Cholesterol and nicotinic acetylcholine receptor: An intimate nanometer-scale spatial relationship spanning the billion year time-scale
Abstract
Once the sterol biosynthetic machinery had progressed over the course of several million years to yield cholesterol, this neutral lipid became an omnipresent and essential component of biomembranes in Eukaryotes. The hopanoids in Prokaryotes and eukaryotic sterols share the ability to provide stability and domain compartmentalization in membranes. Even more important is the intimate association of cholesterol with a wide range of cell-surface membrane proteins, probably responsible for its modulatory effects on neurotransmitter and hormone receptors and ion channels. These effects appear to be exerted via the membrane-embedded segments of essentially all members of the pentameric ligand-gated ion channel and G-protein-coupled receptor superfamilies, which possess consensus linear arrays of amino acid residues recognizing cholesterol with relatively high affinity and specificity, an early evolutionary acquisition already present in ancient bacteria, conserved, and further improved in Eukaryotes. This review focuses on the long-term relationship between cholesterol and these functionally important membrane protein superfamilies, and the ability of cholesterol to induce lateral segregation and ordered domain formation at the nanoscale in cell membranes.
Abbreviations
nAChR | nicotinic acetylcholine receptor |
pLGIC | pentameric ligand-gated ion channels |
GPCR | G-protein coupled receptors |
1.Introduction
This review is part of a series celebrating the 200th anniversary of the discovery of cholesterol. The finding of the neutral lipid in gallstones and the coining of the term “cholesterine” (from the Greek chole for bile and stereos for solid) in 1816 is credited by some authors (e.g. [59,135]) to the French chemist Michel Eugène Chevreul [34], although other authors (e.g. [101]) attribute the discovery, also in gallstones, to the French chemist and physician François Poulletier de la Salle, in 1789. It was only in 1859 that cholesterol was identified as an alcohol by Berthelot [59] and in 1888 F. Reinitzer determined its empirical formula. The 1926 Nobel prize in Chemistry was awarded to Heinrich Wieland for his work on the structure of cholesterol and bile acids – although the tetracyclic ring structure as we know it today was proposed by Wieland only in 1932 [20]. It took the two subsequent decades to clarify the stereochemistry and ring conformations of cholesterol. The 1964 Nobel award in Physiology and Medicine went to Konrad Bloch and Feodor Lynen for their discoveries in the areas of cholesterol biosynthesis and metabolism of fatty acids [135]. In 1985 Michael S. Brown and Joseph L. Goldstein were awarded the Nobel prize for their discoveries concerning the regulation of cholesterol metabolism and the treatment of diseases caused by abnormally elevated cholesterol levels in the blood [26]. The 200th anniversary may prove an interesting opportunity to revisit some of these milestones and to assess how much we have actually learned about this lipid since its “recent” bicentennial discovery – considering that the enzymatic machinery for sterol biosynthesis may have appeared on Earth between 2.7 and 2.4 billion years ago [19], after the emergence of oxygenic photosynthesis with the concomitant oxygenation of the planet [129]. It has been surmised that the early role of sterols in eukaryotes may have been that of protection against oxidative stress when oxygen levels rose [25,56]. High oxygen concentration is required for the generation of sterols [19] and this has been postulated to be at the root of probably one of the most important changes in phylogenetic evolution, i.e. the prokaryote-to-eukaryote transition [32], coincident with the acquisition of new intracellular membrane structures and mechanisms.
A search through Pubmed with the single noun “cholesterol” yields more than 250,000 citations of scientific publications, and today these are clearly outnumbered by references with medical connotations of a dietary, metabolic or physiopathological nature, related to supra-normal levels of cholesterol (e.g. familial or acquired hypercholesterolemia, atherosclerosis and cardiovascular diseases, Alzheimer’s disease, some forms of Niemann–Pick disease) [3,28,45,50,57,76,122,126,130,144], medical conditions associated with sub-normal cholesterol levels (hypocholesterolemia, a relatively common dyslipidemia) or the more rare diseases related to genetic mutations of enzymes involved in the absorption, biosynthesis, transport or metabolism of cholesterol (e.g. chylomicron retention disease, hypobetalipoproteinemia, abetalipoproteinemia, Tangier disease, secondary hypocholesterolemias in hepatic diseases or cancer, malabsorption syndromes, excessive doses of statins, and panic disorder, among several other pathological conditions) [2,60,111,131].
Sterols in general and cholesterol in particular serve key functions in cells, ranging from structural building blocks of cell membranes to dynamic modulators of membrane physicochemical properties, like fluidity, or biological properties, like developmental regulators and precursors of steroid hormones, bile acids, and vitamin D in metazoa. Among the more than 200 lipid species in a cell membrane, cholesterol stands out as a singular lipid in terms of structure, physicochemical properties and unique effects on other lipids and membrane proteins. The mere fact that it accounts for 30–50 mol % of the total lipids of the cell membrane is already quite telling. In the central nervous system, cholesterol is a key component of neuronal and glial membranes, synaptic vesicles and, most importantly, myelin (see recent review in ref. [31]). The principal site of synthesis of cholesterol in brain is the astrocyte [86,101,102]. Although the human brain accounts for about 2% of the body weight, this organ contains almost a quarter of the total pool of cholesterol in the whole organism. Cholesterol controls synapse formation [40,77,86,102,146] and function [23], and alterations of its metabolism have been associated with various neurodevelopmental and neurodegenerative diseases, including psychiatric disorders [15,31,52].
This review will not attempt to cover the clinical aspects associated with cholesterol physiology or its alterations in disease, but will put emphasis on a narrower subject, the influence of cholesterol on membrane proteins belonging to the superfamily of pentameric ligand-gated ion channels (pLGIC). Towards this end, I will use the receptor for the neurotransmitter acetylcholine, namely the nicotinic acetylcholine receptor (nAChR), as a paradigm to illustrate the pleiotropic functional effects of cholesterol on this “founding member” of the pLGIC superfamily. As I develop the subject of the characteristic CARC cholesterol-recognition motif first recognized in these membrane proteins [8], it will become apparent that its presence extends beyond pLGIC, spreading out to the largest class of transmembrane receptors, the G-protein (heterotrimeric guanine nucleotide-binding protein)-coupled receptors (s), also called 7 transmembrane helical (7TM) receptors, and several other proteins, including soluble ones.
Cholesterol binds to and affects the ligand-recognition, gating, and ion permeation properties of a number of hormone and neurotransmitter receptors, as well as ligand- and voltage-gated ion channels [72,73]. In addition, cholesterol modulates the cell-surface organization, exocytic and endocytic trafficking, and various other physiological properties of these membrane proteins. Cholesterol influence on cell-surface organization of membrane proteins is clearly manifested in the cholesterol-mediated dynamic compartmentalization via the recruitment or exclusion of proteins in/from specialized micro- and nano-domains, on the one hand, and in the degree of oligomerization of receptors, on the other, as discussed in two sections of this review. Protein exocytic and endocytic trafficking is also affected by cholesterol homeostatic equilibrium in the cell, as analyzed in another section. Cell-surface receptor function is also subject to regulation – probably the most important of cholesterol effects – in a pleiotropic manner which is highly intertwined with the two other targets of sterol modulation. Cholesterol effects can be exerted in a specific manner involving direct interactions with the membrane-spanning domains of the receptors [8,10,49,105,106] or in a rather diffuse, less-specific manner in the form of changes in the physical state of the bulk lipid bilayer, e.g. on membrane thickness, fluidity [41,69,78] or curvature [70,153]. These two modes of action of cholesterol are widely distributed, being shared by membrane proteins unrelated in structural and evolutionary terms, and have adapted both to rapid responses like the ion channels and receptors in the pLGIC superfamily [14] and to the slow-acting and often intricate metabotropic responses as is the case with the GPCR superfamily [53]. Millions of years ago, before the appearance of the cholesterol-synthesizing enzymatic machinery in eukaryotic organisms, the hopanoids – prokaryotic sterol surrogate compounds – exerted on bacterial ion channels some of the functional effects displayed by cholesterol in Eukaryotes [16]. A section of this review will briefly analyze the intriguing connection between the two regulatory mechanisms.
2.The site of cholesterol–protein contacts: The example of the nAChR
Membrane-embedded proteins necessarily exhibit close and relatively extensive contacts with the surrounding lipids. At the hydrophobic periphery of the protein a first shell [141] of lipids constitutes the “belt” or “boundary” region, the lipid “annulus” [142], a partially ordered but dynamic interface in active exchange with the membrane bilayer bulk lipid [83]. Cholesterol was reported to be excluded from the annular lipid region [142]. Nuclear magnetic resonance (NMR), electron spin resonance (ESR), and fluorescence spectroscopy studies have provided information on the biophysical properties of annular lipids in the immediate periphery of several membrane proteins [82,121] and recently gas-phase mass spectrometry has been applied to explore the annular lipid in great detail, establishing the number of lipid molecules in this membrane subdomain [85]. The fatty acyl chains of the annular lipids are in most cases tightly packed against the hydrophobic surface of the TM region of the protein, and experience a transiently less mobile state relative to that of the bulk bilayer lipid in terms of rotational and translational dynamics [83,116,143]. The rate of exchange between annular shell lipid and the bulk lipid is remarkably fast (
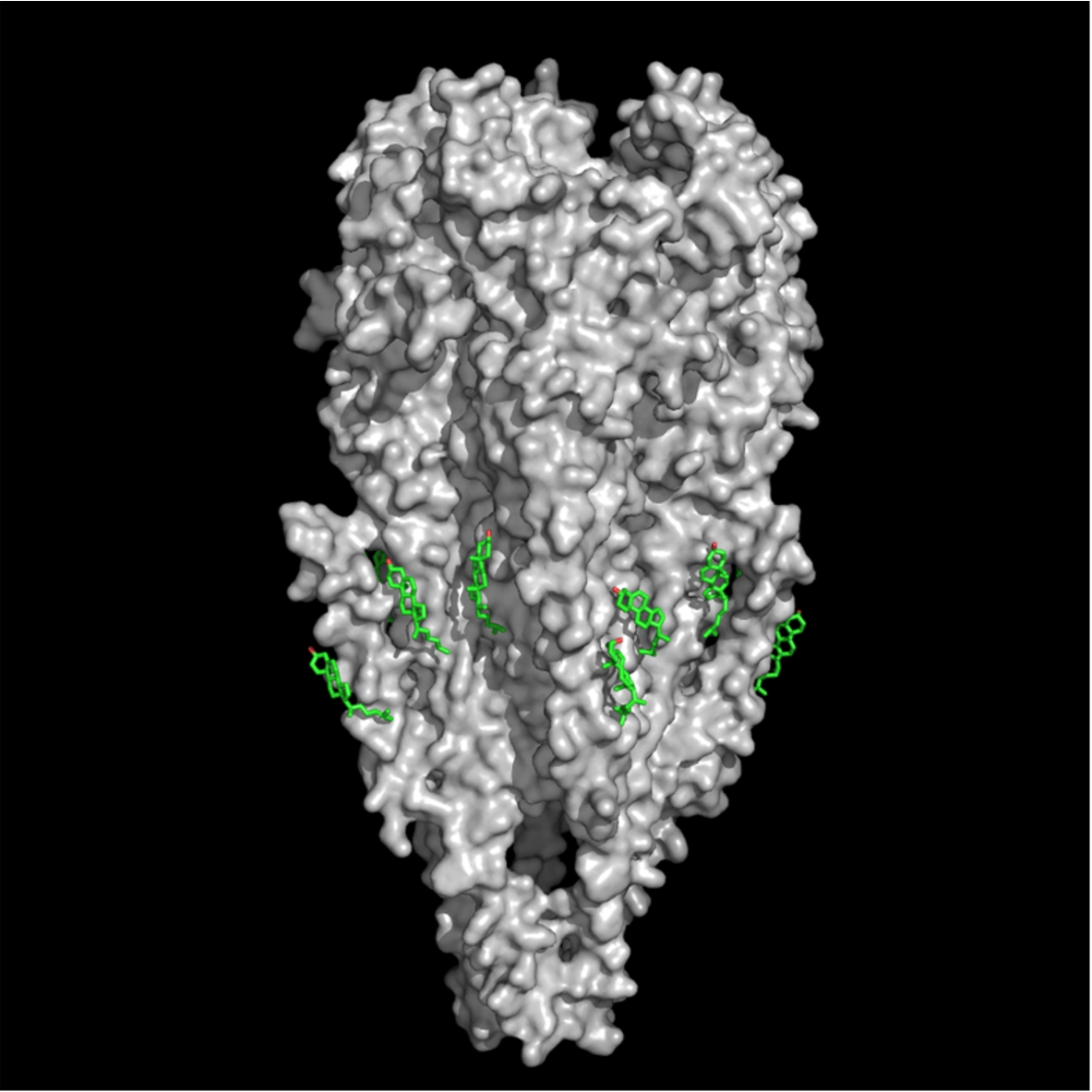
Fig. 1.
CPK molecular model of the nicotinic acetylcholine receptor protein (surface rendered in gray) (MW ca. 270,000 Daltons, PDB 2bg9) and cholesterol molecules (stick rendering, green) (PDB entry 1MT5) at cholesterol-recognition sites located on the rough membrane-embedded surface of the nAChR TM region.
In contrast to annular lipids, non-annular sites in most membrane proteins are heterogeneous in nature, and are located in grooves or cavities between TM helices and/or at the clefts between individual subunits in multi-subunit proteins [70]. Because of their intimate contact with the TM protein surfaces and their limited accessibility to the bulk lipids, non-annular lipids often exhibit higher specificities for the protein than annular sites. Their rate of exchange with the bulk lipid is presumably much slower than that of annular lipids, especially for those sites located deeper in the interior of the protein, farther away from the belt region. In contrast to the early ideas of Warren et al. explicitly indicating that cholesterol was excluded from the annular lipid region [142], subsequent work pointed to non-annular lipids as the sites that excluded cholesterol but accommodated phospholipids (Jones and McNamee, 1988). Free fatty acids resulting from controlled phospholipase A2 enzymatic treatment of Torpedo native nAChR-rich membranes or exogenously added arachidonic acid were found to localize at both annular and non-annular sites [51]. Attempts to identify the location of cholesterol sites using molecular dynamics simulations suggested the occurrence of cholesterol sites deep inside the nAChR TM region [24]. The crosstalk between the two classes of sites provided a plausible explanation for the distinct resting (R) and desensitized (D) states of the nAChR observed in the equilibrium binding studies: transitions between these conformers were interpreted in terms of structural rearrangements of the nAChR TM region involving either the occlusion or decreased availability of the non-annular sites [51].
3.Identifying cholesterol-recognition motifs in membrane proteins
Since the first report of a membrane protein structure in 1985, Stephen White’s laboratory at the University of California at Irvine has compiled 585 new structures solved up to the year 2015 (http://blanco.biomol.uci.edu/mpstruc). At the time of writing this review, the number of membrane proteins of known 3D structure listed in this compilation totals ca. 700 entries. Similarly, the MemProtMD data base created by Samson’s lab in Oxford University lists all known transmembrane proteins embedded in lipid membranes, and the group recently reported that about 2,000 structures have been solved by the combined efforts of X-ray diffraction, single-particle cryoelectron microscopy and solution nuclear magnetic resonance spectroscopy studies [125]. Thus the number of macromolecules solved at the atomic level effectively represents only a minor fraction of the genome coding for membrane proteins, which accounts for no less than 25% of the genes in a cell. It is therefore not surprising that the direct demonstration of cholesterol recognition sites in membrane protein structures is so elusive.
Which alternative approaches can be used to experimentally establish the direct contact between cholesterol and membrane proteins? A spin-labeled cholesterol analogue has not been synthesized to date; cholesterol-like analogues for electron spin resonance spectroscopy studies such as androstanol- or cholestane-spin labels are available, but they differ from cholesterol in some essential structural details. Similarly, cholesterol probes for fluorescence studies devoid of relatively bulky fluorophores are still not available. The smallest of such extrinsic probes is nitrobenzoxadiazole (“NBD”) [94], and this fluorophore adds not only a non-negligible mass to the cholesterol moiety but may also modify its location and orientation in the membrane. An alternative minimally perturbing cholesterol-like probe is dehydroergosterol [113], which inconveniently has low quantum yield, bleaches rapidly, excites in the high-energy UV region of the spectrum in conventional wide-field microscopy [148] and is thus prone to deleterious effects on cell integrity, and requires costly UV-transparent optics for fluorescence microscopy [93]. Dehydroergosterol can also be studied using 2-photon fluorescence microscopy, circumventing one of the above problems, i.e. excitation in the UV region of the spectrum [147,148]. Furthermore, fluorescent cholesterol probes can partition differentially in the plasma membrane as compared to the endogenous lipids [30]. Better extrinsic cholesterol probes, such as BODIPY-cholesterol, or the photoactivatable Ever-Fluor-fluorescein cholesterol have appeared in recent years (reviewed in [30,149]) but the essential difficulties associated with the bulky fluorophores still remain. In other words, the ideal extrinsic cholesterol probe – the bona fide qualitative tracer of ref. [30] for biophysical studies having a non-perturbing reporter group is still not available. For all these reasons other methodologies are needed to identify cholesterol recognition motifs in hormone or neurotransmitter receptors and ion channels without perturbing the sterol recognition region of these proteins. Various groups, including ours, have resorted to analyzing sequences in protein data banks and applying in silico computational methods to explore and detect the presence of putative cholesterol-binding linear domains in the transmembrane (TM) regions of proteins. This methodology resulted in the definition of consensus motifs with predictive value, which can be further applied for identifying cholesterol-binding linear domains [8,42–44,66]. The first such consensus motif to be identified was defined by the linear amino acid sequence (L/V)-X1-5-(Y)-X1-5-(K/R), and termed “cholesterol recognition amino acid consensus” (CRAC) [66]. The linear amino acid array (K/R)-X1-5-(Y/F)-X1-5-(L/V) was found later on in the nAChR and other membrane proteins; it is essentially the mirror version of the CRAC algorithm, and hence referred to as the “CARC” consensus motif [8].
The free energy of interaction between cholesterol molecules and the nAChR amounts to about
Analyses of cholesterol-recognition motifs in membrane [8,49,132] as well as soluble [89–91] proteins have provided strong evidence to sustain the hypothesis that these linear motifs bear functional relevance in cholesterol–protein interactions. Until recently, though, direct experimental demonstration of physical crosstalk between the sterol and the linear peptide sequences was missing for proteins other than GPCRs. We have recently challenged the hypothesis and submitted it to experimental test using two biophysical techniques: the interaction of a prototype CARC domain with cholesterol was studied using lipid monolayer techniques and nuclear magnetic resonance (NMR) spectroscopy. A representative CARC domain for these experimental corroborations was chosen on the basis of previous photolabeling studies of the Torpedo nAChR with the cholesterol analogue probe [3H]azicholesterol, which led to the identification of a (predominant) cholesterol-binding domain in the 4th transmembrane domain (TM4) of the human nAChR γ subunit [62]. As mentioned in preceding paragraphs, our in silico computational approaches [8] led us to identify a typical CARC motif: 455-RVCFLAML-462 (the characteristic Arg, Phe, and Leu amino acid residues outlined bold and underlined) in the human γTM4 that incorporated most of the label in the photoaffinity studies. In addition, molecular modeling simulations (Fig. 2) showed that this TM segment displayed the highest energy of interaction (in the order of
Fig. 2.
Molecular model of the human nAChR γ subunit 4th transmembrane domain (surface rendered in red) with bound cholesterol (CPK rendering, in yellow). This TM segment (Swiss Prot entry P02708), located in the outer membrane leaflet, exhibits the highest energy of interaction with cholesterol, i.e. ca.
![Molecular model of the human nAChR γ subunit 4th transmembrane domain (surface rendered in red) with bound cholesterol (CPK rendering, in yellow). This TM segment (Swiss Prot entry P02708), located in the outer membrane leaflet, exhibits the highest energy of interaction with cholesterol, i.e. ca. −60kJ·mol−1 [8].](https://content.iospress.com:443/media/bsi/2016/5-S1/bsi-5-s1-bsi158/bsi-5-bsi158-g002.jpg)
Additional molecular dynamics studies on the cholesterol derivative-Torpedo γTM4 segment comprising the CARC domain plus a few upstream and downstream amino acid residues (445–460) indicated that azicholesterol did interact tightly with the residues defined by the CARC motif (Lys-449, Phe-452 and Leu-456), although it seemed unlikely that the strength of the interaction sufficed to account for the reduced mobility observed in the molecular simulations; instead, we proposed that the reduced mobility arises from the cholesterol-induced oligomerization of γTM4, a hypothesis consistent with the results of fluorescence studies of γTM4 in POPC vs POPC-cholesterol bilayers [38]. The molecular dynamics studies further indicated that the cholesterol contacts with the CARC motif within γTM4 involved the β-face, leaving the α-face exposed. This suggested that the cholesterol-mediated oligomerization of the peptide is dictated by cholesterol–cholesterol interactions rather than by protein–protein interactions [48]. We next compared the interaction between cholesterol and either wild-type or mutant TM4 peptides. The WT CARC motif was found to exhibit a higher affinity for cholesterol. The three amino acids defining the CARC domain, and the central Phe-452 residue in particular, were found to interact with cholesterol. Replacement of this aromatic residue with alanine (F-452/A mutant) resulted in a significant loss of affinity. This computational finding was challenged by physicochemical lipid monolayer studies using a CARC-carrying peptide and cholesterol. The fact that the F-452/W mutation had no effect led us to suggest that it is the aromatic nature of Phe-452, and not its specific structure, that is required for optimal binding [47], in agreement with previous studies suggesting that CARC motifs could contain any of the three aromatic residues, i.e. Phe, Trp or Tyr [8,49]. Importantly, the monolayer studies also demonstrated the lipid-specificity (CARC recognized cholesterol but not phosphatidylcholine) and the concentration dependency of the binding (saturation was reached for peptide concentrations <10 μM) [47]. NMR spectroscopy experiments (MAS triple resonance magic-angle spinning deuterium NMR using deuterated Ala471) [47] showed that cholesterol addition to phospholipid bilayers containing a synthetic 13C/15N-labeled peptide (Asp464-Val492 in the intact Torpedo γTM4) caused a reduction in the rotational motion of the peptide within the bilayer, further reinforcing the cholesterol-mediated peptide oligomerization hypothesis. It is tempting to extrapolate the results of the isolated peptide experiments to the possible role of the CARC domains in situ, where they could play a role in cholesterol-mediated oligomerization of the receptor. The high affinity, lipid-specific and saturable nature of the converging lipid monolayer and NMR spectroscopy studies, together with the molecular modeling simulations, constitute a much sturdier argument for the direct physical interaction of cholesterol with a CARC cholesterol-recognition motif [47].
4.The same transmembrane domain can accommodate two cholesterol molecules
Probably one of the most valuable aspects of in silico molecular simulation studies is their predictive value. One prognostic outcome of our molecular modelling exercises was the unexpected finding that two cholesterol molecules could actually be accommodated on the same TM domain [47,49]. This inference follows from the vectorial nature of the CRAC and CARC motifs, mirror images on the linear sequence (“apolar” Leu/Val → “basic” Lys/Arg for CRAC and “basic” Lys/Arg → “apolar” Leu/Val for CARC, from the N-terminus to the C-terminus sequence). The CARC sequence starts with a basic residue (Arg or Lys) and this feature makes the CARC motif ideally suited for interaction with cholesterol in the outer leaflet of biological membranes (Fig. 3). The N-terminal domain of type I membrane proteins is extracellular, such that the carbon chain enters the membrane bilayer in the N- to C-terminus direction. A search for cholesterol-recognition motifs over a large series of membrane proteins in combination with molecular dynamics simulations of the whole TM regions showed that, indeed, double cholesterol-recognition motifs occur within the same TM segment in various members of the GPCR superfamily [47]. Interestingly, some commonalities were apparent: a) the three key amino acid residues defining the CARC and CRAC motifs were always involved in the interaction; b) the central aromatic residue could be either Phe or Tyr (and even Trp in the case of CARC). This finding is consistent with the nature of the interaction between cholesterol and aromatic rings in amino acid residues, i.e. the CH–π stacking interaction [96]; c) the branched aliphatic residues (Leu/Val) are well suited to accommodate the protruding methyl groups of cholesterol; d) the terminal basic residue of the motif often forms a hydrogen bond with the oxygen atom of the –OH group of cholesterol [49]; e) the consensus CRAC sequence starts with an aliphatic residue (Leu or Val), and therefore its N-terminal is expected to interact with the apolar groups of cholesterol (sterane, methyl and iso-octyl) in the inner leaflet (Fig. 3). Thus, the selection of CARC and CRAC as cholesterol-binding motifs is justified by robust physicochemical rules [47]. The coexistence of a CARC and a CRAC motif within the same TM segment, one in each leaflet of the membrane, has an important consequence: the host transmembrane protein segment can accommodate two opposite (tail-to-tail) cholesterol molecules (Fig. 3). This ensures that the polar amino acid residues of the motif (Lys/Arg) face the intra- and extracellular milieu, whereas the apolar ends of the motifs (Leu/Val) are deeply buried in the most hydrophobic region of the lipid bilayer. The coexistence of CARC and CARC motifs in the same TM domain is found in type I membrane proteins and in domains 1, 3, 5, and 7 of the GPCRs; the CARC motif is always located in the outer leaflet, whereas the CRAC sequence is found in the inner leaflet. Membrane receptors such as the somatostatin, GABA, serotonin, adenosine, VIP and cannabinoid receptors, as well as the voltage-dependent TRVP1 channel exhibit the mirror cholesterol-recognition motifs. Overall, the mean energy of interaction was in the order of
Fig. 3.
Coexisting CARC and CRAC motifs within the same TM domain. The amino acid residues in the CARC and CRAC motifs exhibit a defined hydrophilic/hydrophobic vectoriality. Furthermore, the predominant occurrence of the CARC motif is in the outer membrane leaflet, and the CRAC array is preferentially located in the inner leaflet. Their vectorial orientation is schematically depicted in the left panel. The cholesterol molecule binding the CARC domain is shown with its –OH group pointing towards the aqueous compartment of the extracellular space. Correspondingly, the hydrophobic/hydrophilic vectoriality of the CRAC motif on the inner leaflet matches the orientation of cholesterol, with its –OH group pointing towards the cytoplasmic compartments. On the right, the example of the CARC/CRAC doublet in the 1st TM domain of the human VIP receptor. The 3D structure of cholesterol was retrieved from PDB entry 1MT5. From ref. [46].
![Coexisting CARC and CRAC motifs within the same TM domain. The amino acid residues in the CARC and CRAC motifs exhibit a defined hydrophilic/hydrophobic vectoriality. Furthermore, the predominant occurrence of the CARC motif is in the outer membrane leaflet, and the CRAC array is preferentially located in the inner leaflet. Their vectorial orientation is schematically depicted in the left panel. The cholesterol molecule binding the CARC domain is shown with its –OH group pointing towards the aqueous compartment of the extracellular space. Correspondingly, the hydrophobic/hydrophilic vectoriality of the CRAC motif on the inner leaflet matches the orientation of cholesterol, with its –OH group pointing towards the cytoplasmic compartments. On the right, the example of the CARC/CRAC doublet in the 1st TM domain of the human VIP receptor. The 3D structure of cholesterol was retrieved from PDB entry 1MT5. From ref. [46].](https://content.iospress.com:443/media/bsi/2016/5-S1/bsi-5-s1-bsi158/bsi-5-bsi158-g003.jpg)
In weighing the possible biological implications of the coexisting CARC-CRAC motifs within the same membrane-embedded peptide we have noticed the special case of the yeast heterologous protein expression system. Functional expression of some human receptors is not always sustained by yeasts [97]. Yeast cells have an essential requirement for ergosterol for their growth, and cholesterol is not an effective substitute. However, Saccharomyces yeast cells transfected with the human μ-opioid receptor increased ligand binding when ergosterol was replaced by cholesterol [68]. In order to account for this observation, we have hypothesized [47] that human CARC/CRAC domains of human origin might exhibit species-specificity for cholesterol as manifested in the yeast expression system due to subtle differences between the two sterol molecules, cholesterol being more flexible due to the presence of several extra-double bonds in ergosterol [7,37].
5.Cholesterol effects on the oligomeric state of G-protein-coupled receptors
GPCRs constitute the largest superfamily of transmembrane proteins, with more than 900 members encoded by the human genome [55]. Their biomedical and economic importance is outlined by the fact that 40% of the marketed drugs produced by the pharmaceutical industry act on GPCRs [35,65,67]. This group of macromolecules is involved in a great variety of physiological processes, including the decoding of signals encoded in neurotransmitters, hormones, chemokines, proteinases, odorant and taste molecules… and photons. They bear in common the ability to transduce external cell surface-targeted stimuli into relatively slow, downstream chemical metabolic cascades inside the cell. The common features also encompass molecular structure: GPCRs are built of seven transmembrane segments (TM1–TM7) and hence are also referred to as 7TM receptors. Upon signal recognition (I avoid the terminology “ligand-recognition” here, because GPCRs recognize not only chemical ligands like hormone peptides or even proteins, but also ions, and even photons as is the case with the GPCR rhodopsin), GPCRs transduce the message to a G-protein at its C-terminal, intracellular domain. The heterotrimeric G-proteins also come in a variety of combinations, transducing inhibitory and excitatory downstream signals. In the context of this review, cholesterol is gaining increasing relevance in GPCR function as a possible modulator of GPCR oligomerization. A matter of debate for more than a decade, it is now more widely accepted that various types of GPCRs undergo dimerization or higher-order oligomerization prior to activation [5], and although monomers in family type A GPCRs are functional, heteromerization appears to be fundamental in receptor trafficking and pharmacology [55]. Current views strongly point to the membrane microenvironment as the key modulator of GPCR oligomerization [54] and more specifically to cholesterol as the dimerization-inducing membrane component [54,104]. The availability of the crystal structure of the β2 adrenergic receptor provided not only the first experimental observation of direct interactions between a GPCR and cholesterol [33,63,114] but also led to elaborating about the importance of the sterol, necessary for the crystallization of the protein, in establishing the dimeric form of this receptor in the crystal lattice [33]. These seminal crystallographic structures of the β2 adrenergic receptor expedited subsequent X-ray studies on binding sites in various other GPCRs like the β1 adrenergic receptor in complex with a G-protein [140] or the human A2A adenosine receptor bound to an agonist [64], and led to the identification of cholesterol sites in several other members of the GPCRs (reviewed e.g. in [133]). Up to the end of 2015, the X-ray structures of more than 60 ligands and 20 receptors had been reported, corresponding to GPCRs class A, B, C and F [35]. Cholesterol-GPCR interactions have been reported to increase the compactness of β-adrenergic and 5-HT1A receptor structures and to enhance the conformational stability towards active or inactive receptor states [58,107]. An additional actor in GPCR dimerization is the fatty acid, palmitic acid [6]. The covalent palmitoylation of the β-adrenergic receptor at two sites appears to recruit cholesterol molecules involved in the stabilization of the receptor dimers (reviewed in [54]) and in the localization of the 5-HT1A serotonin receptor in lipid microdomains [112]. Cholesterol was reported to modulate the membrane partitioning in the plasma membrane [109] and the ligand binding properties [108,120] of this GPCR.
6.Properties of cholesterol in liquid-ordered (Lo) lipid domains
As analyzed in a preceding section, cholesterol can occur in the inner or outer leaflet of the membrane bilayer, or in both leaflets. In general, the outer leaflet possesses a more rigid apolar surface than glycerophospholipids, and this facilitates the preferential interaction with cholesterol. The cholesterol-sphingomyelin domains are also enriched in glycerophospholipids with saturated fatty acyl chains (relative to the average saturation in the rest of the bilayer). The lipid raft hypothesis proposes that these ternary complexes formed by specific self-associated lipid species constitute microdomains or platforms that can intervene in protein partition, signaling and other functional events in cell physiology [4,75,123,124]. Favorable and unfavorable lipid-lipid interactions result in transient lateral heterogeneities that put together or segregate their constituent molecules, respectively. Above/below certain critical lipid and water concentrations and/or temperatures these lateral heterogeneities generate transiently separated lipid phases, the two most prominent of which are the liquid-disordered (Ld) and liquid-ordered (Lo) phases [81,152]. Other factors are also influential in the constitution and transitions between these two (and additional) phases, such as variations in the ionic strength, pH and pressure. Furthermore, the action of lipases can retailor the composition and topography of lipids in the membrane, and hence redesign the Ld and Lo. For instance endogenous flippases (e.g. the scramblases) acting in situ can alter the distribution of lipids across the two leaflets of the membrane whereas other enzymes can modify the fatty acyl chain composition and hence modify the degree of saturation of the membrane phospholipids, a key factor affecting the phase state of the membrane. The temperature and compositional range over which these lateral separations into liquid phases occur is rather large [136–139]. Lipid domains enriched in cholesterol–sphingomyelin-saturated glycerophospholipids are generally considered the “consensus” Lo phase in “test tube” model systems. These ternary systems constitute a more condensed, rigid and thicker bilayer which has been associated with the concept of “lipid raft” [103], although not all rafts appear to contain sphingomyelins. The tighter packing of the phospholipid acyl chains in the presence of the sterol has been termed “condensing effect”. Outside these Lo domains cholesterol associates with other glycerophospholipids (mainly phosphatidylcholines) in a rather loose manner, and at relatively lower concentrations. There is abundant experimental evidence to support the notion that Lo domains exist in binary phospholipid (DPPC)-cholesterol mixtures (see recent review in ref. [119]).
Lipid domains apparently cover a wide range of sizes, from assemblies with <5 nm radius (“utrananodomains” [99]), comprising a couple of hundred lipid molecules per bilayer, to micron-sized platforms with thousands of molecules readily observable by conventional wide-field light microscopy [61,87,110,134]. Functionally, lipid domains play important roles in the cell by way of the lateral separation of chemical species in the plane of the membrane. Membrane-embedded proteins with preferential affinities for Lo or Ld domains could influence both the lifetime and size of the domains in which they are located by selecting their local lipid environment.
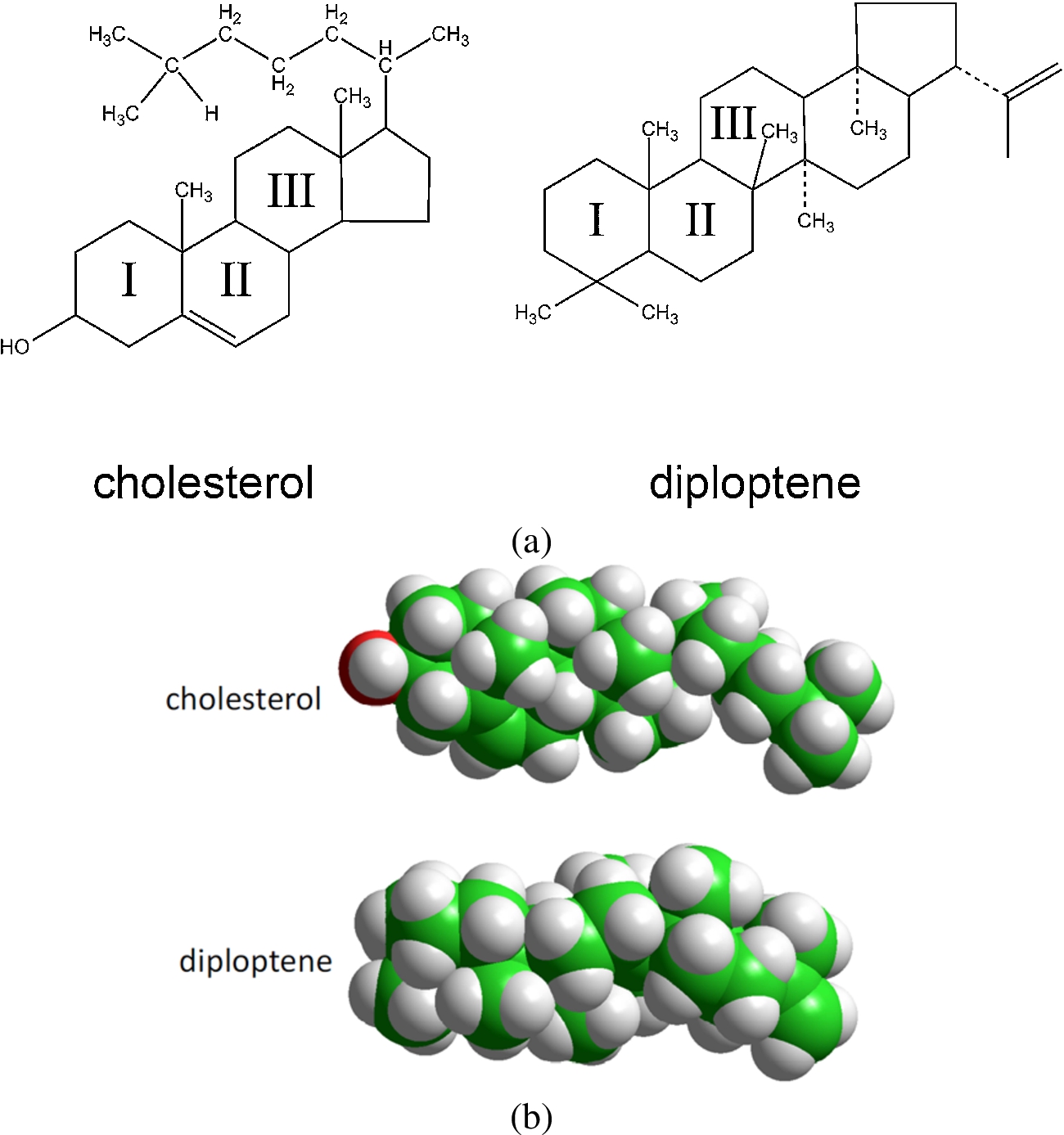
Fig. 4.
Chemical formulae (a) of cholesterol and the hopanoid diploptene and (b) CPK representation of the same molecules.
7.Possible raisons d’être for cholesterol induction of ordered lipid domains
The ability of cholesterol to promote the formation of liquid-ordered domains in a biological membrane stems to a large extent from the long evolutionary process towards optimizing the sterol surface chemistry to two complementary but somewhat contrasting demands: good match with other lipids on the one hand, and with membrane proteins, on the other. The bifacial geometry of cholesterol appears to be the outcome of this struggle [48]. The dissymmetrical nature of the molecule is manifested in the contrast between the rough β face, with methyl substituents in C10 and C13 and an iso-octyl chain in C17, with the featureless, smooth α face. The planar α face is optimally suited to interact with sphingomyelin or the phospholipid fatty acyl chains, a requirement probably driven towards the induction of order in the acyl chain conformations, to ensure tight packing of the sterol with phospholipid saturated acyl chains, a collective property that promotes the formation of Lo domains [150,151]. This capacity varies among sterols; using spin-label fluorescence quenching and DPH polarization techniques we found that most sterols have a tendency to disrupt lipid ordered domains, whereas it is precisely cholesterol and to a lesser extent 25-hydroxycholesterol that exhibit the highest propensity to form and stabilize ordered lipid domains [145]. This obviously implied the tuning of the enzymatic machinery to progress from lanosterol to cholesterol. From in vivo experiments with eukaryotic sterol auxotrophs and microviscosity measurements of sterol-containing artificial membranes with the cholesterol precursor lanosterol (4,4′,14 trimethyl cholastadienol) Bloch has argued that lanosterol is “incompetent for membrane function” [19]. In this sense, lanosterol would not only be the precursor of cholesterol in the sterol biosynthetic pathway but also its predecessor in the molecular evolution to cholesterol; that is, the temporal sequence of cholesterol biosynthesis can be assumed to recapitulate the evolutionary sequence. And what is the purpose of the appearance of cholesterol in the phylogenetic scale? Using a combination of deuterium NMR spectroscopy on multilamellar lipid-sterol systems in combination with Monte Carlo simulations of microscopic models of lipid-sterol interactions, Mouritsen and coworkers [88] concluded that the evolution in the molecular chemistry from lanosterol to cholesterol is manifested in the increase in the ability of the sterols to promote and stabilize lipid order, in the form of the Lo phase in model phospholipid-sterol membranes. However, experimental work using the simplest of bacterial sterol surrogates, the hopanoid diplopterol, demonstrated that this property was also present in bacteria early in evolution [117,118]. The second requirement, i.e. the optimal adjustment of a sterol surface to the complementary surface of a membrane protein [48] is also likely the consequence of evolutionary pressures exerted on both partner molecules in the course of phylogeny [16]. As can be seen in Fig. 4, the main difference between hopanoids and sterols is the loss of a ring from the pentacyclic triterpenoids, the hopanoids, to the tetracyclic triterpenoids, the sterols. This long-term tuning was associated with the stabilization of amino acid motifs in contact with the rough surface of the sterol molecule [48], which is manifested in the strong conservation of amino acid residues in tight contact with membrane lipids exhibited by membrane proteins [1].
8.Occurrence of receptors in postsynaptic ordered lipid domains
The chemical analysis of the postsynaptic apparatus in the peripheral nervous system shows that cholesterol is a very abundant component (see review in [9]) of this specialized membrane. This sterol is an essential partner of the nAChR, affecting its distribution and several of its functional properties in the peripheral synapse, the neuromuscular junction [12,13]. The lateral heterogeneity of lipids in the postsynaptic membranes of the Torpedo electrocyte was an early biophysical finding: protein-associated lipids were shown to be immobilized with respect to bulk membrane lipid [83], and subsequent work has shown that cholesterol-like molecules form part of this protein-immobilized pool [11]. The functional implications of this finding became apparent when it was demonstrated that cholesterol is an essential component for maintaining nAChR agonist-dependent state transitions in the postsynaptic membrane [36]. It has been proposed that there are two cholesterol populations in nAChR-rich membranes from Torpedo: an easily extractable fraction that influences the bulk fluidity of the membrane and a tightly bound receptor-associated fraction [71].
Signaling via the agrin/MuSK complex and interaction between the receptor and rapsyn appears to involve Lo-type lipid platforms [154]. The formation of micron-sized nAChR clusters induced by agrin in muscle cells has been found to be influenced by cholesterol [29]. Laurdan two-photon fluorescence microscopy was used to image nAChR clusters in association with Lo membrane domains [127]. Another study [146] proposed that these cholesterol-rich lipid microdomains and Src-family kinases both contribute to stabilizing nAChRs and the postsynaptic apparatus. A pool of nAChRs has indeed been found in Lo domains in mammalian cells [27,29,80,127,146,154]. In contrast, when reconstituted in a sphingomyelin–cholesterol–POPC (1:1:1) model system, purified nAChR protein from Torpedo was found not to exhibit any preference for Lo domains in vitro [17]. However, inclusion of some sphingomyelin molecular species (brain sphingomyelins, 16:0, 18:0, or 24:1 sphingomyelins) that generate bilayer asymmetry by enriching the sphingolipid content of the outer leaflet of the lipid bilayer appear to favor the partitioning of the nAChR in Lo domains [100]. This can be correlated with the observation that Lo domains in the outer leaflet of a bilayer can induce liquid order in the inner leaflet by a coupling mechanism involving in-register Lo domains in the two halves of the bilayer [74].
9.Evolutionary aspects of hopanoid/sterol-protein interactions in membranes
The appearance in phylogeny of the sterol biosynthetic machinery – that is the enzymes involved in the synthetic pathways of phytosterols in land plants, ergosterol in yeasts, and cholesterol in many other organisms from fungi to vertebrates – is considered a major hallmark in the development of eukaryotic cells [18,19,21]. Although cholesterol is absent from Prokaryotes [19,22,92] a selected group of bacteria such as the myxobacterium Plesiocystis pacifica possess surrogate enzymes of the sterol pathway, a singularity postulated to have been acquired via horizontal gene transfer from eukaryotes [39]. Biosynthesis of cholesterol is an extremely oxygen-intensive process necessitating sufficient terrestrial oxygen to proceed; eleven molecules of oxygen are required for the synthesis of one molecule of cholesterol [129]. Several lines of evidence suggest that cholesterol evolved at least in part as an adaptation to oxygenic conditions on planet Earth following the advent of oxygenic photosynthesis [25,56].
Isopentenyl diphosphate is the universal precursor of the triterpenoids, a class of organic compounds common to Prokaryotes and Eukaryotes. Isopentenyl diphosphate is synthesized by the 2-C-methyl-D-erytriol 4-phosphate pathway in bacteria, and through the mevalonate pathway in Archea and some Eukaryotes [98]. In bacterial cells, isopentenyl diphosphate gives rise to squalene, which is followed via the anaerobic squalene hopane cyclase activity to its cyclization products, the hopanoids (pentacyclic triterpenoids). In eukaryotic cells, isopentenyl pyrophosphate is synthesized starting with the formation of 3-hydroxy-3-methylglutaryl Co A from acetyl-CoA. Initially two molecules of acetyl-CoA condense to form acetoacetyl-CoA, a reaction catalyzed by a thiolase, and acetoacetyl-CoA further condenses with another acetyl-CoA molecule to yield HMG-CoA that is reduced to mevalonate by NADPH catalyzed by HMG-CoA reductase. This is the so-called committed, irreversible and control site in cholesterol biosynthesis. Mevalonate is further converted to 3-isopentenyl pyrophosphate. Six molecules of isopentenyl pyrophosphate, the activated isoprene form, are then condensed to form squalene. Acquisition of oxygen by squalene produces squalene epoxide, subsequently cyclized to lanosterol or to cycloartenol, tetracyclic sterols [95]. Demethylation of the methyl groups of lanosterol yield desmosterol, and the reduction of its side chain double bond finally produces cholesterol. In total, 19 steps are involved to produce cholesterol from its precursor lanosterol, out of the total 37 steps that mediate the elaborate synthesis of cholesterol in animal cells.
The appearance, improvement and maintenance of cholesterol-recognition linear amino acid motifs cover a wide evolutionary span, from Homo sapiens back to the bacterial pentameric channels, structural homologs of the nAChR, e.g. the cyanobacterium Gloebacter violaceous and its orthologue from Erwinia chrysanthemi. The former bacterium expresses hopanoids, which are structurally and functionally similar to sterols. The remarkable preservation of structural features in pLGIC proteins – including lipid recognizing consensus motifs through millions of years has led us to suggest that these domains fulfill important physiological roles [14,16]. In support of this hypothesis is the extensive experimental work showing that mutations in amino acid residues in the TM regions of the nAChR alter channel gating (see review in [11]). Some of these functionally relevant mutations are very close to or within CARC/CARC-like domains. It has been proposed that the hopanoids may have originally participated in the ordering of the membrane bilayer in bacteria [117,118]. We surmise that the counterpart, interacting lipid-recognition moiety in the transmembrane protein segments (the CARC or CRAC recognition domains), may have co-evolved with the lipid biosynthetic apparatus along phylogeny thus facilitating the capacity of some membrane proteins to be recruited into Lo domains in prokaryotic channel-forming and other membrane-embedded proteins; however, upon appearance of oxygenic photosynthesis and the cholesterol synthesizing machinery in the course of phylogeny, this lipid probably acquired protagonism in Eukaryotes and added further functions, such as aiding the process of transducing regulatory signals from the plasma membrane to the protein moiety. Cholesterol-recognizing sequences concomitantly became integrated into the genes coding for many hormone and neurotransmitter receptors as well as channel proteins. This is, in essence, the idea behind the premise that hopanoids in Prokaryotes and cholesterol in Eukaryotes share some common roles; cholesterol added complexity to the protein-lipid interplay.
Acknowledgements
Experimental work quoted in this review was supported by grants PICT 2011-0604 from FONCYT, Ministry of Science and Technology and PIP No. N° 112-201101-01023 from the National Scientific and Technical Research Council of Argentina (CONICET).
References
[1] | L. Adamian, H. Naveed and J. Liang, Lipid-binding surfaces of membrane proteins: Evidence from evolutionary and structural analysis, Biochim Biophys Acta 1808: (4) ((2011) ), 1092–1102. doi:10.1016/j.bbamem.2010.12.008. |
[2] | M.Y. Agargün et al., Low cholesterol level in patients with panic disorder: The association with major depression, Journal of Affective Disorders 50: (1) ((1998) ), 29–32. doi:10.1016/S0165-0327(97)00194-8. |
[3] | C.R. Alving et al., Immunization with cholesterol-rich liposomes induces anti-cholesterol antibodies and reduces diet-induced hypercholesterolemia and plaque formation, J. Lab. Clin. Med. 127: ((1996) ), 40–49. |
[4] | R.G. Anderson and K. Jacobson, A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains, Science 296: (5574) ((2002) ), 1821–1825. doi:10.1126/science.1068886. |
[5] | S. Angers, A. Salahpour and M. Bouvier, Dimerization: An emerging concept for G protein-coupled receptor ontogeny and function, Annu Rev Pharmacol Toxicol 42: ((2002) ), 409–435. |
[6] | S.S. Antollini and F.J. Barrantes, Fatty acid regulation of voltage- and ligand-gated ion channel function, Frontiers in Physiology 7: (573) ((2016) ). |
[7] | M. Baginski, A. Tempczyk and E. Borowski, Comparative conformational analysis of cholesterol and ergosterol by molecular mechanics, Eur Biophys J 17: (3) ((1989) ), 159–166. doi:10.1007/BF00254770. |
[8] | C.J. Baier, J. Fantini and F.J. Barrantes, Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor, Sci Rep 1: ((2011) ), 69. |
[9] | F.J. Barrantes, The lipid environment of the nicotinic acetylcholine receptor in native and reconstituted membranes, Crit. Rev. Biochem. Mol. Biol. 24: (5) ((1989) ), 437–478. |
[10] | F.J. Barrantes, Structural basis for lipid modulation of nicotinic acetylcholine receptor function, Brain Res Brain Res Rev 47: (1–3) ((2004) ), 71–95. |
[11] | F.J. Barrantes, Cholesterol effects on nicotinic acetylcholine receptor, J. Neurochem. 103: (Suppl 1) ((2007) ), 72–80. |
[12] | F.J. Barrantes, Cholesterol effects on nicotinic acetylcholine receptor: Cellular aspects, in: Subcell Biochem., Springer Verlag, (2010) , pp. 467–487. |
[13] | F.J. Barrantes, Regulation of the nicotinic acetylcholine receptor by cholesterol as a boundary lipid, in: Cholesterol Regulation of Ion Channels and Receptors, I. Levitan and F.J. Barrantes, eds, John Wiley & Sons, (2012) , pp. 183–204. |
[14] | F.J. Barrantes, Phylogenetic conservation of protein–lipid motifs in pentameric ligand-gated ion channels, Biochimica et Biophysica Acta (BBA) – Biomembranes 1848: (9) ((2015) ), 1796–1805. doi:10.1016/j.bbamem.2015.03.028. |
[15] | F.J. Barrantes, V. Borroni and S. Vallés, Neuronal nicotinic acetylcholine receptor–cholesterol crosstalk in Alzheimer’s disease, FEBS Letters 584: (9) ((2010) ), 1856–1863. |
[16] | F.J. Barrantes and J. Fantini, From hopanoids to cholesterol: Molecular clocks of pentameric ligand-gated ion channels, Prog Lipid Res 63: ((2016) ), 1–13. |
[17] | V. Bermudez et al., Partition profile of the nicotinic acetylcholine receptor in lipid domains upon reconstitution, J Lipid Res 51: (9) ((2010) ), 2629–2641. |
[18] | K. Bloch, The biological synthesis of cholesterol, Science 150: (3692) ((1965) ), 19–28. doi:10.1126/science.150.3692.19. |
[19] | K.E. Bloch, Speculations on the evolution of sterol structure and function, CRC Crit Rev Biochem 7: (1) ((1979) ), 1–5. doi:10.3109/10409237909102566. |
[20] | K.E. Bloch, The structure of cholesterol and bile acids, Trends Biochem Sci 7: ((1982) ), 334–336. doi:10.1016/0968-0004(82)90267-5. |
[21] | K.E. Bloch, Sterol structure and membrane function, CRC Crit Rev Biochem 14: (1) ((1983) ), 47–92. doi:10.3109/10409238309102790. |
[22] | M. Bloom and O.G. Mouritsen, The evolution of membranes, Can. J. Chem. 66: ((1988) ), 706–712. |
[23] | M.V. Borroni, A.S. Vallés and F.J. Barrantes, The lipid habitats of neurotransmitter receptors in brain, Biochimica et Biophysica Acta (BBA) – Biomembranes 1858: (11) ((2016) ), 2662–2670. doi:10.1016/j.bbamem.2016.07.005. |
[24] | G. Brannigan et al., Embedded cholesterol in the nicotinic acetylcholine receptor, Proc. Natl. Acad. Sci. U.S.A. ((2008) ). |
[25] | A.J. Brown and A.M. Galea, Cholesterol as an evolutionary response to living with oxygen, Evolution 64: (7) ((2010) ), 2179–2183. |
[26] | M.S. Brown and J.L. Goldstein, How LDL receptors influence cholesterol and atherosclerosis, Scientific American 251: ((1984) ), 52–60. |
[27] | J. Bruses, N. Chauvet and U. Rutishauser, Membrane lipid rafts are necessary for the maintenance of the (alpha)7 nicotinic acetylcholine receptor in somatic spines of ciliary neurons, J. Neurosci. 21: (2) ((2001) ), 504–512. |
[28] | M. Burns and K. Duff, Cholesterol in Alzheimer’s disease and tauopathy, Ann. N.Y. Acad. Sci. 977: ((2002) ), 367–375. |
[29] | J.A. Campagna and J. Fallon, Lipid rafts are involved in C95 (4,8) agrin fragment-induced acetylcholine receptor clustering, Neuroscience 138: (1) ((2006) ), 123–132. |
[30] | M. Carquin et al., Recent progress on lipid lateral heterogeneity in plasma membranes: From rafts to submicrometric domains, Prog Lipid Res, 62: ((2016) ), 1–24. |
[31] | V. Cartocci et al., Can cholesterol metabolism modulation affect brain function and behavior? J Cell Physiol 232: (2) ((2017) ), 281–286. |
[32] | L.-L. Chen, G.-Z. Wang and H.-Y. Zhang, Sterol biosynthesis and prokaryotes-to-eukaryotes evolution, Biochemical and Biophysical Research Communications 363: (4) ((2007) ), 885–888. doi:10.1016/j.bbrc.2007.09.093. |
[33] | V. Cherezov et al., High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor, Science 318: (5854) ((2007) ), 1258–1265. doi:10.1126/science.1150577. |
[34] | M.E. Chevreul, Recherches chimiques sur les corps gras, et particulièrement sur leurs combinaisons avec les alcalis. Sixième mémoire. Examen des graisses d’homme, de mouton, de boeuf, de jaguar et d’oie, Annales de Chimie et de Physique 2: ((1816) ), 339–372. |
[35] | R.M. Cooke et al., Structures of G protein-coupled receptors reveal new opportunities for drug discovery, Drug Discovery Today 20: (11) ((2015) ), 1355–1364. |
[36] | M. Criado, H. Eibl and F.J. Barrantes, Effects of lipids on acetylcholine receptor. Essential need of cholesterol for maintenance of agonist-induced state transitions in lipid vesicles, Biochemistry 21: (15) ((1982) ), 3622–3629. |
[37] | J. Czub and M. Baginski, Comparative molecular dynamics study of lipid membranes containing cholesterol and ergosterol, Biophys J 90: (7) ((2006) ), 2368–2382. doi:10.1529/biophysj.105.072801. |
[38] | R.F.M. de Almeida et al., Cholesterol modulates the organization of the gamma M4 transmembrane domain of the muscle nicotinic acetylcholine receptor, Biophysical Journal 86: (4) ((2004) ), 2261–2272. doi:10.1016/S0006-3495(04)74284-8. |
[39] | E. Desmond and S. Gribaldo, Phylogenomics of sterol synthesis: Insights into the origin, evolution, and diversity of a key eukaryotic feature, Genome Biol Evol, 1: ((2009) ), 364–381. |
[40] | J.M. Dietschy and S.D. Turley, Thematic review series: Brain lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal, J. Lipid Res. 45: (8) ((2004) ), 1375–1397. doi:10.1194/jlr.R400004-JLR200. |
[41] | A. el Battari et al., Modification of HT 29 cell response to the vasoactive intestinal peptide (VIP) by membrane fluidization, Biochimie, 67: (12) ((1985) ), 1217–1223. |
[42] | R.F. Epand et al., Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains, Biochemistry 45: (19) ((2006) ), 6105–6114. |
[43] | R.M. Epand, Proteins and cholesterol-rich domains, Biochim Biophys Acta 1778: (7–8) ((2008) ), 1576–1582. |
[44] | R.M. Epand et al., Cholesterol interaction with proteins that partition into membrane domains: An overview, Subcell Biochem 51: ((2010) ), 253–278. |
[45] | J. Fantini et al., Lipid rafts: Structure, function and role in HIV, Alzheimer’s and prion diseases, Expert Rev. Mol. Med. 4: (27) ((2002) ), 1–22. |
[46] | J. Fantini et al., Molecular mechanisms of protein–cholesterol interactions in plasma membranes: Functional distinction between topological (tilted) and consensus (CARC/CRAC) domains, Chemistry and Physics of Lipids 199: ((2016) ), 52–60. doi:10.1016/j.chemphyslip.2016.02.009. |
[47] | J. Fantini et al., A mirror code for protein-cholesterol interactions in the two leaflets of biological membranes, Scientific Reports 6: ((2016) ), 21907. |
[48] | J. Fantini and F.J. Barrantes, Sphingolipid/cholesterol regulation of neurotransmitter receptor conformation and function, Biochim Biophys Acta 1788: (11) ((2009) ), 2345–2361. doi:10.1016/j.bbamem.2009.08.016. |
[49] | J. Fantini and F.J. Barrantes, How cholesterol interacts with membrane proteins: An exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains, Front Physiol 4: ((2013) ), 31. |
[50] | D. Fass et al., Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module, Nature 388: ((1997) ), 691–693. doi:10.1038/41798. |
[51] | G.A. Fernandez Nievas, F.J. Barrantes and S.S. Antollini, Conformation-sensitive steroid and fatty acid sites in the transmembrane domain of the nicotinic acetylcholine receptor, Biochemistry 46: (11) ((2007) ), 3503–3512. |
[52] | C. Frank et al., Cholesterol depletion inhibits synaptic transmission and synaptic plasticity in rat hippocampus, Exp. Neurol. 212: (2) ((2008) ), 407–414. |
[53] | S.G. Furness et al., Ligand-dependent modulation of G protein conformation alters drug efficacy, Cell 167: (3) ((2016) ), 739–749, e11. |
[54] | S. Gahbauer and R.A. Böckmann, Membrane-mediated oligomerization of G protein coupled receptors and its implications for GPCR function, Frontiers in Physiology 7: (494) ((2016) ). |
[55] | S.A. Gaitonde and J. Gonzalez-Maeso, Contribution of heteromerization to G protein-coupled receptor function, Curr Opin Pharmacol 32: ((2016) ), 23–31. |
[56] | A.M. Galea and A.J. Brown, Special relationship between sterols and oxygen: Were sterols an adaptation to aerobic life? Free Radic Biol Med 47: (6) ((2009) ), 880–889. |
[57] | P. Gamba et al., Oxidized cholesterol as the driving force behind the development of Alzheimer’s disease, Frontiers in Aging Neuroscience 7: ((2015) ). |
[58] | G. Gimpl, Interaction of G protein coupled receptors and cholesterol, Chem Phys Lipids 199: ((2016) ), 61–73. |
[59] | D.S. Goodman, Cholesterol ester metabolism, Physiol Rev 45: (4) ((1965) ), 747–839. |
[60] | P. Grieb et al., Serum cholesterol in cerebral malignancies, Journal of Neuro-Oncology 41: (2) ((1999) ), 175–180. doi:10.1023/A:1006131418126. |
[61] | J. Griffie, G. Burn and D.M. Owen, The nanoscale organization of signaling domains at the plasma membrane, Curr Top Membr 75: ((2015) ), 125–165. doi:10.1016/bs.ctm.2015.03.004. |
[62] | A.K. Hamouda et al., Cholesterol interacts with transmembrane alpha-helices M1, M3, and M4 of the Torpedo nicotinic acetylcholine receptor: Photolabeling studies using [3H]Azicholesterol, Biochemistry 45: (3) ((2006) ), 976–986. |
[63] | M.A. Hanson et al., A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor, Structure 16: (6) ((2008) ), 897–905. |
[64] | V.P. Jaakola et al., The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist, Science 322: (5905) ((2008) ), 1211–1217. doi:10.1126/science.1164772. |
[65] | K.A. Jacobson, New paradigms in GPCR drug discovery, Biochem Pharmacol 98: (4) ((2015) ), 541–555. doi:10.1016/j.bcp.2015.08.085. |
[66] | N. Jamin et al., Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor, Mol Endocrinol 19: (3) ((2005) ), 588–594. |
[67] | P. Kumari, E. Ghosh and A.K. Shukla, Emerging approaches to GPCR ligand screening for drug discovery, Trends Mol Med 21: (11) ((2015) ), 687–701. |
[68] | B. Lagane et al., Role of sterols in modulating the human mu-opioid receptor function in Saccharomyces cerevisiae, J Biol Chem 275: ((2000) ), 33197–33200. |
[69] | D.F. Lazar and F. Medzihradsky, Altered microviscosity at brain membrane surface induces distinct and reversible inhibition of opioid receptor binding, J Neurochem 59: (4) ((1992) ), 1233–1240. |
[70] | A.G. Lee, How lipids affect the activities of integral membrane proteins, Biochim Biophys Acta 1666: (1–2) ((2004) ), 62–87. |
[71] | W.S. Leibel et al., Two pools of cholesterol in acetylcholine recptor-rich membranes from Torpedo, Biochim. Biophys. Acta 8987: ((1987) ), 249–260. |
[72] | I. Levitan and F.J. Barrantes, Cholesterol Regulation of Ion Channels and Receptors, John Wiley & Sons, Hoboken, NJ, (2012) , pp. 1–289. |
[73] | I. Levitan, D.K. Singh and A. Rosenhouse-Dantsker, Cholesterol binding to ion channels, Frontiers in Physiology 5: ((2014) ). |
[74] | Q. Lin and E. London, Ordered raft domains induced by outer leaflet sphingomyelin in cholesterol-rich asymmetric vesicles, Biophysical Journal 108: (9) ((2015) ), 2212–2222. doi:10.1016/j.bpj.2015.03.056. |
[75] | D. Lingwood and K. Simons, Lipid rafts as a membrane-organizing principle, Science 327: (5961) ((2010) ), 46–50. doi:10.1126/science.1174621. |
[76] | H. Mabuchi et al., Molecular genetics of cholesterol transport and cholesterol reverse transport disorders (familial hypercholesterolemia and CETP deficiency) and coronary heart disease, Ann. NY Acad. Sci. 748: ((1995) ), 333–341. |
[77] | C. Madwar, G. Gopalakrishnan and R.B. Lennox, Lipid microdomains in synapse formation, ACS Chemical Neuroscience ((2016) ). |
[78] | P.A. Maguire and M.J. Druse, The influence of cholesterol on synaptic fluidity, dopamine D1 binding and dopamine-stimulated adenylate cyclase, Brain Res Bull 23: (1–2) ((1989) ), 69–74. |
[79] | S.B. Mantipragada et al., Lipid-protein interactions and effect of local anesthetics in acetylcholine receptor-rich membranes from Torpedo marmorata electric organ, Biochemistry 42: (30) ((2003) ), 9167–9175. |
[80] | S. Marchand et al., Rapsyn escorts the nicotinic acetylcholine receptor along the exocytic pathway via association with lipid rafts, J Neurosci 22: (20) ((2002) ), 8891–8901. |
[81] | D. Marsh, General features of phospholipid phase transitions, Chemistry and Physics of Lipids 57: ((1991) ), 109–120. doi:10.1016/0009-3084(91)90071-I. |
[82] | D. Marsh et al., ESR spin-label studies of lipid–protein interactions in membranes, Biophys J 37: ((1982) ), 265–274. |
[83] | D. Marsh and F.J. Barrantes, Immobilized lipid in acetylcholine receptor-rich membranes from Torpedo marmorata, Proc. Natl. Acad. Sci. U.S.A. 75: (9) ((1978) ), 4329–4333. |
[84] | D. Marsh, A. Watts and F.J. Barrantes, Phospholipid chain immobilization and steroid rotational immobilization in acetylcholine receptor-rich membranes from Torpedo marmorata, Biochim Biophys Acta 645: (1) ((1981) ), 97–101. doi:10.1016/0005-2736(81)90516-2. |
[85] | M.T. Marty et al., Probing the lipid annular belt by gas-phase dissociation of membrane proteins in nanodiscs, Angew Chem Int Ed Engl 55: (2) ((2016) ), 550–554. doi:10.1002/anie.201508289. |
[86] | D.H. Mauch et al., CNS synaptogenesis promoted by glia-derived cholesterol, Science 294: (5545) ((2001) ), 1354–1357. doi:10.1126/science.294.5545.1354. |
[87] | F.R. Maxfield, Plasma membrane microdomains, Curr. Opin. Cell Biol. 14: (4) ((2002) ), 483–487. |
[88] | L. Miao et al., From lanosterol to cholesterol: Structural evolution and differential effects on lipid bilayers, Biophys. J. 82: (3) ((2002) ), 1429–1444. doi:10.1016/S0006-3495(02)75497-0. |
[89] | G.A. Morrill and A.B. Kostellow, Molecular properties of globin channels and pores: Role of cholesterol in ligand binding and movement, Frontiers in Physiology 7: (360) ((2016) ). |
[90] | G.A. Morrill, A.B. Kostellow and R.K. Gupta, The pore-lining regions in cytochrome c oxidases: A computational analysis of caveolin, cholesterol and transmembrane helix contributions to proton movement, Biochim Biophys Acta 1838: (11) ((2014) ), 2838–2851. doi:10.1016/j.bbamem.2014.07.011. |
[91] | G.A. Morrill, A.B. Kostellow and R.K. Gupta, The role of receptor topology in the vitamin D3 uptake and Ca2+ response systems, Biochemical and Biophysical Research Communications ((2016) ). |
[92] | O.G. Mouritsen and M.J. Zuckermann, What’s so special about cholesterol? Lipids 39: (11) ((2004) ), 1101–1113. |
[93] | S. Mukherjee et al., Cholesterol distribution in living cells: Fluorescence imaging using dehydroergosterol as a fluorescent cholesterol analog, Biophys J 75: (4) ((1998) ), 1915–1925. doi:10.1016/S0006-3495(98)77632-5. |
[94] | S. Mukherjee and A. Chattopadhyay, Membrane organization at low cholesterol concentrations: A study using 7-nitrobenz-2-oxa-1,3-diazol-4-yl-labeled cholesterol, Biochemistry 35: ((1996) ), 1311–1322. |
[95] | Y. Nakatani et al., Search for the most primitive membranes: Some remaining problems, Orig Life Evol Biosph 42: (5) ((2012) ), 497–501. |
[96] | M. Nishio et al., CH–π hydrogen bonds in biological macromolecules, Physical Chemistry Chemical Physics ((2014) ). |
[97] | M. Opekarova and W. Tanner, Specific lipid requirements of membrane proteins – a putative bottleneck in heterologous expression, Biochim Biophys Acta 1610: (1) ((2003) ), 11–22. doi:10.1016/S0005-2736(02)00708-3. |
[98] | G. Ourisson, M. Rohmer and K. Poralla, Prokaryotic hopanoids and other polyterpenoid sterol surrogates, Annu Rev Microbiol 41: ((1987) ), 301–333. doi:10.1146/annurev.mi.41.100187.001505. |
[99] | P. Pathak and E. London, The effect of membrane lipid composition on the formation of lipid ultrananodomains, Biophys J 109: (8) ((2015) ), 1630–1638. doi:10.1016/j.bpj.2015.08.029. |
[100] | V.L. Perillo et al., Transbilayer asymmetry and sphingomyelin composition modulate the preferential membrane partitioning of the nicotinic acetylcholine receptor in Lo domains, Arch Biochem Biophys 591: ((2016) ), 76–86. |
[101] | F.W. Pfrieger, Cholesterol homeostasis and function in neurons of the central nervous system, Cell Mol Life Sci 60: (6) ((2003) ), 1158–1171. |
[102] | F.W. Pfrieger, Role of cholesterol in synapse formation and function, Biochim Biophys Acta 1610: (2) ((2003) ), 271–280. |
[103] | L.J. Pike, The challenge of lipid rafts, J Lipid Res 50: (Suppl) ((2009) ), S323–S328. |
[104] | K. Pluhackova et al., Dynamic cholesterol-conditioned dimerization of the G protein coupled chemokine receptor type 4, PLoS Comput Biol 12: (11) ((2016) ), e1005169. |
[105] | J.L. Popot et al., Preferential affinity of acetylcholine receptor protein for certain lipids studied using monolayer cultures, C R Acad Sci Hebd Seances Acad Sci D, 285: (9) ((1977) ), 1005–1008. |
[106] | I.M. Posada et al., A cholesterol recognition motif in human phospholipid scramblase 1, Biophys J 107: (6) ((2014) ), 1383–1392. doi:10.1016/j.bpj.2014.07.039. |
[107] | X. Prasanna, D. Sengupta and A. Chattopadhyay, Cholesterol-dependent conformational plasticity in GPCR dimers, Sci Rep 6: ((2016) ), 31858. |
[108] | T.J. Pucadyil and A. Chattopadhyay, Cholesterol modulates ligand binding and G-protein coupling to serotonin(1A) receptors from bovine hippocampus, Biochim. Biophys. Acta 1663: (1–2) ((2004) ), 188–200. |
[109] | T.J. Pucadyil and A. Chattopadhyay, Cholesterol depletion induces dynamic confinement of the G-protein coupled serotonin(1A) receptor in the plasma membrane of living cells, Biochim. Biophys. Acta 1768: (3) ((2007) ), 655–668. doi:10.1016/j.bbamem.2007.01.002. |
[110] | M. Rao and S. Mayor, Active organization of membrane constituents in living cells, Curr Opin Cell Biol 29: ((2014) ), 126–132. |
[111] | L.M. Refolo et al., A cholesterol-lowering drug reduces beta-amyloid pathology in a transgenic mouse model of Alzheimer’s disease, Neurobiol. Dis. 8: (5) ((2001) ), 890–899. |
[112] | U. Renner et al., Localization of the mouse 5-hydroxytryptamine(1A) receptor in lipid microdomains depends on its palmitoylation and is involved in receptor-mediated signaling, Mol Pharmacol 72: (3) ((2007) ), 502–513. |
[113] | J. Rogers, A.G. Lee and D.C. Wilton, The organisation of cholesterol and ergosterol in lipid bilayers based on studies using non-perturbing fluorescent sterol probes, Biochim Biophys Acta 552: (1) ((1979) ), 23–37. doi:10.1016/0005-2736(79)90243-8. |
[114] | D.M. Rosenbaum et al., GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function, Science 318: (5854) ((2007) ), 1266–1273. doi:10.1126/science.1150609. |
[115] | N.P. Rotstein et al., Composition of lipids in elasmobranch electric organ and acetylcholine receptor membranes, J Neurochem 49: (5) ((1987) ), 1333–1340. doi:10.1111/j.1471-4159.1987.tb00996.x. |
[116] | A. Rousselet, P.F. Devaux and K.W. Wirtz, Free fatty acids and esters can be immobilized by receptor rich membranes from Torpedo marmorata but not phospholipid acyl chains, Biochem. Biophys. Res. Commun. 90: ((1979) ), 871–877. doi:10.1016/0006-291X(79)91908-9. |
[117] | J.P. Saenz et al., Functional convergence of hopanoids and sterols in membrane ordering, Proc Natl Acad Sci U S A 109: (35) ((2012) ), 14236–14240. doi:10.1073/pnas.1212141109. |
[118] | J.P. Saenz et al., Hopanoids as functional analogues of cholesterol in bacterial membranes, Proc Natl Acad Sci U S A 112: (38) ((2015) ), 11971–11976. doi:10.1073/pnas.1515607112. |
[119] | F. Schmid, Physical mechanisms of micro- and nanodomain formation in multicomponent lipid membranes, Biochim Biophys Acta ((2016) ). |
[120] | S. Shrivastava et al., Chronic cholesterol depletion using statin impairs the function and dynamics of human Serotonin1A receptors, Biochemistry ((2010) ). |
[121] | A.C. Simmonds et al., Annular and non-annular binding sites on the ( |
[122] | K. Simons and J. Gruenberg, Jamming the endosomal system: Lipid rafts and lysosomal storage diseases, Trends Cell Biol. 10: (11) ((2000) ), 459–462. doi:10.1016/S0962-8924(00)01847-X. |
[123] | K. Simons and E. Ikonen, Functional rafts in cell membranes, Nature 387: (6633) ((1997) ), 569–572. |
[124] | K. Simons and G. van Meer, Lipid sorting in epithelial cells, Biochemistry 27: (17) ((1988) ), 6198–6202. |
[125] | P.J. Stansfeld et al., MemProtMD: Automated insertion of membrane protein structures into explicit lipid membranes, Structure 23: (7) ((2015) ), 1350–1361. |
[126] | M. Stefani and G. Liguri, Cholesterol in Alzheimer’s disease: Unresolved questions, Curr. Alzheimer Res. 6: (1) ((2009) ), 15–29. |
[127] | F. Stetzkowski-Marden et al., Agrin elicits membrane lipid condensation at sites of acetylcholine receptor clusters in C2C12 myotubes, J Lipid Res. 47: (10) ((2006) ), 2121–2133. doi:10.1194/jlr.M600182-JLR200. |
[128] | E. Strandberg and J.A. Killian, Snorkeling of lysine side chains in transmembrane helices: How easy can it get? FEBS Lett 544: (1–3) ((2003) ), 69–73. |
[129] | R.E. Summons et al., Steroids, triterpenoids and molecular oxygen, Philos Trans R Soc Lond B Biol Sci 361: (1470) ((2006) ), 951–968. |
[130] | X. Sun et al., Niemann–Pick C variant detection by altered sphingolipid trafficking and correlation with mutations within a specific domain of NPC1, Am. J. Hum. Genet. 68: (6) ((2001) ), 1361–1372. |
[131] | C.B. Taylor and K.J. Ho, A review of human cholesterol metabolism, Arch Pathol. 84: (1) ((1967) ), 3–14. |
[132] | R. Tripathy, D. Mishra and V.B. Konkimalla, A novel fuzzy C-means approach for uncovering cholesterol consensus motif from human G-protein coupled receptors (GPCR), Karbala International Journal of Modern Science 1: (4) ((2015) ), 212–224. doi:10.1016/j.kijoms.2015.11.006. |
[133] | N. Vaidehi, S. Bhattacharya and A.B. Larsen, Structure and dynamics of G-protein coupled receptors, Adv Exp Med Biol 796: ((2014) ), 37–54. |
[134] | T.S. van Zanten and S. Mayor, Current approaches to studying membrane organization, F1000Res 4: ((2015) ). |
[135] | D.E. Vance and H. Van den Bosch, Cholesterol in the year 2000, Biochim Biophys Acta 1529: (1–3) ((2000) ), 1–8. |
[136] | S.L. Veatch et al., Liquid domains in vesicles investigated by NMR and fluorescence microscopy, Biophys. J. 86: (5) ((2004) ), 2910–2922. |
[137] | S.L. Veatch and S.L. Keller, Organization in lipid membranes containing cholesterol, Phys. Rev. Lett. 89: (26) ((2002) ), 268101. |
[138] | S.L. Veatch and S.L. Keller, Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol, Biophys. J. 85: (5) ((2003) ), 3074–3083. |
[139] | S.L. Veatch and S.L. Keller, A closer look at the canonical ‘raft mixture’ in model membrane studies, Biophys. J. 84: (1) ((2003) ), 725–726. |
[140] | T. Warne et al., Structure of a beta1-adrenergic G-protein-coupled receptor, Nature 454: (7203) ((2008) ), 486–491. doi:10.1038/nature07101. |
[141] | G.B. Warren et al., Reversible lipid titrations of the activity of pure adenosine triphosphatase-lipid complexes, Biochemistry 13: (27) ((1974) ), 5501–5507. |
[142] | G.B. Warren et al., Cholesterol is excluded from the phospholipid annulus surrounding an active calcium transport protein, Nature 255: (5511) ((1975) ), 684–687. doi:10.1038/255684a0. |
[143] | A. Watts, I.D. Volotovski and D. Marsh, Rhodopsin–lipid associations in bovine rod outer segment membranes. Identification of immobilized lipid by spin-labels, Biochemistry 18: ((1979) ), 5006–5013. |
[144] | C.L. Wellington, Cholesterol at the crossroads: Alzheimer’s disease and lipid metabolism, Clin. Genet. 66: (1) ((2004) ), 1–16. |
[145] | J.J. Wenz and F.J. Barrantes, Steroid structural requirements for stabilizing or disrupting lipid domains, Biochemistry 42: (48) ((2003) ), 14267–14276. |
[146] | R. Willmann et al., Cholesterol and lipid microdomains stabilize the postsynapse at the neuromuscular junction, EMBO J 25: (17) ((2006) ), 4050–4060. |
[147] | D. Wüstner, Fluorescent sterols as tools in membrane biophysics and cell biology, Chem Phys Lipids 146: ((2007) ). |
[148] | D. Wüstner et al., Potential of ultraviolet widefield imaging and multiphoton microscopy for analysis of dehydroergosterol in cellular membranes, Microsc Res Tech 74: ((2011) ). |
[149] | D. Wüstner et al., Imaging approaches for analysis of cholesterol distribution and dynamics in the plasma membrane, Chemistry and Physics of Lipids 199: ((2016) ), 106–135. doi:10.1016/j.chemphyslip.2016.03.003. |
[150] | X. Xu et al., Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts). Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide, J. Biol. Chem. 276: (36) ((2001) ), 33540–33546. |
[151] | X. Xu and E. London, The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation, Biochemistry 39: (5) ((2000) ), 843–849. |
[152] | P.L. Yeagle, Lipid regulation of cell membrane structure and function, FASEB J 3: ((1989) ), 1833–1842. |
[153] | S.O. Yesylevskyy et al., Cholesterol induces uneven curvature of asymmetric lipid bilayers, Scientific World Journal 2013: ((2013) ), 965230. |
[154] | D. Zhu, W.C. Xiong and L. Mei, Lipid rafts serve as a signaling platform for nicotinic acetylcholine receptor clustering, J. Neurosci. 26: (18) ((2006) ), 4841–4851. doi:10.1523/JNEUROSCI.2807-05.2006. |




