-3 (Cul3), a protein involved in ubiquitination as overexpressed in T24T. Silencing of this protein attenuated proliferation and migration of T24T. Along the same lines, IHC of Cul3 in tissue microarrays demonstrated association of its expression levels with tumor stage, metastasis and disease-specific survival [57].
Shrinivasan et al. [58] employed antibody microarrays to identify proteins differentially expressed between patients with and without local recurrence. The group identified 255 proteins with significantly differential abundance between the two groups (19 tumor samples from patients with and 6 samples from patients without recurrence). Among the most significant were an upregulation of prelamin-A/C (LMNA), transcription factor AP-1 (JUN), and nuclease-sensitive element-binding protein 1 (YBOX1) and downregulation of L-selectin (LYAM1), cyclin-dependent kinase inhibitor 1 (CDN1A) and mothers against decapentaplegic homolog 3 (SMAD3). A classifier based on 20 proteins could predict recurrence with 91% accuracy [58]. Based on reports in breast cancer, Hemdan et al. [59] investigated stathmin 1 in the context of bladder cancer and found increased expression in tissue associated with unfavourable outcome. Attenuation of stathmin 1 expression reduced growth and migration of the T24 cells [59].
Chen et al. [60] employed laser microdissection to compare the proteome of bladder cancer and adjacent non-tumorous tissue. Three proteins, 4F2 cell-surface antigen heavy chain (SLC3A2), stathmin (STMN1) and transgelin-2 (TAGLN2), could be further verified as upregulated in cancer cells using IHC. Both, STMN1 and TAGLN2 were also found significantly increased in urine of bladder cancer patients [60].
Wu et al. [61] investigated the correlation of galectin-1 overexpression (previously identified by the group) with bladder cancer pathology and prognosis using IHC of a set of 185 primary cases. Galectin-1increased expression was significantly correlated with tumor grade (p = 0.037), vascular invasion (p = 0.02), nodal status (p = 0.0012), and significantly predicted disease specific survival (p = 0.0002) [61].
PLASMA PROTEOMICS
In the case of bladder cancer, proteomic studies using blood are scarce in comparison to urine analysis. Besides the aforementioned study of Orenes-Pinero et al. [28], Bansal et al. [62] investigated serum from low-grade (n = 33) and high-grade (n = 32) bladder cancer and healthy controls (n = 25) using 2DE. The authors identified five differentially expressed proteins, two of which, S100A8 and S100A9, distinguishing bladder cancer (low and high grade) from healthy controls with an AUC of 0.946 [62]. In a later study, the same authors confirmed these initial findings investigating 160 sera samples from 52 healthy controls, 55 pre-operative, and 53 post-operative bladder cancer patients [63]. Using ELISA the reduced abundance of S100A8, S100A9, S100A4, and CA-1 could be shown; concomitantly, annexin V was progressively and significantly increased in post-operative compared to pre-operative bladder cancer sera samples [63].
CONCLUSION
Evidently, multiple proteomics studies aiming at discovering biomarkers for detection of bladder cancer and disease prognosis have been performed the past ten years, and a huge number of potential biomarkers for these two contexts of use have been reported. It is interesting to note that proteomics studies have not been applied towards the investigation of predictive markers; likely attributed to the incompatibility between number of tested variables and available sample sizes, becoming particularly pronounced when investigating treatment response. Instead, prognostic associations are more commonly addressed, also reflected in the presented studies. The predictive value of alpha 1-acid glycoprotein for outcome of patients with metastatic cancer, following treatment with Docetaxel -based chemotherapy, is currently being addressed in a clinical trial (NCT01814150), as well as of various cytokines as predictors of response to intravesical therapy (NCT01007058). It is noteworthy that DNA/RNA-based markers are currently more commonly been investigated in clinical trials [64, 65] in comparison to protein markers, to a good extent attributed to difficulties in protein assay development and lack of enrichment strategies for proteins (similar to PCR); however it is expected that this will gradually change following advancements in MS-based multiplex targeted quantification strategies [5, 9].
Unfortunately the experimental design in many of the reported biomarker discovery studies, involving healthy controls, is suboptimal; nevertheless, efforts towards designing clinically relevant validation studies (including high risk groups at primary diagnosis and groups under surveillance) can also be observed (i.e. [13, 41]. Variability in protein findings between individual studies are evident, to some extent reflecting the plethora of applied experimental approaches and complexity of the proteome, without ruling out the possibility of false associations due to suboptimal power of individual studies. Nevertheless, reproducible findings reported in various independent studies also exist (summarized in Table 2; several members of the apolipoprotein family, fibrinogen chains, alpha-1-antitrypsin, alpha-2-macroglobulin, carbonic anhydrase 1, profilin-1, uromodulin, basement membrane-specific heparan sulfate proteoglycan), hence representing markers of higher existing evidence. With no doubt, a major concern is that most of these biomarkers may be reflective of hematuria and are also highly significantly associated with kidney disease (e.g. apolipoproteins, uromodulin, or alpha-1-antitrypsin). Nevertheless, their incorporation into panels of biomarkers appears to be of value (as shown in Table 1), as clearly these proteins are changing as a result of the disease. Two main avenues of research can now be foreseen: on one hand, target integration of the existing disparate data towards a better understanding of disease pathophysiology. Initial efforts in this direction become evident [66–68], including generation of bladder cancer-specific databases compiling existing findings [68]. Special emphasis should be given in placing these findings in the context (and thus refining) existing knowledge on bladder cancer subtypes [69, 70]. Currently, this is clearly missing in the vast majority of reported proteomics studies, especially those investigating urine biomarkers, in part due to the applied context of use (study of non invasive disease). In the case of tissue markers, such correlations to other molecular data were investigated, in a relatively well powered study, in the case of BLCAP [55] where expression of various proteins involved in differentiation (uroplakin and 15-prostaglandin dehydrogenase) as well as in oncogenic pathways (p53, c-Jun, and ErbB2) was investigated. Such coordinated studies placing biomarker data in the context of existing phenotypes are needed in order to refine current bladder cancer phenotypes, but also attribute biological significance to urinary findings and potentially guide the development of non invasive prognostic/predictive tests. A centralized deposit of verified biomarker findings following manuscript acceptance could assist in this direction of organizing, at least from now on, new findings and facilitating future integrative analyses. At the same time and in parallel, accumulation of clinical samples to support proper validation studies is urgently needed. Such a resource would substantially support investigation of diagnostic, prognostic as well as predictive contexts of use, the latter also including complications from treatment, such as thromboembolism, not addressed by now. Any trial design should allow for investigating the added value of the biomarker, which in the case of bladder cancer may correspond to a decrease in the number of cystoscopies (for low risk disease), increase its accuracy rates, as well as guiding treatment selection. Existing guidelines for reporting biomarker discovery data as well as adoption of the REMARK criteria [71] are important safeguards ensuring that biomarker research is conducted in an implementation – focused manner.
Establishing the needed clinical resources becomes increasingly challenging, requiring among others, establishment of a fine balance between the ever-increasing demands on ethical issues and at the same time the ethical and moral obligation to maximize and make best use of available clinical resources in order to accelerate improvement in patient management. In this context, the bladder cancer community could gain by following examples of existing centralized resources of well characterized clinical samples, available to support biomarker research in the context of other diseases (for example: the resources and mechanisms established by the National Institute of Diabetes and Digestive and Kidney Diseases; https://www.niddkrepository.org/home/). Promising in this direction are two currently active clinical trials (NCT02379429; Care of the Urothelial Cancer Patient and Prospective Procurement of Urothelial Cancer Tissue; NCT02012699; Cancer Research Repository for Individuals With Cancer Diagnosis, High Risk Individuals, and Individuals With No History of Cancer (Control) (iCaRe2)) targeting generation of biorepositories to support biomarker and in general molecular studies in bladder cancer. It seems a competitive application procedure for use of such samples, based on existing evidence (supporting biomarker data), sample (volume) requirements, as well as assay availability including platform analytical validation data, would be the most appropriate approach towards making optimal use of this promising resource. Such resources, in combination to the adoption of an open minded collaborative approach between investigators could clearly be a major step forward, organizing research in bladder cancer and catalysing multi-parametric validation studies and subsequent steps of biomarker implementation. Easier said than done but clearly feasible.
CONFLICT OF INTEREST
Maria Frantzi is employed by Mosaiques Diagnostics that established the CE-MS clinical proteomics platform.
ACKNOWLEDGMENTS
Supported by funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreements no. 601933 (TransBioBC).
REFERENCES
[1] | Schmitz-Dräger BJ , Droller M , Lokeshwar VB , Lotan Y , Hudson MA , van Rhijn BW , Marberger MJ , Fradet Y , Hemstreet GP , Malmstrom PU , Ogawa O , Karakiewicz PI , Shariat SF . Molecular markers for bladder cancer screening, early diagnosis, and surveillance: The WHO/ICUD consensus. Urol Int (2015) ;94: (1):1–24. |
[2] | Kamat AM , Hahn NM , Efstathiou JA , Lerner SP , Malmström PU , Choi W , Guo CC , Lotan Y , Kassouf W . Bladder cancer. Lancet (2016) . pii: S0140-6736(16)30512-8.. |
[3] | van Rhijn BW , Catto JW , Goebell PJ , Knüchel R , Shariat SF , van der Poel HG , Sanchez-Carbayo M , Thalmann GN , Schmitz-Dräger BJ , Kiemeney LA . Molecular markers for urothelial bladder cancer prognosis: Toward implementation in clinical practice. Urol Oncol (2014) ;32: (7):1078–87. |
[4] | Lotan Y , Shariat SF , Schmitz-Dräger BJ , Sanchez-Carbayo M , Jankevicius F , Racioppi M , Minner SJ , Stöhr B , Bassi PF , Grossman HB . Considerations on implementing diagnostic markers into clinical decision making in bladder cancer. Urol Oncol (2010) ;28: (4):441–8. |
[5] | Frantzi M , Latosinska A , Fluhe L , Hupe MC , Critselis E , Kramer MW , et al. Developing proteomic biomarkers for bladder cancer: Towards clinical application. Nature Reviews Urology (2015) ;12: (6):317–30. |
[6] | D’Costa JJ , Goldsmith JC , Wilson JS , Bryan RT , Ward DG . A systematic review of the diagnostic and prognostic value of urinary protein biomarkers in urothelial bladder cancer. Bl Cancer (2016) ;2: (3):301–17. |
[7] | Santucci L , Bruschi M , Candiano G , Lugani F , Petretto A , Bonanni A , Ghiggeri GM . Urine Proteome Biomarkers in Kidney Diseases. I. Limits, Perspectives, and First Focus on Normal Urine Biomark Insights (2016) ;11: :41–8. |
[8] | Mischak H , Kolch W , Aivaliotis M , Bouyssie D , Court M , Dihazi H , et al. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteomics Clinical Applications (2010) ;4: (4):464–78. |
[9] | Mischak H , Critselis E , Hanash S , Gallagher WM , Vlahou A , Ioannidis JP . Epidemiologic design and analysis for proteomic studies: A primer on -omic technologies. Am J Epidemiol (2015) ;181: (9):635–47. |
[10] | Filip S , Zoidakis J , Vlahou A , Mischak H . Advances in urinary proteome analysis and applications in systems biology. Bioanalysis (2014) ;6: (19):2549–69. |
[11] | Theodorescu D , Wittke S , Ross MM , Walden M , Conaway M , Just I , et al. Discovery and validation of new protein biomarkers for urothelial cancer: A prospective analysis. The Lancet Oncology (2006) ;7: (3):230–40. |
[12] | Schiffer E , Vlahou A , Petrolekas A , Stravodimos K , Tauber R , Geschwend JE , et al. Prediction of muscle-invasive bladder cancer using urinary proteomics. Clinical Cancer Research (2009) ;15: (15):4935–43. |
[13] | Frantzi M , van Kessel KE , Zwarthoff EC , Marquez M , Rava M , Malats N , et al. Development and Validation of Urine-based Peptide Biomarker Panels for Detecting Bladder Cancer in a Multi-center Study. Clinical Cancer Research (2016) ;22: (16):4077–86. |
[14] | Kreunin P , Zhao J , Rosser C , Urquidi V , Lubman DM , Goodison S . Bladder cancer associated glycoprotein signatures revealed by urinary proteomic profiling. Journal of Proteome Research (2007) ;6: (7):2631–9. |
[15] | Yang N , Feng S , Shedden K , Xie X , Liu Y , Rosser CJ , et al. Urinary glycoprotein biomarker discovery for bladder cancer detection using LC/MS-MS and label-free quantification. Clinical Cancer Research (2011) ;17: (10):3349–59. |
[16] | Linden M , Lind SB , Mayrhofer C , Segersten U , Wester K , Lyutvinskiy Y , et al. Proteomic analysis of urinary biomarker candidates for nonmuscle invasive bladder cancer. Proteomics (2012) ;12: (1):135–44. |
[17] | Lei T , Zhao X , Jin S , Meng Q , Zhou H , Zhang M . Discovery of potential bladder cancer biomarkers by comparative urine proteomics and analysis. Clinical Genitourinary Cancer (2013) ;11: (1):56–62. |
[18] | Li H , Li C , Wu H , Zhang T , Wang J , Wang S , et al. Identification of Apo-A1 as a biomarker for early diagnosis of bladder transitional cell carcinoma. Proteome science (2011) ;9: (1):21. |
[19] | Li F , Chen DN , He CW , Zhou Y , Olkkonen VM , He N , et al. Identification of urinary Gc-globulin as a novel biomarker for bladder cancer by two-dimensional fluorescent differential gel electrophoresis (2D-DIGE). Journal of Proteomics (2012) ;77: :225–36. |
[20] | Li C , Li H , Zhang T , Li J , Liu L , Chang J . Discovery of Apo-A1 as a potential bladder cancer biomarker by urine proteomics and analysis. Biochemical and Biophysical Research Communications (2014) ;446: (4):1047–52. |
[21] | Chen YT , Chen CL , Chen HW , Chung T , Wu CC , Chen CD , et al. Discovery of novel bladder cancer biomarkers by comparative urine proteomics using iTRAQ technology. Journal of Proteome Research (2010) ;9: (11):5803–15. |
[22] | Chen YT , Chen HW , Domanski D , Smith DS , Liang KH , Wu CC , et al. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. Journal of Proteomics (2012) ;75: (12):3529–45. |
[23] | Zoidakis J , Makridakis M , Zerefos PG , Bitsika V , Esteban S , Frantzi M , et al. Profilin 1 is a potential biomarker for bladder cancer aggressiveness. Molecular & Cellular Proteomics: MCP (2012) ;11: (4):M111 009449. |
[24] | Frantzi M , Zoidakis J , Papadopoulos T , Zurbig P , Katafigiotis I , Stravodimos K , et al. IMAC fractionation in combination with LC-MS reveals H2B and NIF-1 peptides as potential bladder cancer biomarkers. Journal of Proteome Research (2013) ;12: (9):3969–79. |
[25] | Tyan YC , Yang MH , Chen SC , Jong SB , Chen WC , Yang YH , et al. Urinary protein profiling by liquid chromatography/tandem mass spectrometry: ADAM28 is overexpressed in bladder transitional cell carcinoma. Rapid Communications in Mass Spectrometry: RCM (2011) ;25: (19):2851–62. |
[26] | Shimwell NJ , Bryan RT , Wei W , James ND , Cheng KK , Zeegers MP , et al. Combined proteome and transcriptome analyses for the discovery of urinary biomarkers for urothelial carcinoma. British Journal of Cancer (2013) ;108: (9):1854–61. |
[27] | Taylor JM , Yaneva M , Velasco K , Philip J , Erdjument-Bromage H , Ostrovnaya I , et al. Aminopeptidase activities as prospective urinary biomarkers for bladder cancer. Proteomics Clinical Applications (2014) ;8: (5-6):317–26. |
[28] | Orenes-Pinero E , Barderas R , Rico D , Casal JI , Gonzalez-Pisano D , Navajo J , et al. Serum and tissue profiling in bladder cancer combining protein and tissue arrays. Journal of Proteome Research (2010) ;9: (1):164–73. |
[29] | Kawanishi H , Matsui Y , Ito M , Watanabe J , Takahashi T , Nishizawa K , et al. Secreted CXCL1 is a potential mediator and marker of the tumor invasion of bladder cancer. Clinical Cancer Research (2008) ;14: (9):2579–87. |
[30] | Chen CL , Lin TS , Tsai CH , Wu CC , Chung T , Chien KY , et al. Identification of potential bladder cancer markers in urine by abundant-protein depletion coupled with quantitative proteomics. Journal of Proteomics (2013) ;85: :28–43. |
[31] | Chen CL , Lai YF , Tang P , Chien KY , Yu JS , Tsai CH , et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. Journal of Proteome Research (2012) ;11: (12):5611–29. |
[32] | Tan LB , Chen KT , Yuan YC , Liao PC , Guo HR . Identification of urine PLK2 as a marker of bladder tumors by proteomic analysis. World Journal of Urology (2010) ;28: (1):117–22. |
[33] | Tsui KH , Tang P , Lin CY , Chang PL , Chang CH , Yung BY . Bikunin loss in urine as useful marker for bladder carcinoma. The Journal of Urology (2010) ;183: (1):339–44. |
[34] | Lu CM , Lin JJ , Huang HH , Ko YC , Hsu JL , Chen JC , et al. A panel of tumor markers, calreticulin, annexin A2, and annexin A3 in upper tract urothelial carcinoma identified by proteomic and immunological analysis. BMC Cancer (2014) ;14: :363. |
[35] | Urquidi V , Chang M , Dai Y , Kim J , Wolfson ED , Goodison S , et al. IL-8 as a urinary biomarker for the detection of bladder cancer. BMC Urology (2012) ;12: :12. |
[36] | Ebbing J , Mathia S , Seibert FS , Pagonas N , Bauer F , Erber B , et al. Urinary calprotectin: A new diagnostic marker in urothelial carcinoma of the bladder. World Journal of Urology (2014) ;32: (6):1485–92. |
[37] | Kumar P , Nandi S , Tan TZ , Ler SG , Chia KS , Lim WY , et al. Highly sensitive and specific novel biomarkers for the diagnosis of transitional bladder carcinoma. Oncotarget (2015) ;6: (15):13539–49. |
[38] | Goodison S , Chang M , Dai Y , Urquidi V , Rosser CJ . A multi-analyte assay for the non-invasive detection of bladder cancer. PLoS One (2012) ;7: (10):e47469. |
[39] | Rosser CJ , Ross S , Chang M , Dai Y , Mengual L , Zhang G , et al. Multiplex protein signature for the detection of bladder cancer in voided urine samples. The Journal of Urology (2013) ;190: (6):2257–62. |
[40] | Urquidi V , Kim J , Chang M , Dai Y , Rosser CJ , Goodison S . CCL18 in a multiplex urine-based assay for the detection of bladder cancer. PLoS One (2012) ;7: (5):e37797. |
[41] | Rosser CJ , Chang M , Dai Y , Ross S , Mengual L , Alcaraz A , et al. Urinary protein biomarker panel for the detection of recurrent bladder cancer. Cancer Epidemiology, Biomarkers & Prevention (2014) ;23: (7):1340–5. |
[42] | Shimizu Y , Furuya H , Greenwood PB , Chan O , Dai YF , Thornquist MD , Goodison S , Rosser CJ . A multiplex immunoassay for the non-invasive detection of bladder cancer. Journal of Translational Medicine (2016) )14–31. |
[43] | Huang S , Kou L , Furuya H , Yu C , Goodison S , Kattan MW , et al. A nomogram derived by combination of demographic and biomarker data improves the non-invasive evaluation of patients at risk for bladder cancer. Cancer Epidemiology, Biomarkers & Prevention (2016) ;25: (9):1361–6. |
[44] | Soukup V , Kalousova M , Capoun O , Sobotka R , Breyl Z , Pesl M , et al. Panel of urinary diagnostic markers for non-invasive detection of primary and recurrent urothelial urinary bladder carcinoma. Urologia Internationalis (2015) ;95: (1):56–64. |
[45] | Vlahou A . Back to the future in bladder cancer research. Expert Review of Proteomics (2011) ;8: (3):295–7. |
[46] | Mischak H , Ioannidis JP , Argiles A , Attwood TK , Bongcam-Rudloff E , Broenstrup M , et al. Implementation of proteomic biomarkers: Making it work. European Journal of Clinical Investigation (2012) ;42: (9):1027–36. |
[47] | Lin CY , Tsui KH , Yu CC , Yeh CW , Chang PL , Yung BY . Searching cell-secreted proteomes for potential urinary bladder tumor markers. Proteomics (2006) ;6: (15):4381–9. |
[48] | Makridakis M , Roubelakis MG , Bitsika V , Dimuccio V , Samiotaki M , Kossida S , et al. Analysis of secreted proteins for the study of bladder cancer cell aggressiveness. Journal of Proteome Research (2010) . 4;9: (6):3243–59. |
[49] | Welton JL , Khanna S , Giles PJ , Brennan P , Brewis IA , Staffurth J , et al. Proteomics analysis of bladder cancer exosomes. Molecular & Cellular Proteomics (2010) ;9: (6):1324–38. |
[50] | Tolson JP , Flad T , Gnau V , Dihazi H , Hennenlotter J , Beck A , et al. Differential detection of S100A8 in transitional cell carcinoma of the bladder by pair wise tissue proteomic and immunohistochemical analysis. Proteomics (2006) ;6: (2):697–708. |
[51] | Peng XC , Gong FM , Chen Y , Qiu M , Cheng K , Tang J , et al. Proteomics identification of PGAM1 as a potential therapeutic target for urothelial bladder cancer. Journal of Proteomics (2016) ;132: :85–92. |
[52] | Sheng KH , Yao YC , Chuang SS , Wu H , Wu TF . Search for the tumor-related proteins of transition cell carcinoma in Taiwan by proteomic analysis. Proteomics (2006) ;6: (3):1058–65. |
[53] | Feldman AS , Banyard J , Wu CL , McDougal WS , Zetter BR . Cystatin B as a tissue and urinary biomarker of bladder cancer recurrence and disease progression. Clinical Cancer Research (2009) ;15: (3):1024–31. |
[54] | Barboro P , Rubagotti A , Orecchia P , Spina B , Truini M , Repaci E , et al. Differential proteomic analysis of nuclear matrix in muscle-invasive bladder cancer: Potential to improve diagnosis and prognosis. Cellular Oncology (2008) ;30: (1):13–26. |
[55] | Moreira JM , Ohlsson G , Gromov P , Simon R , Sauter G , Celis JE , et al. Bladder cancer-associated protein, a potential prognostic biomarker in human bladder cancer. Molecular & Cellular Proteomics (2010) ;9: (1):161–77. |
[56] | Chung H , Kim B , Jung SH , Won KJ , Jiang X , Lee CK , et al. Does phosphorylation of cofilin affect the progression of human bladder cancer? BMC Cancer (2013) ;13: :45. |
[57] | Grau L , Luque-Garcia JL , Gonzalez-Peramato P , Theodorescu D , Palou J , Fernandez-Gomez JM , et al. A quantitative proteomic analysis uncovers the relevance of CUL3 in bladder cancer aggressiveness. PLoS One (2013) ;8: (1):e53328. |
[58] | Srinivasan H , Allory Y , Sill M , Vordos D , Alhamdani MS , Radvanyi F , et al. Prediction of recurrence of non muscle-invasive bladder cancer by means of a protein signature identified by antibody microarray analyses. Proteomics (2014) ;14: (11):1333–42. |
[59] | Hemdan T , Linden M , Lind SB , Namuduri AV , Sjostedt E , de Stahl TD , et al. The prognostic value and therapeutic target role of stathmin-1 in urinary bladder cancer. British Journal of Cancer (2014) ;111: (6):1180–7. |
[60] | Chen CL , Chung T , Wu CC , Ng KF , Yu JS , Tsai CH , et al. Comparative tissue proteomics of microdissected specimens reveals novel candidate biomarkers of bladder cancer. Molecular & Cellular Proteomics (2015) ;14: (9):2466–78. |
[61] | Wu TF , Li CF , Chien LH , Shen KH , Huang HY , Su CC , et al. Galectin-1 dysregulation independently predicts disease specific survival in bladder urothelial carcinoma. The Journal of Urology (2015) ;193: (3):1002–8. |
[62] | Bansal N , Gupta A , Sankhwar SN , Mahdi AA . Low- and high-grade bladder cancer appraisal via serum-based proteomics approach. Clinica Chimica Acta (2014) ;436: :97–103. |
[63] | Bansal N , Gupta AK , Gupta A , Sankhwar SN , Mahdi AA . Serum-based protein biomarkers of bladder cancer: A pre- and post-operative evaluation. Journal of Pharmaceutical and Biomedical Analysis (2016) ;124: :22–5. |
[64] | Li Q , Bagrodia A , Cha EK , Coleman JA . Prognostic genetic signatures in upper tract urothelial carcinoma Curr Urol Rep (2016) ;17: (2):12. |
[65] | Sweis RF , Galsky MD . Emerging role of immunotherapy in urothelial carcinoma— Immunobiology/biomarkers. Urol Onc (2016) ;34: :556–65. |
[66] | Feng J , He W , Song Y , Wang Y , Simpson RJ , Zhang X , et al. Platelet-derived growth factor receptor beta: A novel urinary biomarker for recurrence of non-muscle-invasive bladder cancer. PLoS One (2014) ;9: (5):e96671. |
[67] | Bhat A , Heinzel A , Mayer B , Perco P , Muhlberger I , Husi H , et al. Protein interactome of muscle invasive bladder cancer. PLoS One (2015) ;10: (1):e0116404. |
[68] | Bhat A , Mokou M , Zoidakis J , Jankowski V , Vlahou A , Mischak H . BcCluster: A bladder cancer database at the molecular level. Bl Cancer (2016) ;2: (1):65–76. |
[69] | Lerner SP , Robertson AG . Molecular Subtypes of Non-muscle Invasive Bladder Cancer. Cancer Cell (2016) ;30: (1):1–3. |
[70] | Lerner SP , McConkey DJ , Hoadley KA , Chan KS , Kim WY , Radvanyi F , Höglund M , Real FX . Bladder cancer molecular taxonomy: Summary from a consensus meeting. Bl Cancer (2016) ;2: (1):37–47. |
[71] | McShane LM , Altman DG , Sauerbrei W , Taube SE , Gion M , Clark GM . REporting recommendations for tumour MARKer prognostic studies (REMARK). British Journal of Cancer (2005) ;93: (4):387–91. |
Figures and Tables
Fig.1
Main applications of proteomics research reflected in manuscripts from the last ten years. Special emphasis has been placed on urine analysis as a source of diagnostic markers, as well as tissue for the identification of prognostic/predictive markers.
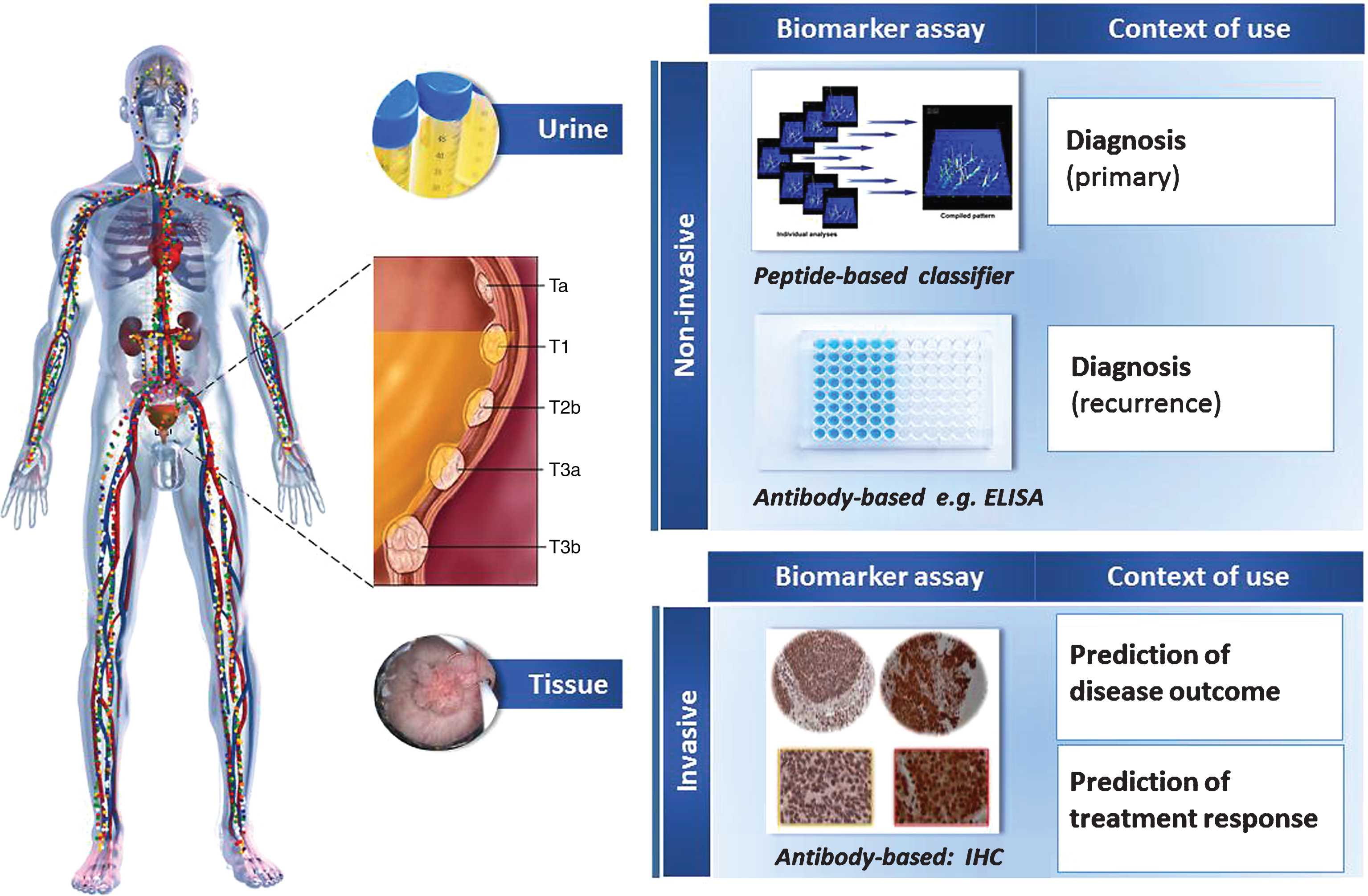
Table 1
Panels of protein biomarkers and their reported performance in bladder cancer detection
| Study | N Biomarkers | Proteins/ | Study Cohort | Number of | Clinical | Proteomics | AUC | Overall | Overall | PPV | NPV | Reference |
| Peptides | Characteristics | Subjects | Context | Technique | (95% CI) | Sensitivity | Specificity | (%) | (%) | Test | ||
| (%) | (%) | (sens/spec) | ||||||||||
| Rosser et al. 201441 | 10-biomarker panel | IL8, MMP9, MMP10, SERPINA1, VEGFA, ANG, CA9, APOE, SERPINE1, SDC1 | Retrospective, multi-center (n = 2) | n= 125; 53 BC recurrence/72 Negative for Recurrence/ | Prior BC history (recurrence) | ELISA | 0.904 (0.853–0.956) | 79 | 88 | 82 | 85 | Urovysion: 42% /94% |
| Prediction rule = 0.223(IL-8) –1.871(MMP9) + 2.239(SERPINA1) + 0.356(ANG) – 0.216(VEGFA) 0.330 (CA9) + 0.689(MMP10) + 6.889(APOE) – 0.408(SERPINE1) + 1.021(SDC1) – 0.141 | Urinary cytology: 33% /90% | |||||||||||
| Shimizu et al. 201642 | 10-biomarker panel | IL8, MMP9, MMP10, ANG, APOE, SDC1, A1AT, PAI1, CA9, VEGFA | Case-control | n = 262; Training Phase 29 BC, 33 benign/healthy controls | BC vs benign and healthy | Multiplex ELISA | 0.888 (0.842–0.934) | 85 | 81 | 82 | 84 | N/A |
| Validation Phase 100 BC, 100 benign/healthy controls | ||||||||||||
| Urquidi et al. 201240 | 3 Biomarkers | CCL18, PAI-1, CD44 | Case-control | n = 127; | BC vs healthy | ELISA | 0.938 (0.871–0.967) | 86 | 89 | 86 | 87 | BTA Trak: 80% /84% |
| 64 BC 63 healthy controls | ||||||||||||
| Urinary cytology: 28% /98% | ||||||||||||
| Goodison et al. 201238 | 8-biomarker panel | IL-8, MMP-9, PAI-1, VEGF, ANG, CA-9, APOE, MMP-10 | Case-control | n = 127; | BC vs healthy | ELISA | 0.980 | 92 | 97 | 97 | 92 | BTA Trak: 79% /83% |
| 64 BC 63 benign/healthy controls | ||||||||||||
| Urinary cytology: 33% /98% | ||||||||||||
| Huang et al. 201643 | Nomogram (biomarkers and demographic data) | MMP9, MMP10, IL8, VEGFA, SERPINE1, SERPINA1, CA9, APOE, ANG and SCD1 | Case-control, mulicenter (n = 10) | n = 686; | BC vs benign | ELISA | 0.891 (0.86–0.92) | 78 | 86 | 80 | 85 | N/A |
| 394 BC 292 benign urologic controls | ||||||||||||
| Theodorescu et al. 200611 | 22-biomarker panel | Fibrinopeptide A | Prospective BC Cohort | TrainingPhase 46 BC 33 healthy controls | BC vs benign and healthy | CE-MS | N/A | 100 | 100 -validation phase | N/A | N/A | N/A |
| 73- specificity analysis | ||||||||||||
| ValidationPhase 31 BC 11 healthy controls | ||||||||||||
| SpecificityAnalysis 281 benign 33 healthy controls | ||||||||||||
| 45 Non BC diseases | ||||||||||||
| Schiffer et al. 200912 | Nomogram (biomarkers and grade) | Membrane associated progesterone receptor component 1, Collagen α-1 (I) chain, Uromodulin, Collagen α-1 (III) chain | Prospective BC Cohort | Training Phase n = 424 71 NMIBC 56 MIBC 297 control set | NMIBC vs MIBC | CE-MS | 0.87 (0.79–0.92) | 92 | 68 | N/A | N/A | N/A |
| ValidationPhase n = 130 68 NMIBC 62 MIBC | ||||||||||||
| Frantzi et al. 201613 | 116 peptide biomarkers | Collagen fragments, Hemoglobin A, Apolipoprotein A, fibrinogen A, B2-microglobulin,small proline-rich protein 3,insulin, and histidine-rich glycoprotein. | Retrospective, multicenter (n = 5) | TrainingPhase N = 425 Validation Phase N = 270 | BC vs patients with suspicious symptoms for BC | CE-MS | 0.87 (0.83–0.91) | 91 | 68 | 83 | 81 | N/A |
| 106-peptide biomarkers | Collagen fragments, Apolipoprotein A-I, HSPG2, ADAMTS1, ADAM22 | Retrospective, mulicenter (n = 2) | TrainingPhase N = 425 Validation Phase N = 211 | Prior BC history (recurrence) | CE-MS | 0.75 (0.68–0.80) | 87 | 51 | 94 | 32 | Urinary cytology: 31% /100% |
Abbreviations: A1T1-Alpha-1-antitrypsin; ANG- Angiotensinogen; APOE-Apolipoprotein E; BC-Bladder Cancer; CA9: Carbonic anhydrase 9; IL8- Interleukin 8; MMP9-Matrix metalloproteinase-9; MMP10- Matrix metalloproteinase-10, MIBC-Muscle invasive bladder cancer; NMIBC-Non-muscle invasive bladder cancer; VEGFA-Vascular endothelial growth factor APAI1-Plasminogen activator inhibitor 1; SCD1-Syndecan-1.
Table 2
Commonly reported biomarkers identified by proteomics experiments
| Study | Proteins | Biol. Fluid | Discovery | Verification | AUC | |||||
| Platform | Subjects | Comparison/Clinical Context | Regulation | Platform | Subjects | Comparison/Clinical Context | Or % sens./spec. | |||
| Li et al. 201420 | Apolipoprotein A1 | Urine | 2DE-MALDI-TOF | N/A | BC vs HC | Upregulated | WB; ELISA¥; ELISA± | 6 BC/6 HC; 40 BC/40 HC 223 BC/156 HC | BC vs healthy controls | 89/86 |
| Chen et al. 201021 | Urine | iTRAQ LC-MS/MS | 14 hernia/6 LG Ta/10 HG NMIBC/7 HG MIBC* | BC vs hernia | Upregulated | ELISA, WB | ELISA: 50 hernia/21 LG Ta/9 HG NMIBC/16 HG MIBC | BC vs hernia | 0.982 | |
| Li et al. 201118 | Urine | 2DE-MALDI-TOF | 40 BC/24 healthy controls* | BC vs HC | Upregulated | ELISA | 40 BC (24 LG; 16 HG)/24 HC/8 benign | BC vs HC or benign; LG vs HG | 0.928; 0.875 | |
| Chen et al. 201231 | Urine | Isotopic dimethylation labeling/LC-MS/MS | 9 hernia/2 LG Ta/5 HG NMIBC/2 HG MIBC* | BC vs hernia | Upregulated | LC-MRM/MS | 12 hernia/7 LG Ta/17 HG NMIBC/4 HG MIBC/8 HU-UTI | BC vs hernia | 0.830 | |
| Chen et al. 201330 | Urine | ProSpectrum/MARS- Depletion, iTRAQ LC-MS/MS | 7 hernia/4 LG Ta/5 HG NMIBC/4 HG MIBC* | BC vs hernia | Upregulated | Apolipoprotein Kit (Multiplex) | 49 hernia/20 LG Ta/33 HG NMIBC/10 HG MIBC | BC vs hernia | 0.875 | |
| Frantzi et al. 201613 | Urine | CE-MS | 341 primary BC/110 urological controls | BC vs urol. controls | Upregulated | CE-MS | 168 primary BC/102 urological controls | BC vs suspicious patients | 0.870 (Ipanel) | |
| Urine | CE-MS | 109 recurrent BC/316 negative for recurrence | prior BC history | Upregulated | CE-MS | 55 recurrent BC/156 negative for recurrence | Prior BC history (recurrence) | 0.720 (Ipanel) | ||
| Chen et al. 201330 | Apolipoprotein A2 | Urine | ProSpectrum/MARS- Depletion, iTRAQ LC-MS/MS | 7 hernia/4 LG Ta/5 HG NMIBC/4 HG MIBC* | BC vs hernia | Upregulated | Apolipoprotein Kit (Multiplex) | 49 hernia/20 LG Ta/33 HG NMIBC/10 HG MIBC | BC vs hernia | 0.864 |
| Chen et al. 201231 | Urine | Isotopic dimethylation labeling/LC-MS/MS | 9 hernia/2 LG Ta/5 HG NMIBC/2 HG MIBC* | BC vs hernia | Upregulated | LC-MRM/MS | 12 hernia/7 LG Ta/17 HG NMIBC/4 HG MIBC/8 HU-UTI | BC vs hernia | 0.756 | |
| Chen et al. 201021 | Urine | iTRAQ LC-MS/MS | 14 hernia/6 LG Ta/10 HG NMIBC/7 HG MIBC* | BC vs hernia | Upregulated | ELISA, WB | ELISA: 48 hernia/19 LG Ta/40 HG NMIBC/16 HG MIBC | BC vs hernia | 0.631 | |
| Chen et al. 201330 | Apolipoprotein E | Urine | ProSpectrum/MARS- Depletion, iTRAQ LC-MS/MS | 7 hernia/4 LG Ta/5 HG NMIBC/4 HG MIBC* | BC vs hernia | Upregulated | Apolipoprotein Kit (Multiplex) | 49 hernia/20 LG Ta/33 HG NMIBC/10 HG MIBC | BC vs hernia | 0.745 |
| Linden et al. 201216 | Urine | Albumin and IgG depletion kit/LC-MS/MS | 6 BC/6 HC*; 5 BC/5 HC | BC vs HC | Upregulated | WB | 17 BC/26 HC | BC vs healthy controls | 89/31 | |
| Bansal et al. 20148 | S100A8 | Serum | 2DE-MALDI-TOF | 33 LG/32 HG/25 HC | BC (HG/LG) vs HC | Upregulated | WB; ELISA | 33 LG/32 HG/25 HC | BC vs HC | 0.856 |
| Bansal et al. 20169 | Serum | MARS- Depletion | 55 BC pre-OP/53 BC post-OP/52 HC | BC pre-OP/post-OP vs HC | Upregulated | ELISA | 55 BC pre-OP/52 HC | BC pre-OP vs HC | 0.850 | |
| Chen et al. 201231 | Urine | Isotopic dimethylation labeling/LC-MS/MS | 9 Hernia/2 LG Ta/5 HG NMIBC/2 HG MIBC* | BC vs hernia | Upregulated | LC-MRM/MS | 12 hernia/7 LG Ta/17 HG NMIBC/4 HG MIBC/8 HU-UTI | BC vs hernia | 0.836 | |
| Bansal et al. 201462 | S100A9 | Serum | 2DE-MALDI-TOF | 33 LG/32 HG/25 HC | BC (HG/LG) vs HC | Upregulated | WB; ELISA | 33 LG/32 HG/25 HC | BC vs healthy controls | 0.902 |
| Bansal et al. 201663 | Serum | MARS- Depletion | 55 BC pre-OP/53 BC post-OP/52 HC | BC pre-OP/BC post-OP vs HC | Upregulated | ELISA | 55 BC pre-OP/52 HC | BC pre-OP vs healthy controls | 0.957 | |
| Chen et al. 201231 | Urine | Isotopic dimethylation labeling/LC-MS/MS | 9 Hernia/2 LG Ta/5 HG NMIBC/2 HG MIBC* | BC vs hernia | Upregulated | LC-MRM/MS | 12 hernia/7 LG Ta/17 HG NMIBC/4 HG MIBC/8 HU | BC vs hernia | 0.815 | |
| Bansal et al. 201462 | Carbonic anhydrase I | Serum | 2DE-MALDI-TOF | 33 LG/32 HG/25 HC | BC (HG/LG) vs HC | Upregulated | WB; ELISA | 33 LG/32 HG/25 HC | BC vs healthy controls | 0.891 |
| Bansal et al. 201663 | Serum | MARS- Depletion | 55 BC pre-OP/53 BC post-OP/52 HC | BC pre-OP/BC post-OP vs HC | Upregulated | ELISA | 55 BC pre-OP/52 HC | BC pre-OP vs healthy controls | 0.908 | |
| Chen et al. 201231 | Urine | Isotopic dimethylation labeling/LC-MS/MS | 9 hernia/2 LG Ta/5 HG NMIBC/2 HG MIBC* | BC vs hernia | Upregulated | LC-MRM/MS | 12 hernia/7 LG Ta/17 HG NMIBC/4 HG MIBC/8HU | BC vs hernia | 0.837 | |
| Chen et al. 201021 | Heparin cofactor 2 | Urine | iTRAQ LC-MS/MS | 14 hernia/6 LG Ta/10 HG NMIBC/7 HG MIBC * | BC vs hernia | Upregulated | ELISA, WB | 54 hernia/19 LG Ta/40 HG NMIBC/16 HG MIBC | BC vs hernia | 0.819 |
| Chen et al. 201231 | Isotopic dimethylation labeling/LC-MS/MS | 9 hernia/2 LG Ta/5 HG NMIBC/2 HG MIBC* | BC vs hernia | Upregulated | LC-MRM/MS | 12 hernia/7 LG Ta/17 HG NMIBC/4 HG MIBC/8 HU | BC vs hernia | 0.854 | ||
| Chen et al. 201231 | Alpha-1 antitrypsin | Urine | Isotopic dimethylation labeling/LC-MS/MS | 9 hernia/2 LG Ta/5 HG NMIBC/2 HG MIBC* | BC vs hernia | Upregulated | LC-MRM/MS | 12 hernia/7 LG Ta/17 HG NMIBC/4 HG MIBC/8 HU-UTI | BC vs hernia | 0.729 |
| Linden et al. 201216 | Urine | Albumin and IgG depletion kit/LC-MS/MS | 6 BC/6 HC*; 5 BC/5 HC | BC vs HC | Upregulated | WB | 17 BC/26 HC | BC vs healthy controls | 66/85 | |
| Yang et al. 201110 | Urine | Dual-lectin affinity chromatography/LC-MS/MS | 54 BC/46 HC | BC vs HC | Upregulated | ELISA | 35 BC/35 HC | BC vs healthy controls | 74/80 | |
| Schiffer et al. 200912 | Membrane-associated progesterone receptor component 1 | Urine | CE-MS | 71 NMIBC; 56 MIBC; 297 control set | Stage; Progression | Downregula-ted | CE-MS | 68 NMIBC; 62 MIBC | Stage; Progression | 0.870 (Ipanel) |
| Frantzi et al. 201613 | Urine | CE-MS | 341 primary BC/110 urological controls | prior BC history | CE-MS | 168 primary BC/102 urological controls | BC vs suspicious patients | 0.870 (Ipanel) | ||
| CE-MS | 109 rec BC/316 negative for recurrence | CE-MS | 55 recurrent BC/156 negative for recurrence | prior BC history | 0.720 (Ipanel) | |||||
| Frantzi et al. 201613 | Fibrinogen Beta | Urine | CE-MS | 341 primary BC/110 urological controls | BC vs UC | Upregulated | CE-MS | 168 primary BC/102 UC | BC vs suspicious patients | 0.870 (Ipanel) |
| Linden et al. 201216 | Urine | CE-MS | 109 rec BC/316 negative for recurrence | Prior BC history | Upregulated | CE-MS | 55 recurrent BC/156 negative for recurrence | prior BC history | 0.720 (Ipanel) | |
| Chen et al. 201231 | Urine | Albumin and IgG depletion kit/LC-MS/MS | 6 BC/6 HC*; 5 BC/5 HC | BC vs HC | Upregulated | WB | 17 BC/26 HC | BC vs healthy controls | 77/77 | |
| Urine | Isotopic dimethylation labeling/LC-MS/MS | 9 hernia/2 LG Ta/5 HG NMIBC/2 HG MIBC* | BC vs hernia | Upregulated | LC-MRM/MS | 12 hernia/7 LG Ta/17 HG NMIBC/4 HG MIBC/8 HU-UTI | BC vs hernia | 0.831 | ||
Abbreviations: AUC- Area under the ROC curve; BC-Bladder Cancer; HC- Healthy Controls; HG- High Grade; HU- Hematuria; iTRAQ- Isobaric tag for relative and absolute quantitation; LC- Liquid Chromatography; LG- Low Grade; MALDI- Matrix-assisted laser desorption/ionization; MIBC-Muscle Invasive Bladder Cancer; MS- Mass Spectrometry; NMIBC- Non Muscle Invasive Bladder Cancer; pre-OP- pre- operative samples; post-OP- post-operative samples, TOF- Time of Flight; UC; Urological Controls; WB-Western Blot. *Samples pooled per category/¥ ELISA Training Phase/±ELISA Validation Phase. The presented proteins were shortlisted based on the following criteria: a) protein biomarkers independently verified by at least two groups or in two different study settings (methodologies, type of comparison), b) biomarkers with diagnostic value as proven by statistically significant results at the verification phase, c) verification phase including at least 40 samples.




