Independent Replication of Published Germline Polymorphisms Associated with Urinary Bladder Cancer Prognosis and Treatment Response
Abstract
Background:
Many studies investigated the prognostic or predictive relevance of single nucleotide polymorphisms (SNPs) in biologically plausible genes in urinary bladder cancer (UBC) patients. Most published SNP associations have never been replicated in independent patient series.
Objective:
To independently replicate all previously reported associations between germline SNPs and disease prognosis or treatment response in UBC.
Methods:
A Pubmed search was performed to identify studies published by July 1, 2014 reporting on germline SNP associations with UBC prognosis or treatment response. For the replication series, consisting of 1,284 non-muscle-invasive bladder cancer (NMIBC) and 275 muscle-invasive or metastatic bladder cancer (MIBC) patients recruited through the Netherlands Cancer Registry, detailed clinical data were retrieved from medical charts. Patients were genotyped using a genome-wide SNP array. SNP association with recurrence-free, progression-free, and overall survival (OS) within specific patient and treatment strata was tested using Cox regression analyses.
Results:
For only six of the 114 evaluated SNPs, the association with either UBC prognosis or treatment response was replicated at the p < 0.05 level: rs1799793 (ERCC2) and rs187238 (IL18) for BCG recurrence; rs6678136 (RGS4) and rs11585883 (RGS5) for NMIBC progression; rs12035879 (RGS5) and rs2075786 (TERT) for MIBC OS.
Conclusions:
Non-replicated genetic associations in the literature require cautious interpretation. This single replication does not provide definitive proof of association for the six SNPs, and non-replication of other SNPs may result from population-specific effects or the retrospective patient enrollment.
INTRODUCTION
Urinary bladder cancer (UBC) is a common disease which poses a significant burden on patients and healthcare systems. In non-muscle-invasive bladder cancer (NMIBC), the tendency to recur requires intensive surveillance and repeated treatment. In muscle-invasive or metastatic bladder cancer (MIBC), mortality is high and major surgery and systemic chemotherapy are part of standard care. There is large interpatient variability in prognosis and treatment response in both NMIBC and MIBC. Traditional clinicopathological factors account for only part of this variability. The risk group stratification using EORTC predictors is recommended by the EAU guidelines for decision-making in clinical practice. However, even though these tools aid in discrimination of NMIBC recurrence and progression risk at the group level, they do not allow for precise prediction of the fate of an individual patient [1–3]. In MIBC, there is an urgent need for better tools to identify which patients would benefit most from neo-adjuvant or adjuvant chemotherapy and predict who will likely benefit from a cystectomy or should be offered systemic therapy [4]. This emphasizes the importance of identifying new prognostic and predictive (bio)markers to facilitate more accurate outcome prediction in the individual patient, and of research that fuels our mechanistic insight in urothelial carcinogenesis and progression.
The last decade, many studies have investigated the association between germline variants in biologically plausible genes and UBC prognosis or treatment response [5–7]. Among these candidate genes were those implicated in well-known cancer-related pathways, such as DNA repair, cell-cycle control, and inflammation. A previous literature review by our group indicated that most of the candidate-gene studies were underpowered [6]. Even for NMIBC, the largest studies to date included no more than 400 patients. Only a small set of variants was evaluated by more than one study; for these variants mostly inconsistent results were reported. No more than two of the several dozen studies that claimed an association, reported on successful replication of findings in independent patient series [8, 9]. Extensive external validation of reported associations is however essential to distinguish true from false-positive associations.
This study aims to independently replicate all reported prognostic and predictive single nucleotide polymorphisms (SNPs) among 1,559 UBC patients of the Nijmegen Bladder Cancer Study, currently one of the largest UBC patient series worldwide.
MATERIALS AND METHODS
Literature search and SNP selection
As an update to our previous literature review [6], a Pubmed search was performed to identify all studies published by July 1, 2014 on the association between germline genetic variants and UBC prognosis or treatment response. Titles and abstracts of all identified studies published in English-language journals were manually reviewed for relevance according to the selection criteria presented in Fig. 1. We focused on single-SNP associations, and finally evaluated 114 SNPs mapping to 83 genes for which association was claimed with at least one of the endpoints listed in Table 1.
Patient population and outcome definitions
We used data of the Nijmegen Bladder Cancer Study (NBCS) [10]. In the NBCS, UBC patients diagnosed in one of seven hospitals in the mid-eastern part of the country were identified through the population-based Netherlands Cancer Registry (NCR) held by the Netherlands Comprehensive Cancer Organization (IKNL). Patients diagnosed under the age of 75 years were invited to the NBCS by IKNL on behalf of the treating physicians. The NBCS started in May 2007 with the invitation of UBC patients diagnosed in 1995–2006 who were still alive. Later, the NBCS was expanded with three more recently diagnosed patient cohorts (2006–2008, 2008-2009, 2009-2010) in three phases (January 2009, November 2010, February 2012, respectively). Of all the invitees, 66% agreed to participate. Participants were sent a lifestyle questionnaire to fill out and blood samples were collected by local Thrombosis Service centers. All participants gave written informed consent and the study was approved by the institutional review board of Radboud university medical center, Nijmegen, the Netherlands. For this study, we only used data of participants included in the first two enrollment phases of the NBCS (diagnosis between 1995 and 2008) for which genome-wide genotyping data were available.
Follow-up regarding vital status of NBCS patients until December 31st, 2012 was obtained through linkage of NCR data with the Dutch Municipal Personal Records Database. Detailed data on clinicopathological tumor characteristics, treatment, and clinical outcome were abstracted from medical charts. From the 1,601 genotyped NBCS participants (see ‘Genotyping and imputation’), we excluded: three participants with an unconfirmed UBC diagnosis based on medical chart review; nine NMIBC patients with incomplete follow-up information; 30 patients with previous or synchronous upper urinary tract cancer. Finally, the study population consisted of 1,559 UBC patients (1,284 NMIBC/275 MIBC).
For replication of published prognostic and/or predictive SNPs in NMIBC, we assessed the endpoints recurrence and progression in the overall patient group and also within relevant treatment strata (i.e., treated with transurethral resection (TUR)-only (n = 463), bacillus Calmette-Gu
To replicate postulated SNP associations in MIBC, the endpoint overall survival (OS) was evaluated (Table 1). OS of patients with primary MIBC was defined as the time from the initial TUR until death (from all causes) or date of censoring (i.e., December 31st, 2012 or date of five-year follow-up), whichever came first.
Genotyping and imputation
As part of a genome-wide association study (GWAS) for UBC risk led by Radboud university medical center and deCODE Genetics (Reykjavik, Iceland) [10], germline DNA of NBCS participants diagnosed with UBC in the period 1995–2008 was genotyped using the Illumina HumanHapCNV370 BeadChip. Imputation with IMPUTE v2 software was performed to increase marker density by using the 1000 Genomes phase 1 integrated version 3 haplotypes (released March 2012, 1,092 individuals) as a reference panel (https://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html). Imputation was performed separately for NBCS participants diagnosed from 1995–2006 (NBCS1) and from 2006–2008 (NBCS2). Pre-imputation quality control (QC) was performed based on the following exclusion criteria: SNP yield <96% , minor allele frequency (MAF) <1% , Hardy-Weinberg equilibrium (HWE) p-value <10–5, sample yield <96% , European ancestry <90% (based on Structure analysis), duplicate samples based on a relatedness check (using genome-wide complex trait analysis [GCTA] software), and gender mismatch with phenotypic data. This resulted in a total set of 333,068 and 333,920 markers (325,019 overlapping) used as input for imputation in 1273 NBCS1 and 328 NBCS2 samples, respectively.
For evaluation of reported SNP associations, genotype data of the two imputation series was combined (total n = 1,601; 38,037,363 overlapping markers). From replication, we excluded reported SNPs (n = 2) that did not pass the following post-imputation QC criteria: HWE p-value >10–3 and IMPUTE info score >0.4 (in both imputation series) (Fig. 1).
Statistical analysis
Each SNP was assessed for association with the respective outcome measure based on an additive, dominant, and recessive inheritance model. In each model, the most common (‘major’) allele in the NBCS was used as reference allele. SNP association parameters were obtained by Cox regression analyses (using the estimated genotypic probabilities) performed with ProbABEL v0.1-3 [12]. Independent prognostic/predictive value of SNPs was evaluated by multivariable analyses including relevant prognostic variables. For NMIBC these were: age at diagnosis, gender, tumor stage, tumor grade, tumor focality (solitary versus multifocal; exact tumor number was poorly documented in medical files), presence of concomitant carcinoma in situ, and treatment (TURT only±1 postoperative (p.o.) chemotherapy (CT) instillation; adjuvant intravesical (i.v.) CT; adjuvant i.v. immunotherapy (IT); both adjuvant i.v. CT and IT). Treatment was not included as a variable in the analyses for TUR-only-treated, BCG-treated and i.v. chemotherapy-treated NMIBC patients. Tumor size was not included due to absence of this information in medical files for a large fraction of patients. For MIBC these were: age at diagnosis, gender and an aggregate measure based on TNM classification (tumors of stage T2-T4a with N0/NX and M0/MX vs. tumor of stage T4(b) ór any T with N ≥1/N+ and/or M1). Statistical significance was tested using a one degree-of-freedom (df) Wald test. As our goal was to replicate previously identified SNP associations, we did not correct for multiple testing. A two-sided significance threshold of p < 0.05 was used. In some cases, the strand orientation for strand ambiguous SNPs (A/T; C/G) could not be inferred from the original report, complicating assessment of consistency in direction of SNP effect. In these cases, we assumed strand consistency of the allele in the published report (e.g., C) and allele in the NBCS (e.g., G) if both had frequency <0.45.
RESULTS
Baseline patient and tumor characteristics of the study population are given in Table 2. Median follow-up time of NMIBC and MIBC patients (i.e., time from the initial TUR until last update of outcome status) was 5.3 (interquartile range [IQR]: 3.7–8.7) and 9.8 (IQR: 6.7–13) years, respectively. Five-year Kaplan-Meier risk of recurrence, progression (NMIBC), and overall death (MIBC) was 50.5% , 17.2% , and 32.4% , respectively. Number of recurrence and progression events among the total of 1284 NMIBC patients was 598 and 197, respectively; among the total of 275 MIBC patients, 87 deceased.
Associations with recurrence in NMIBC
Among the 23 tested SNPs, only rs3795617 (RGS13) was associated with NMIBC recurrence at the p < 0.05 threshold (CT/TT vs. CC: hazard ratio [HR] = 1.26 [95% CI: 1.04–1.53]), but with conflicting direction of effect compared to the original study (Tables 3, 4, S1) [13]. Borderline significant evidence (0.05 < p < 0.10) for a directionally consistent association with recurrence was demonstrated for rs1800795 (IL6) and rs25487 (XRCC1) [14, 15].
Associations with recurrence in TUR-only-treated NMIBC
A statistically significant decrease in recurrence risk (HR = 0.23 [95% CI: 0.07–0.72]) was found for homozygous T allele carriers of rs11594179 (SUFU) compared to patients with the CT/CC genotype (Table 3). Direction of effect was however conflicting with the association presented in the original report (Table 4), and the variant failed previous external validation in the Spanish EPICURO cohort [8]. None of the other 13 evaluated variants showed association at the p < 0.05 threshold (Table S2). Yet, a borderline significant risk increase was observed with each G allele copy of rs1233560 (SHH), which confirms the previously reported and already replicated association [8].
Associations with recurrence in BCG-treated NMIBC
The SNP rs1799793 in ERCC2 was found to be statistically significantly associated with recurrence in BCG-treated patients (TT vs. CT/CC: HR = 2.76 [95% CI: 1.53–4.99]) (Table 3). This is in line with the previously reported 3-fold increased recurrence risk among homozygous A allele carriers compared to patients with the GG genotype, with no significant difference between patients with genotype GA and GG (Table 4) [16]. Furthermore, we replicated the association of rs187238 (IL18): GG vs. CG/CC: HR = 2.35 (95% CI: 1.20–4.59) [17]. We could not confirm association with BCG recurrence of any of the other 35 reported SNPs (Table S3).
Associations with progression in NMIBC
From the 17 evaluated SNPs, the previously postulated association with NMIBC progression of two variants in regulator of G-protein signaling (RGS) genes could be replicated (Tables 3, 4) [13]. The SNP rs6678136 (RGS4) correlated with a risk increase for progression in both heterozygous and homozygous A allele carriers (dominant model: HR = 1.39 [95% CI: 1.02–1.90]). Besides, homozygous carriers (n = 3) of rs11585883 [C] (RGS5) experienced an elevated risk of progression (recessive model: HR = 10.0 [95% CI: 2.47–40.6]). Also, we observed a trend towards recessive association with progression of rs2297518 (NOS2), consistent with the original report [18]. A borderline significant and directionally consistent association with NMIBC progression was found as well for two SNPs originally associated with cancer-specific survival (CSS) in a combined NMIBC and MIBC patient series: rs1042522 (TP53) and rs9302752 (NOD2) (Table S4) [19, 20].
Associations with progression in chemotherapy-treated NMIBC
For six SNPs originally associated with CSS in (intravesical/systemic) chemotherapy-treated UBC [21], we evaluated association with NMIBC progression after initial intravesical chemotherapy treatment. None of the SNPs showed association (Table S5).
Associations with overall survival in MIBC
In accordance with the original study, we observed a recessive association with OS in MIBC for rs12035879 [A] in RGS5 (HR = 2.79 [95% CI: 1.40–5.56]) and rs2075786 [A] in TERT (HR = 1.93 [95% CI: 1.01–3.68]) (Tables 3, 4) [13, 22]. For three of the other 42 evaluated SNPs (i.e., rs2227983 [EGFR], rs613120 [PGR], and rs1131341 [NQO1]), we identified a borderline significant association with patient survival that confirms the previously reported direction of effect (Table S6) [22–24].
In conclusion, association of only six SNPs could be replicated (i.e., directionally consistent association at p < 0.05): rs1799793 (ERCC2), rs187238 (IL18), rs6678136 (RGS4), rs11585883 (RGS5), rs12035879 (RGS5), and rs2075786 (TERT). Directionally consistent association at borderline significance was observed for nine SNPs, among which rs1233560 (SHH) was replicated before. Multivariable analyses indicated independent prognostic/predictive value for all replicated SNPs, except for rs6678136 (RGS4) (Table 3). It did not reveal additional, directionally consistent SNP associations at p < 0.05 (data not shown).
DISCUSSION
We conducted an independent replication analysis of 114 SNPs previously reported to be associated with UBC outcome based on candidate-gene studies, within one of the largest, population-based UBC patient series. We restricted replication to SNPs only, as other types of genetic variants (n = 8) were insufficiently covered by our genotyping array. We could confirm association of only six of the proposed prognostic or predictive SNPs at the p < 0.05 level and with directionally consistent effects with the original publication. In addition, one of the three earlier replicated SNP associations, i.e., rs1233560 (SHH) with NMIBC recurrence after TUR, was confirmed with borderline significance [8].
Non-replication of a large proportion of the published SNP associations is not surprising given that the candidate-gene studies were small and evaluated most candidates just once [25]. The large sample size and long follow-up of the NBCS patient cohort gave us higher power to detect the reported associations, limiting the chance of false-negative findings. In the overall NMIBC group, our study had >80% power at α= 0.05 to replicate association of a SNP conferring a relative hazard per risk allele (frequency: 0.20) of at least 1.2 for recurrence (five-year risk: 50%) and 1.4 for progression (five-year risk: 20%). Although our MIBC patient series was relatively small, our study still had >80% power at α= 0.05 to detect a risk allele relative hazard of at least 1.7 and 1.5 for overall mortality (five-year risk: 30%), assuming a risk allele frequency of 0.20 and 0.40, respectively.
We can however not exclude that lack of replicating the other reported SNP associations can be attributed to differences in genetic or environmental background between the discovery and NBCS population, or to heterogeneity in patient characteristics, therapeutic schemes, or endpoint definitions. In order to exemplify this, we redid our analysis of the 17 SNPs that were reported to be associated with disease progression. As an alternative definition for progression we now used a shift to muscle invasive disease (see Supplementary Table S9). Of course, the confidence intervals of the effect estimates are generally wider because of this much stricter definition (i.e. less endpoints). Because of that, the prognostic effect of the RGS4 SNP lost its statistical significance. Also, the effect estimates itself are sometimes quite different. For example, the prognostic effect of the RGS5 SNP in the dominant model is totally different. Also, the NOD2 SNP has now statistically significant prognostic value in the recessive model. In general, however, the effect estimates are not higher (i.e. stronger prognostic effect) with this stricter definition as might have been expected if they really had prognostic value for progression to MIBC.
Also, even though this study replicated SNP associations identified through a hypothesis-driven approach, not all variants were selected because of predicted or known functional consequences but also as representative, haplotype-tagging SNPs for selected candidate genes. Only part of the reported SNP associations may therefore actually capture the causal variant. Possible differences in allele frequencies and in linkage-disequilibrium (LD) structure between our European population and the discovery population may therefore affect the strength of association for non-causal SNPs and, consequently, our power for replication. This emphasizes the need for additional replication initiatives in patients of similar ethnic background, and identification of causal variation through fine-mapping efforts in the original discovery population. Reporting of causal rather than associated genetic variants would increase potential for clinical utility. Given that fine-mapping and functional validation of associations are currently feasible using relatively affordable technologies, perhaps publication standards should be raised accordingly.
The NBCS patient cohort was retrospectively enrolled, with a time lag between diagnosis and study enrollment up to 12 years. The absence of prevalent patients that failed to survive until the sampling date has resulted in a study population biased towards favorable survival. This may have biased the effect size estimates to some extent and thereby the ability of our study to reproduce the reported associations. Assessment of the association of the six replicated SNPs within the subset of our patient cohort with a maximum time between diagnosis and study enrollment of 3 years showed, however, only marginal differences in the effect size estimates compared to the total patient cohort (data not shown).
Genotype data for the large majority of evaluated SNPs in our NBCS replication series was based on imputation; for 27 (24%) of the 114 SNPs included in this study the patients’ genotype was measured through the Illumina chip. Nevertheless, the IMPUTE info score that expresses the level of imputation reliability was at least 0.8 for 79 (91%) and even >0.9 for 71 (82%) of the 87 imputed SNPs. Also given the fact that our analyses took into account the uncertainty in genotype estimation, we expect that the influence of potential genotype misclassification among study participants on effect estimation is negligible.
On the other hand, single replication by our study does not provide definitive proof of the six SNP associations. The prior evidence supporting these candidate associations was limited (Table 4). None of the six replicated SNP associations was reported more than once. Only association of rs17999793 (ERCC2) with recurrence after BCG was previously evaluated by more than one study, but was identified exclusively by Gangwar et al. [16, 26]. Also, if we would have tested association for a random selection of 114 (non-candidate) variants at an alpha level of 0.05, we would a priori expect to find association for six of them purely by chance. Replication in additional patient cohorts is needed to confirm true association. For this reason, we will only shortly touch upon the biological background of the replicated variants.
The strongest association signal (p = 7.88×10–4) was found in relation to BCG recurrence for rs1799793 (Asp312Asn) in the excision repair cross-complementation group 2 (ERCC2) gene, which encodes a nucleotide excision repair (NER) protein. NER may diminish reactive oxygen species (ROS)-induced DNA damage in the inflammatory response that is evoked by BCG [16, 26]. In addition, we replicated association with BCG response of a promoter variant (rs187238) in interleukin-18 (IL18), which exerts its anti-tumor activity by augmenting interferon-γ production, promoting T-helper 1 differentiation, and enhancing cytotoxic activity [27]. Furthermore, our study strengthened the evidence for implication in NMIBC progression and MIBC survival of two RGS genes, which may influence processes such as tumor neovascularization through modulation of G protein-coupled receptor (GPCR) signaling [28]. We also confirmed association with MIBC survival of the intronic SNP rs2075786 in telomerase (TERT) involved in maintenance of telomere ends and thereby in escape of cellular senescence in tumor cells [29]. Finally, we added to the evidence for association of a 3’ UTR variant (rs1233560) of sonic hedgehog (SHH), encoding the ligand that initiates one of the major signaling pathways in cancer stem cells, with recurrence after TUR [8].
The fact that we do not observe more extreme significance levels, even at the relatively high degree of statistical power, indicates that if true associations exist at all, most of the evaluated genetic variants are probably only weakly associated with patient outcome in our population. It remains therefore questionable whether the currently reported markers will ever markedly contribute to more accurate outcome prediction at the individual patient-level in order to tailor treatment and surveillance plans.
This first replication study of all previously reported prognostic and predictive SNP markers in UBC suggests that non-replicated genetic associations in the literature require a cautious interpretation. It supports the notion that many published prognostic and predictive SNPs that lack a replication may represent false-positives (25), also in UBC. Additional large-scale replication initiatives are needed to confirm this statement. Based on our replication in the NBCS, the SNPs rs1799793 (ERCC2), rs187238 (IL18), rs2075786 (TERT), rs1233560 (SHH), rs6678136 (RGS4), rs11585883 and rs12035879 (RGS5) are especially worth pursuing in further replication and functional follow-up studies. The low number of replicated SNP associations underscores the need to apply more stringent criteria when reporting associations, in particular replicating identified associations as part of the original study. Also, to allow the reader to judge the usefulness of a study and understand why conclusions may be dissimilar for different studies, transparent and complete reporting should be encouraged [30]. Importantly, the initiation of a strong international consortium of bladder cancer prognostic studies is highly called for to facilitate the replication of genetic association findings across populations. In the future, an agnostic GWAS approach, which is not biased by a priori assumption of relevant candidate loci and includes extensive replication, may lead to elucidation of new disease mechanisms and prognostic or predictive biomarkers.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
ACKNOWLEDGMENTS
We would like to thank all the participants in the study for their willingness to provide blood samples for our research. We thank our collaborators from deCODE Genetics in Reykjavik for the genotyping, and Freerk van Dijk and Morris Swertz from GoNL (http://www.nlgenome.nl/) for imputation. Anne Grotenhuis was supported by a PhD Scholarship of the Radboud Institute for Health Sciences. This work was sponsored by the Stichting Nationale Computerfaciliteiten (National Computing Facilities Foundation, NCF) for the use of supercomputer facilities, with financial support from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organization for Scientific Research, NWO).
Appendices
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/BLC-150027.
REFERENCES
[1] | Babjuk M , Burger M , Zigeuner R , Shariat SF , van Rhijn BW , Comperat E , Sylvester RJ , Kaasinen E , Bohle A , Palou Redorta J , Roupret M . European Association of Urology. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update Eur Urol (2013) ;64: (4):639–653. |
[2] | Sylvester RJ . How well can you actually predict which non-muscle-invasive bladder cancer patients will progress? Eur Uro (2011) ;60: (3):431–434; discussion 433-4. |
[3] | Xylinas E , Kent M , Kluth L , Pycha A , Comploj E , Svatek RS , Lotan Y , Trinh QD , Karakiewicz PI , Holmang S , Scherr DS , Zerbib M , Vickers AJ , Shariat SF . Accuracy of the EORTC risk tables and of the CUETO scoring model to predict outcomes in non-muscle-invasive urothelial carcinoma of the bladder. Br J Cancer (2013) ;109: (6):1460–1466. |
[4] | Witjes JA , Comperat E , Cowan NC , De Santis M , Gakis G , Lebret T , Ribal MJ , Van der Heijden AG , Sherif A . European Association of Urology. EAU guidelines on muscle-invasive and metastatic bladder cancer: Summary of the guidelines.. Eur Urol (2014) ;65: (4):778–792. |
[5] | Chang DW , Gu J , Wu X . Germline prognostic markers for urinary bladder cancer: Obstacles and opportunities. Urol Oncol (2012) ;30: (4):524–532. |
[6] | Grotenhuis AJ , Vermeulen SH , Kiemeney LA . Germline genetic markers for urinary bladder cancer risk, prognosis and treatment response. Future Oncol (2010) ;6: (9):1433–1460. |
[7] | Gu J , Wu X . Genetic susceptibility to bladder cancer risk and outcome. Per Med (2011) ;8: (3):365–374. |
[8] | Chen M , Hildebrandt MA , Clague J , Kamat AM , Picornell A , Chang J , Zhang X , Izzo J , Yang H , Lin J , Gu J , Chanock S , Kogevinas M , Rothman N , Silverman DT , Garcia-Closas M , Grossman HB , Dinney CP , Malats N , Wu X . Genetic variations in the sonic hedgehog pathway affect clinical outcomes in non-muscle-invasive bladder cancer. Cancer Prev Res (Phila) (2010) ;3: (10):1235–1245. |
[9] | Ke HL , Chen M , Ye Y , Hildebrandt MA , Wu WJ , Wei H , Huang M , Chang DW , Dinney CP , Wu X . Genetic variations in micro-RNA biogenesis genes and clinical outcomes in non-muscle-invasive bladder cancer. Carcinogenesis (2013) ;34: (5):1006–1011. |
[10] | Rafnar T , Vermeulen SH , Sulem P , Thorleifsson G , Aben KK , Witjes JA , Grotenhuis AJ , Verhaegh GW , Hulsbergen-van de Kaa CA , Besenbacher S , Gudbjartsson D , Stacey SN , Gudmundsson J , Johannsdottir H , Bjarnason H , Zanon C , Helgadottir H , Jonasson JG , Tryggvadottir L , Jonsson E , Geirsson G , Nikulasson S , Petursdottir V , Bishop DT , Chung-Sak S , Choudhury A , Elliott F , Barrett JH , Knowles MA , de Verdier PJ , Ryk C , Lindblom A , Rudnai P , Gurzau E , Koppova K , Vineis P , Polidoro S , Guarrera S , Sacerdote C , Panadero A , Sanz-Velez JI , Sanchez M , Valdivia G , Garcia-Prats MD , Hengstler JG , Selinski S , Gerullis H , Ovsiannikov D , Khezri A , Aminsharifi A , Malekzadeh M , van den Berg LH , Ophoff RA , Veldink JH , Zeegers MP , Kellen E , Fostinelli J , Andreoli D , Arici C , Porru S , Buntinx F , Ghaderi A , Golka K , Mayordomo JI , Matullo G , Kumar R , Steineck G , Kiltie AE , Kong A , Thorsteinsdottir U , Stefansson K , Kiemeney LA . European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum Mol Genet (2011) ;20: (21):4268–4281. |
[11] | Grotenhuis AJ , Dudek AM , Verhaegh GW , Witjes JA , Aben KK , van der Marel SL , Vermeulen SH , Kiemeney LA . Prognostic relevance of urinary bladder cancer susceptibility loci. PLoS One (2014) ;9: (2):e89164. |
[12] | Aulchenko YS , Struchalin MV , van Duijn CM . ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics (2010) ;11: , 134. |
[13] | Lee EK , Ye Y , Kamat AM , Wu X . Genetic variations in regulator of G-protein signaling (RGS) confer risk of bladder cancer. Cancer (2013) ;119: (9):1643–1651. |
[14] | Ahirwar D , Kesarwani P , Manchanda PK , Mandhani A , Mittal RD . Anti- and proinflammatory cytokine gene polymorphism and genetic predisposition: Association with smoking, tumor stage and grade, and bacillus Calmette-Guerin immunotherapy in bladder cancer. Cancer Genet Cytogenet (2008) ;184: (1):1–8. |
[15] | Sanyal S , De Verdier PJ , Steineck G , Larsson P , Onelov E , Hemminki K , Kumar R . Polymorphisms in XPD, XPC and the risk of death in patients with urinary bladder neoplasms. Acta Oncol (2007) ;46: (1):31–41. |
[16] | Gangawar R , Ahirwar D , Mandhani A , Mittal RD . Impact of nucleotide excision repair ERCC2 and base excision repair APEX1 genes polymorphism and its association with recurrence after adjuvant BCG immunotherapy in bladder cancer patients of North India. Med Oncol (2010) ;27: (2):159–166. |
[17] | Jaiswal PK , Singh V , Srivastava P , Mittal RD . Association of IL-12, IL-18 variants and serum IL-18 with bladder cancer susceptibility in North Indian population. Gene (2013) ;519: (1):128–134. |
[18] | Ryk C , Wiklund NP , Nyberg T , De Verdier PJ . Ser608Leu polymorphisms in the nitric oxide synthase-2 gene may influence urinary bladder cancer pathogenesis. Scand J Urol Nephrol (2011) ;45: (5):319–325. |
[19] | Guirado M , Gil H , Saenz-Lopez P , Reinboth J , Garrido F , Cozar JM , Ruiz-Cabello F , Carretero R . Association between C13ORF31, NOD2, RIPK2 and TLR10 polymorphisms and urothelial bladder cancer. Hum Immunol (2012) ;73: (6):668–672. |
[20] | Horikawa Y , Nadaoka J , Saito M , Kumazawa T , Inoue T , Yuasa T , Tsuchiya N , Nishiyama H , Ogawa O , Habuchi T . Clinical implications of the MDM2 SNP309 and p53 Arg72Pro polymorphisms in transitional cell carcinoma of the bladder. Oncol Rep (2008) ;20: (1):49–55. |
[21] | Sacerdote C , Guarrera S , Ricceri F , Pardini B , Polidoro S , Allione A , Critelli R , Russo A , Andrew AS , Ye Y , Wu X , Kiemeney LA , Bosio A , Casetta G , Cucchiarale G , Destefanis P , Gontero P , Rolle L , Zitella A , Fontana D , Vineis P , Matullo G . Polymorphisms in the XRCC1 gene modify survival of bladder cancer patients treated with chemotherapy. Int J Cancer (2013) ;133: (8):2004–2009. |
[22] | Andrew AS , Gui J , Sanderson AC , Mason RA , Morlock EV , Schned AR , Kelsey KT , Marsit CJ , Moore JH , Karagas MR . Bladder cancer SNP panel predicts susceptibility and survival. Hum Genet (2009) ;125: (5-6):527–539. |
[23] | Mason RA , Morlock EV , Karagas MR , Kelsey KT , Marsit CJ , Schned AR , Andrew AS . EGFR pathway polymorphisms and bladder cancer susceptibility and prognosis. Carcinogenesis (2009) ;30: (7):1155–1160. |
[24] | Sanyal S , Ryk C , De Verdier PJ , Steineck G , Larsson P , Onelov E , Hemminki K , Kumar R . Polymorphisms in NQO1 and the clinical course of urinary bladder neoplasms. Scand J Urol Nephrol (2007) ;41: (3):182–90. |
[25] | Ioannidis JP . Why most published research findings are false. PLoS Med (2005) ;2: (8):e124. |
[26] | Gu J , Zhao H , Dinney CP , Zhu Y , Leibovici D , Bermejo CE , Grossman HB , Wu X . Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin Cancer Res (2005) ;11: (4):1408–1415. |
[27] | Luo Y , Yamada H , Chen X , Ryan AA , Evanoff DP , Triccas JA , O’Donnell MA . Recombinant Mycobacterium bovis bacillus Calmette-Guerin (BCG) expressing mouse IL-18 augments Th1 immunity and macrophage cytotoxicity. Clin Exp Immunol (2004) ;137: (1):24–34. |
[28] | Hurst JH , Hooks SB . Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol (2009) ;78: (10):1289–1297. |
[29] | Gomez DE , Armando RG , Farina HG , Menna PL , Cerrudo CS , Ghiringhelli PD , Alonso DF . Telomere structure and telomerase in health and disease (review). Int J Oncol (2012) ;41: (5):1561–1569. |
[30] | McShane LM , Altman DG , Sauerbrei W , Taube SE , Gion M , Clark GM . Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. REporting recommendations for tumour MARKer prognostic studies (REMARK). Eur J Cancer (2005) ;41: (12):1690–1696. |
Figures and Tables
Fig.1
Flowchart visualizing the selection steps leading to the final set of single nucleotide polymorphisms included in this replication study. Abbreviations: BCG: bacillus Calmette-Guérin; CNVs: copy number variations; MIBC: muscle-invasive or metastatic bladder cancer; NMIBC: non-muscle-invasive bladder cancer; SNP: single nucleotide polymorphism; UBC: urinary bladder cancer; VNTRs: variable number tandem repeats; QC: quality control.
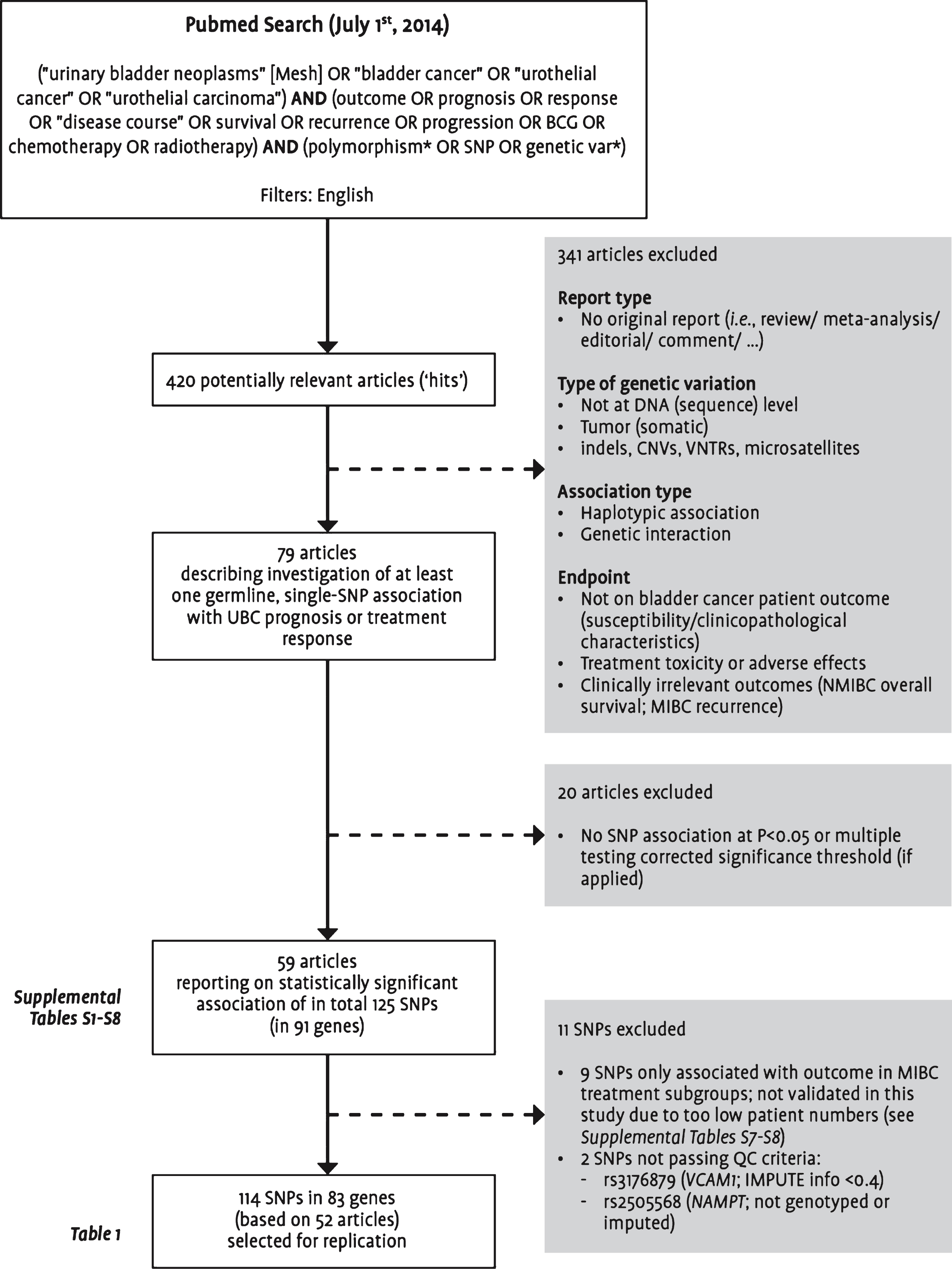
Table 1
Overview of the number of publications and postulated SNP associations per endpoint as reported and used for replication
| Reported endpoint | Reported patient (sub)group | No. publications | No. SNPs | No. genes | Endpoint and (sub)population used for replication | Study details + replication results |
| Recurrence | NMIBC | 14 | 21 | 16 | Recurrence in NMIBC | Table S1 |
| Recurrence | NMIBC+MIBC | 2 | 2 | 2 | ||
| Recurrence | TUR-only-/non-BCG-treated NMIBC | 5 | 14 | 10 | Recurrence in TUR-only-treated NMIBC | Table S2 |
| Recurrence | BCG-treated NMIBC | 27 | 37 | 32 | Recurrence in BCG-treateda NMIBC | Table S3 |
| Progression | NMIBC | 5 | 9 | 7 | Progression in NMIBC | Table S4 |
| Progression and/or cancer-specific survival | NMIBC+MIBC | 7 | 8 | 7 | Progression in NMIBC/ Overall survival in MIBC | Tables S4/S6 |
| Cancer-specific survival | Intravesical/systemic chemotherapy-treated NMIBC+MIBC | 1 | 6 | 2 | Progression in intravesical chemotherapy-treateda NMIBC (Reproducibility not evaluated in MIBC) | Table S5 |
| Progression | MIBC | 2 | 1 | 1 | Overall survival in MIBC | Table S6 |
| Cancer-specific survival | MIBC | 4 | 6 | 5 | ||
| Overall survival | MIBC | 3 | 14 | 9 | ||
| Overall survival | NMIBC+MIBC | 5 | 17 | 16 |
aTreated with at least six intravesical instillations. Please note: Numbers do not add up to the total of 114 evaluated SNPs, 83 genes, and 52 publications due to the existing overlap between the listed endpoints. Abbreviations: BCG: bacillus Calmette-Gu
Table 2
Baseline patient and tumor characteristics of the NBCS study population used for replication
| n (%) | NMIBC | MIBC | |
| n = 1,284 | n = 275 | ||
| Male gender | 1,078 (84) | 198 (72) | |
| Median age (range; in yrs) | 64 (25–92) | 64 (27–93) | |
| Smoking status | Never cigarette smoker | 200 (16) | 30 (11) |
| Ever cigarette smoker | 955 (74) | 166 (60) | |
| Unknown | 129 (10) | 79 (29) | |
| Tumor stage | 0a | 865 (67) | – |
| 0is | 50 (3.9) | – | |
| I | 352 (27) | – | |
| II | – | 136 (49) | |
| III | – | 51 (19) | |
| IV | – | 88 (32) | |
| Unknown | 17 (1.3) | – | |
| Concomitant CIS | No | 1157 (90) | 121 (44) |
| Yes | 113 (8.8) | 71 (26) | |
| Unknown | 14 (1.1) | 83 (30) | |
| Tumor gradea | Low grade | 774 (60) | 15 (5.5) |
| High grade | 497 (39) | 238 (86) | |
| Unknown | 13 (1.0) | 22 (8.0) | |
| Tumor histology | UCC | 1,272 (99) | 242 (88) |
| SCC | – | 11 (4.0) | |
| AC | 1 (0.1) | 9 (3.3) | |
| Other | 2 (0.2) | 11 (4.0) | |
| Unknown | 9 (0.7) | 2 (0.7) | |
| Tumor size | <3 cm | 182 (14) | 15 (5.5) |
| ≥ 3 cm | 94 (7.3) | 39 (14) | |
| Unknown | 1008 (79) | 221 (80) | |
| Tumor focality | Solitary | 700 (55) | 179 (65) |
| Multifocal | 504 (39) | 69 (25) | |
| Unknown | 80 (6.2) | 27 (9.8) | |
| Initial treatment NMIBCb | TUR only (±one immediate p.o. i.v. CT instillation) | 586 (46) | – |
| TUR + adjuvant i.v. CT | 366 (29) | – | |
| TUR + adjuvant i.v. IT | 243 (19) | – | |
| TUR + both adjuvant i.v. CT and IT | 23 (1.8) | – | |
| Immediate radical cystectomy | 19 (1.5) | – | |
| Other | 1 (0.1) | – | |
| Unknown | 46 (3.6) | – | |
| Median time at risk (IQR; in yrs) | Recurrencec | 2.9 (1.2–5.1) | – |
| Progressionc | 4.9 (3.0–7.6) | – | |
| Overall deathc | – | 6.2 (3.0–9.7) |
aLow grade: WHO 1973 differentiation grade 1 or 2, WHO/ISUP 2004 low-grade, or Malmström (Modified Bergkvist) grade 1 or 2a; High grade: WHO 1973 differentiation grade 3, WHO/ISUP 2004 high-grade, or Malmström (Modified Bergkvist) grade 2b or 3. bReplication among NMIBC treatment subgroups was performed among 463 patients treated with TUR only (without one p.o. i.v. CT instillation), and 192 and 311 patients initially treated with ≥6 intravesical instillations of BCG or chemotherapy, respectively. cFive-year Kaplan-Meier risk of recurrence, progression (NMIBC), and overall death (MIBC) is 50.5% , 17.2% , and 32.4% , respectively. Abbreviations: AC: adenocarcinoma; CIS: carcinoma in situ; CT: chemotherapy; IT: immunotherapy; i.v.: intravesical; MIBC: muscle-invasive bladder cancer; NMIBC: non-muscle-invasive bladder cancer; p.o.: post-operative; SCC: squamous cell carcinoma; TUR: transurethral resection; UCC: urothelial cell carcinoma.
Table 3
Summary of significant associations (p < 0.05) between candidate SNPs and prognostic and predictive endpoints in the NBCS patient series
| Endpoint | Gene | SNP | M/ I | Info | HWE pa | A1b | A2b | Genotype counts | Model | |||||
| (NBCS1/ NBCS2) | (A1A1/A1A2/A2A2)c | Unadjusted | Adjustedd | |||||||||||
| Event | No event | HR (95% CI) | pe | HR (95% CI) | pe | |||||||||
| NMIBC –recurrence | RGS13 | rs3795617 | M | M | 1 | C | T | 136/318/144 | 204/335/147 | ADD | 1.13 (1.01–1.27) | 0.033 | 1.12 (0.99–1.26) | 0.080 |
| DOM | 1.26 (1.04–1.53) | 0.016 | 1.20 (0.98–1.47) | 0.085 | ||||||||||
| REC | 1.10 (0.91–1.33) | 0.308 | 1.12 (0.92–1.37) | 0.268 | ||||||||||
| BCG-treated | ERCC2 | rs1799793f | I | 0.927 0.904 | 0.74 | C | T | 27/31/15 | 49/62/8 | ADD | 1.49 (1.03–2.15) | 0.033 | 1.55 (1.05–2.31) | 0.029 |
| NMIBC –recurrence | DOM | 1.21 (0.73–2.00) | 0.462 | 1.16 (0.67–2.01) | 0.594 | |||||||||
| REC | 2.76 (1.53–4.99) | 7.88×10–4 | 3.41 (1.79–6.47) | 1.80×10–4 | ||||||||||
| BCG-treated | IL18 | rs187238f | I | 0.999 0.998 | 0.54 | C | G | 35/28/10 | 65/48/6 | ADD | 1.38 (0.97–1.96) | 0.073 | 1.74 (1.18–2.57) | 5.49×10–3 |
| NMIBC –recurrence | DOM | 1.25 (0.79–1.98) | 0.340 | 1.59 (0.97–2.62) | 0.066 | |||||||||
| REC | 2.35 (1.20–4.59) | 0.013 | 3.57 (1.65–7.72) | 1.20×10–3 | ||||||||||
| TUR-only-treated | SUFU | rs11594179 | M | M | 0.49 | C | T | 150/90/3 | 143/62/15 | ADD | 0.95 (0.76–1.18) | 0.635 | 0.99 (0.79–1.24) | 0.929 |
| NMIBC –recurrence | DOM | 1.09 (0.84–1.42) | 0.497 | 1.17 (0.89–1.53) | 0.255 | |||||||||
| REC | 0.23 (0.07–0.72) | 0.012 | 0.24 (0.08–0.75) | 0.014 | ||||||||||
| NMIBC | RGS4 | rs6678136f | M | M | 1 | G | A | 55/105/37 | 389/506/192 | ADD | 1.18 (0.97–1.43) | 0.103 | 1.15 (0.93–1.44) | 0.205 |
| –progression | DOM | 1.39 (1.02–1.90) | 0.038 | 1.31 (0.92–1.85) | 0.130 | |||||||||
| REC | 1.08 (0.75–1.54) | 0.677 | 1.09 (0.74–1.63) | 0.653 | ||||||||||
| NMIBC | RGS5 | rs11585883f | I | 0.984 0.986 | 0.06 | T | C | 166/29/2 | 936/150/1 | ADD | 1.20 (0.83–1.75) | 0.339 | 1.21 (0.80–1.81) | 0.367 |
| –progression | DOM | 1.13 (0.76–1.66) | 0.548 | 1.13 (0.74–1.72) | 0.585 | |||||||||
| REC | 10.0 (2.47–40.6) | 1.27×10–3 | 6.76 (1.57–29.2) | 0.010 | ||||||||||
| MIBC –overall | RGS5 | rs12035879f | I | 0.619 0.607 | 4.7×10–3 | G | A | 23/44/20 | 62/101/24 | ADD | 1.72 (1.13–2.61) | 0.011 | 1.49 (0.97–2.29) | 0.066 |
| survival | DOM | 1.76 (0.88–3.54) | 0.110 | 1.63 (0.78–3.43) | 0.195 | |||||||||
| REC | 2.79 (1.40–5.56) | 3.47×10–3 | 2.10 (1.01–4.37) | 0.048 | ||||||||||
| MIBC –overall | TERT | rs2075786f | I | 0.719 0.721 | 0.19 | G | A | 34/36/17 | 81/86/21 | ADD | 1.27 (0.90–1.80) | 0.171 | 1.56 (1.09–2.24) | 0.015 |
| survival | DOM | 1.18 (0.71–1.97) | 0.527 | 1.47 (0.88–2.47) | 0.142 | |||||||||
| REC | 1.93 (1.01–3.68) | 0.045 | 2.97 (1.50–5.90) | 1.81×10–3 | ||||||||||
aHWE p-value was calculated based on measured or best-guess genotypes in the total group of study subjects included for imputation (n = 1,601). bA1: major/reference allele; A2: minor/predictive allele (both according to plus (+) strand orientation). cExpected genotype counts for imputed SNPs were calculated using SNPTEST v2 based on the sum of probabilities across all individuals in the respective patient subgroup, and rounded to the nearest whole number. dIn NMIBC (subgroups): adjusted for gender (male/female), age (<60/60–70/>70 yrs), tumor stage (Ta/T1/CIS), tumor grade (low/ high grade), concomitant CIS (no/yes), tumor focality (solitary/multifocal), and in case of analysis of the total NMIBC group, treatment (TURT only±1 p.o. CT instillation/ adjuvant i.v. CT/ adjuvant i.v. IT/ both adjuvant i.v. CT and IT); In MIBC: adjusted for gender (male/female), age (continuous), and aggregate measure based on TNM classification (i.e. tumors of stage T2-T4a with N0/NX and M0/MX vs. tumor of stage T4(b) ór any T with N ≥1/N+ and/or M1). eValues in boldface type indicate significance at P < 0.05. fSNPs with directionally consistent association compared with original publication. Abbreviations: ADD: additive; BCG: bacillus Calmette-Guérin; CI: confidence interval; DOM: dominant; HR: hazard ratio; HWE: Hardy-Weinberg equilibrium; I: imputed; M: measured (genotyped); MIBC: muscle-invasive or metastatic bladder cancer; NMIBC: non-muscle-invasive bladder cancer; REC: recessive; SNP: single nucleotide polymorphism; TUR: transurethral resection.
Table 4
Summary of previous publications reporting associations with SNPs that meet the p < 0.05 threshold in the current replication study
| Reference | Gene | SNP | N (n events) | Allelesa | Genotype counts | Model | HR (95% CI) | p | Directionally consistent replication in current study |
| Lee EK et al. [13] | RGS13 | rs3795617 | 421 NMIBC (232 R) | G/A | NR | ADD | 0.79 (0.65–0.96) | 0.019 | N |
| RGS4 | rs6678136 | 421 NMIBC (85 P) | G/A | NR | DOM | 2.07 (1.20–3.57) | 9.4×10–3 | Y | |
| RGS5 | rs11585883 | 421 NMIBC (85 P) | A/G | NR | DOM | 1.93 (1.12–3.32) | 0.018 | Y | |
| RGS5 | rs12035879 | 325 MIBC (144 OD) | G/A | NR | REC | 1.65 (1.02–2.66) | 0.039 | Y | |
| Gangawar R et al. | ERCC2 | rs1799793 | 74 BCG-NMIBC (35 R) | G/A | 31/28/15 | GENO | 0.64 (0.25–1.64) | 0.356 | Y |
| [16] | 3.07 (1.22–7.68) | 0.016 | |||||||
| Jaiswal PK et al. | IL18 | rs187238 | 78 BCG-NMIBC (34 R) | G/C | 36/39/3 | GENO | 2.35 (1.09–5.10) | 0.030 | Y |
| [17] | 2.43 (0.50–11.79) | 0.269 | |||||||
| Chen M et al. | SUFU | rs11594179 | 141 TUR-only-NMIBC | G/A | GG: 93 | DOM | 1.57 (1.00–2.45) | 0.05 | N |
| [8] | (92 R) | GA+AA: 48 | |||||||
| Andrew AS et al. | TERT | rs2075786 | 410 MIBC (172 OD) | T/C | 164/198/57 | GENO | 0.8 (0.6–1.1) | NR | Y |
| [22] | DOM | 0.5 (0.3–1.0) | (Logrank | ||||||
| 0.8 (0.5–1.0) | p = 0.1) |
aFirst allele is reference/major allele, and second allele is predictor/minor allele. Abbreviations: ADD: additive; BCG: bacillus Calmette-Guérin; CI: confidence interval; DOM: dominant; GENO: genotypic; HR: hazard ratio; MIBC: muscle-invasive or metastatic bladder cancer; NMIBC: non-muscle-invasive bladder cancer; NR: not reported; OD: overall death; P: progression; R: recurrence; REC: recessive; SNP: single nucleotide polymorphism; TUR: transurethral resection.




