The presence of herpesviruses in malignant but not in benign or recurrent pleomorphic adenomas
Abstract
BACKGROUND:
The etiology of salivary gland tumors is mainly unknown. The anatomical location of the salivary glands, with the mucosal pathway to the oral cavity and its rich microbiome, raises the question of potential viral background.
OBJECTIVE:
This study focuses on the potential presence of herpes-, polyoma- and parvoviruses in pleomorphic adenoma (PA), recurrent pleomorphic adenoma (RPA) and carcinoma ex pleomorphic adenoma (CaxPA).
METHODS:
Thirty different viruses were analyzed by PCR-based assays in 68 formalin-fixed paraffin-embedded salivary gland tumors (25 PA, 31 RPA and 12 CaxPA).
RESULTS:
Virus DNA was detected altogether in 19/68 (28%) tumor samples. Human herpesviruses 6B and 7 (HHV-6B and HHV-7) and Epstein-Barr virus (EBV) were frequently and almost exclusively found in CaxPA (5/12, 7/12, and 3/12, respectively). Within the 7 CaxPA that were virus-positive, 3 samples contained 3, and 1 sample even 4, different viruses. Infrequent viral positivity was shown for parvovirus B19 and cutavirus, as well as Merkel cell and Malawi polyomaviruses.
CONCLUSIONS:
Our unexpected finding of herpesvirus DNA almost exclusively in CaxPA tissues deserves further in-depth studies.
1Introduction
Pleomorphic adenoma (PA) is the most common benign tumor of the salivary glands with an annual incidence of 4.2–4.9 /100,000. Surgical removal is usually curative, but benign recurrence occurs in 1–6.7% depending on the surgical method and follow-up time (5–20 years) [1, 2]. A small proportion, 0.15% of PAs [1] and 3.2–6% of recurrent PAs (RPA) [1, 3, 4] may undergo malignant transformation to carcinoma ex pleomorphic adenoma (CaxPA).
The etiology of PAs remains unknown but smoking and alcohol consumption seem insignificant factors in this disease [5]. The anatomical location of the salivary glands, with the mucosal pathway to the oral cavity and its rich microbiome, raises the question of potential viral background. Indeed, already in the 1950s Ludwik Gross was able to induce salivary gland tumor growth in mice by infiltrating an agent, later known as polyomavirus, into the glands [6, 7]. In PAs, some positivity for polyomaviruses, Epstein-Barr virus (EBV), human herpesviruses 6B and 7 (HHV-6 and -7) and human papillomaviruses (HPV) have been discovered [8–10]. However, no consensus exists on the etiologic role of the different viruses found in PAs. The presence of viruses in RPA and CaxPA remains an unexplored field.
Viruses and other microbes are estimated to cause 15% of all cancers worldwide [11]. Oncogenic viruses are found within several DNA-virus families such as Papillomaviridae, Polyomaviridae, Hepadnaviridae, and Herpesviridae [12–18], whereas oncolytic viruses are found in the Parvoviridae family [19]. Within Herpesviridae, human herpesvirus 8 (HHV-8) causes Kaposi‘s sarcoma [18, 20], whereas EBV is strongly associated with eg. Burkitt‘s lymphoma and nasopharyngeal carcinoma [21]. Other herpesviruses, such as herpes simplex (HSV-1 and -2) and varicella zoster (VZV) are neurotropic and widespread, but without clear association to malignancies. HHV-6B and -7 infect the general population during early childhood, mainly via saliva, whereas HHV-6A is an infrequent finding [22]. The infection of HHV-6 and 7 is often symptomless but can present with fever, infantile seizures or rash.
Human polyomaviruses comprise a group of 14 viruses, most with a high seroprevalence among humans. Primary infections are typically asymptomatic or with unknown symptoms, and these viruses persist in the human body, mainly in the urogenital tract or the lymphatic system [23, 24]. Merkel cell virus (MCPyV) is known to be oncogenic and can cause Merkel cell carcinoma in humans [13]. Among head and neck tumors (juvenile angiofibroma, head and neck squamous cell carcinoma) the presence of polyomaviruses varies from sporadic MCPyV to up to 37% of JC polyomavirus (JCPyV) [25, 26] but their role in carcinogenesis is yet to be identified.
Parvovirus B19 (B19V) causes erythema infectiosum (fifth disease), aplastic crisis, anemias, and fetal death [27]. After the primary infection, B19V seems to enter and persist in various non-permissive tissues with unknown clinical significance [28, 29]. In the past two decades, many new parvoviruses have been discovered: human bocaviruses 1-4 (HBoV 1–4), bufa- (BuV), tusa- (TuV), and cutavirus (CuV) [30]. HBoV1 causes respiratory tract infections in children, whereas HBoV2–4 are enteric, but more severe symptoms like encephalitis may be present [27]. BuV, TuV, and CuV were originally found in diarrheic stool samples from children [31–33]. CuV has further been associated with cutaneous T-cell lymphoma (CTCL) and it has been detected in other skin cancers, including melanoma [33–36]. The clinical role of these recently discovered viruses needs further studies.
This study was carried out in order to explore the presence of herpes-, polyoma-, and parvoviruses in PA, RPA and CaxPA. The results could give insight into the presence of viral infections in these tumor types and their potential role in malignant transformation.
2Materials and methods
2.1Ethics
A research ethics committee approval and research permission were obtained at the Helsinki University Hospital (§31/07.03.2019, HUS/332/2019).
2.2Patients and clinical specimens
This study is a retrospective cross-sectional cohort study. The patients were treated at the Department of Otorhinolaryngology - Head and Neck Surgery, Helsinki University Hospital, Helsinki, Finland. Clinical information was recorded from hospital charts. Three different tumor groups were constituted: “ordinary” PA, RPA and CaxPA (Fig. 1, Supplemental Table 1). For the PA group we selected 25 consecutive patients (25 samples) treated during 2005–2006 with no known history of recurrence within a 12-year follow-up. RPA and CaxPA groups included all patients with available samples diagnosed between 2000 and 2018. The CaxPA group included 12 patients (12 samples). For the RPA group we included primary tumor samples and all samples from recurrences that were available. Only recurrent tumors that had appeared after adequate surgery, i.e., with superficial or partial parotidectomy and with no report of capsule rupture were accepted. The 31 RPA samples (8 primary and 23 recurrent tumors) fulfilling our criteria were from 21 individuals. Altogether 68 FFPE tumor samples were collected from the Helsinki Biobank (permission no §73/15.05.2019, HUS/118/2019).
Fig. 1
Tumor and sample types with respective viral analyses. PA, pleomorphic adenoma; RPA recurrent pleomorphic adenoma; CaxPA carcinoma ex pleomorphic adenoma.
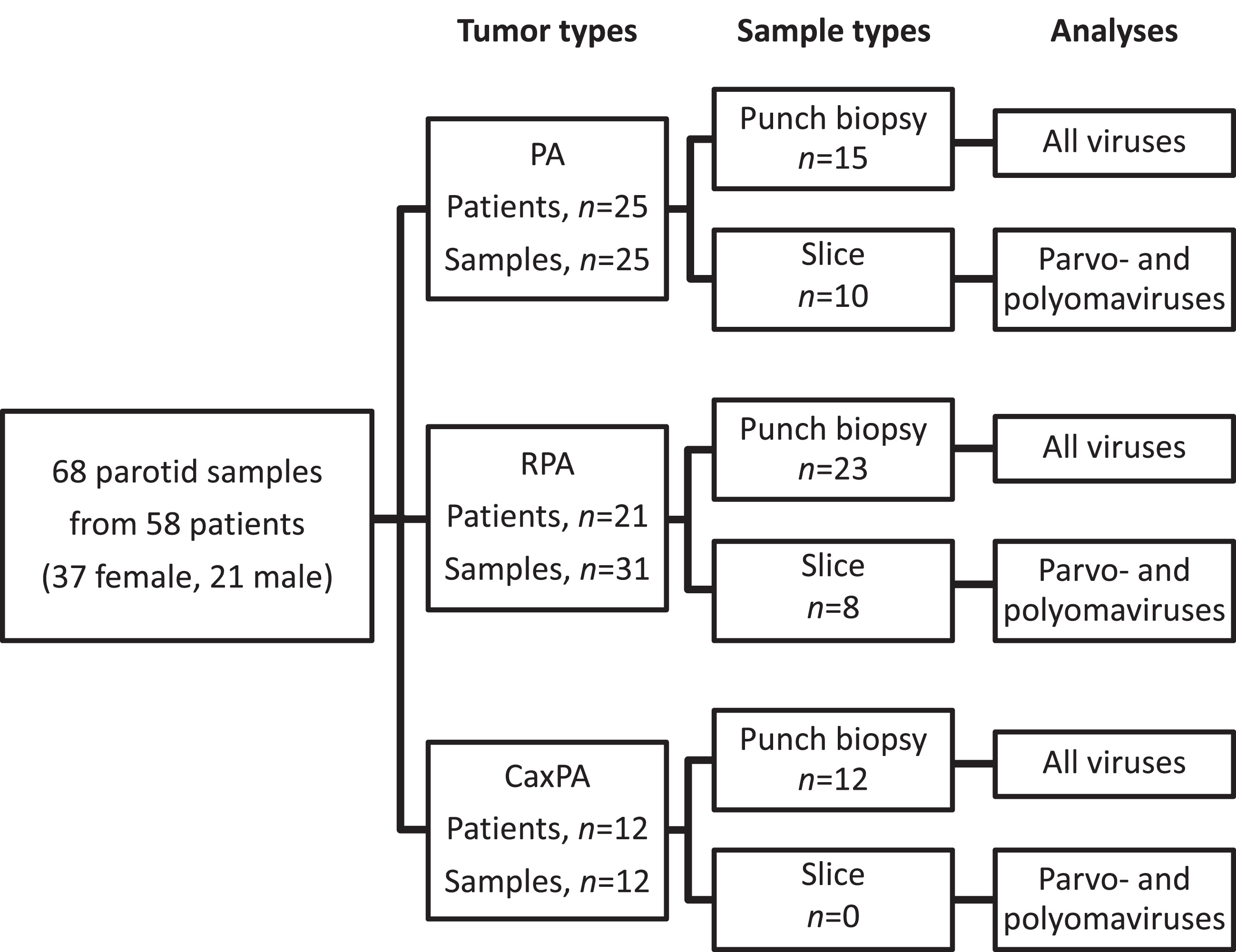
The majority of the samples (15 PAs, 23 RPAs, 12 CaxPAs) were collected from FFPE tissue blocks as 2 mm punch biopsies (Fig. 1) in a PCR-sterile manner from the site of the tumor tissue defined by an experienced oral pathologist. Due to scant tumor material, punch biopsies were not available from additional 18 samples (10 PAs, 8 RPAs), and thus, from these tumors the samples were collected as 10μm-thick slices (3-4 sections per sample) from the tumor-containing FFPE blocks. However, due to the known potential presence of herpesvirus DNA in normal salivary gland tissue [9] only punch biopsies from the actual tumor were included in the herpesvirus analyses (Fig. 1). Polyoma- and parvovirus analyses were performed with both the punch biopsy and slice samples. All samples were collected in 1.5 ml microcentrifuge tubes and stored at –20°C until DNA extraction.
2.3DNA extraction and viral DNA assays
Paraffin was dissolved from the tissue samples with xylene at room temperature for 30 minutes, repeated once. Tissue was collected by centrifugation and total DNA was extracted using QIAamp DNA mini kit (Qiagen, Heiden, Germany) according to the manufacturer’s protocol, with the exception of using 40μl of proteinase K. The DNA preps were eluted in 100μl AE buffer and stored at –20° C.
Evaluation of DNA quality and human cell quantity was done by subjecting the samples to the reference gene RNase-P qPCR, as described [37].
The viruses analyzed in this study are listed in Table 2 (Supplemental Table 2). For members of Herpesviridae, a three-tube multiplex qPCR assay [38] was applied to target each virus (HSV-1, HSV-2 and VZV; HHV-6A, HHV-6B and HHV-7; EBV, cytomegalovirus (CMV) and HHV-8). For the 13 human virus members of Polyomaviridae, BK polyomavirus (BKPyV), JCPyV, Karolinska Institute polyomavirus (KIPyV), Washington University polyomavirus (WUPyV), MCPyV, human polyomavirus 6 (HPyV6), human polyomavirus 7 (HPyV7), Trichodysplasia spinulosa polyomavirus (TSPyV), human polyomavirus 9 (HPyV9), Malawi polyomavirus (MWPyV), St. Louis polyomavirus (STLPyV), human polyomavirus 12 (HPyV12), and New Jersey polyomavirus (NJPyV), the VP1 genes were detected by a multiplex PCR assay coupled with a Luminex-based detection system [24, 39]. As for the members of Parvoviridae, B19V was detected and quantified with a pan-B19 qPCR, targeting the NS1 genes of all 3 genotypes [37], whereas HBoV1-4 were detected by a multiplex qPCR, targeting the NS1 regions of all 4 bocaviruses [40]. CuV, BuV, and TuV were detected and quantified by a multiplex qPCR [36, 41] targeting the NS1 region of BuV and VP2 regions of TuV and CuV. The PCR amplicons were cloned into pSTblue vector and sequenced. All qPCR reactions consisted of Maxima probe qPCR Master Mix (Thermo Scientific) with or without ROX as passive reference dye or 2xTaqpath Proamp Multiplex Master Mix (Fischer), each primer (forward and reverse), probe and template (2, 5–5μl), and molecular biology-grade H2O to a final volume of 20–25μl. All real-time qPCR assays were performed with AriaMx Realtime PCR System (Agilent Technologies, Santa Clara, CA).
Table 2
Heat map indicating number (and % in parenthesis) of PA, RPA and CaxPA samples containing DNA of each virus type
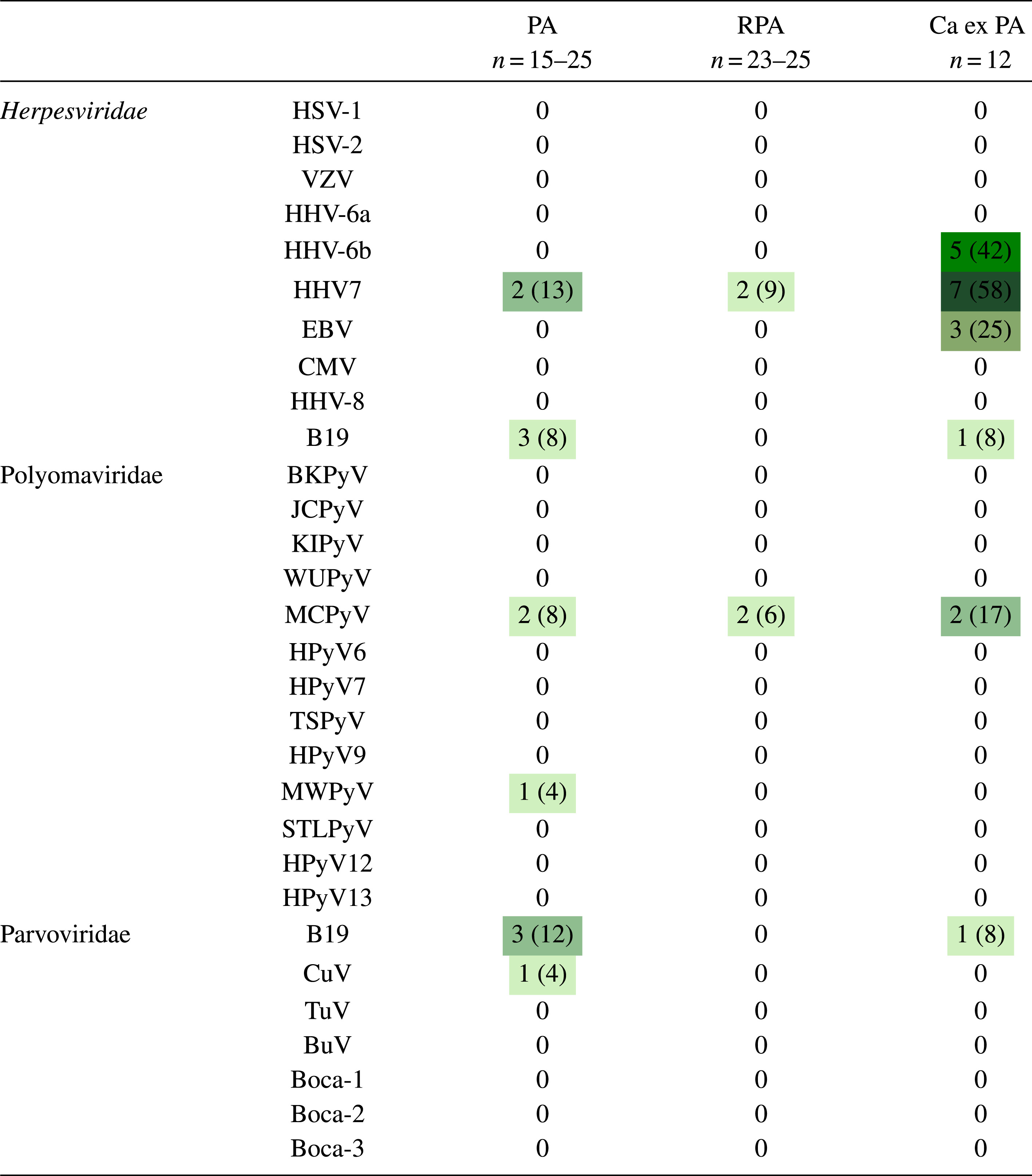 |
PA, pleomorphic adenoma; RPA recurrent pleomorphic adenoma; CaxPA carcinoma ex pleomorphic adenoma; HSV-1, herpes simplex virus 1; HSV-2 herpes simplex virus 2; VZV, varicella zoster virus, HHV-6A, human herpesvirus 6A; HHV-6B, human herpesvirus 6B; HHV-7, human herpesvirus 7; EBV, Epstein-Barr virus; CMV, cytomegalovirus; HHV-8, human herpesvirus -8; BKPyV, BK polyomavirus; JCPyV, JC polyomavirus; KIPyV, Karolinska Institute polyomavirus; WUPyV, Washington University polyomavirus; MCPyV, Merkel cell virus; HPyV -6, human polyomavirus 6; HPyV-7, human polyomavirus 7; TSPyV, Trichodysplasia spinulosa polyomavirus, HPyV-9, human polyomavirus 9; MWPyV, Malawi polyomavirus; STLPyV, St Louis polyomavirus; HPyV12, human polyomavirus 12; NJPyV, New Jersey polyomavirus; B19V, B19 parvovirus; CuV, cutavirus; TuV, Tusavirus; BuV, bufavirus; Boca 1-4, bocavirus 1-4.
Molecular biology grade water was included in all PCR runs as a non-template control. Ten-fold diluted plasmids (101–106), containing each viral target amplicon, served as PCR standards and as positive controls. To avoid contamination, the master mix components, samples and plasmids were handled in separate rooms. Laminar hoods were used when preparing and handling the samples and plasmids.
For the viruses detected by qPCR, a positive result from duplicate wells or sequencing was defined as positive. For the polyomavirus Luminex-based multiplex assay, single positive findings were accepted.
2.4Statistics
Statistical analyses were performed using GraphPad Prims 8 software (GraphPad, San Diego, CA) and Fisher’s exact test when comparing virus prevalence between different tumor types.
3Results
3.1Patient characteristics
We analyzed altogether 68 samples from 58 patients having PA, RPA or CaxPA. All 68 samples were included in polyoma- and parvovirus analyses, and the 50 punch-biopsy samples were included in the herpesvirus analyses (Fig. 1). On average, the patients with CaxPA were roughly 10 years older (57.6±13.3, mean±SD) at the time of surgery than those with PA (45.5±16.1) and RPA (46.0±15.0, Table 1). There were 8 males and 4 females in the CaxPA group in contrast to the PA and RPA groups, where females predominated.
Table 1
Patients, samples and positive viral DNA
| Herpesviridae | PA | RPA | CaxPA | |
| Patients | 15 | 15 | 12 | |
| Samples | 15 | 23 | 12 | |
| Gender: female/male | 10/5 | 11/4 | 4/8 | |
| Median age at operation x years (mean x, range x-y) years | 45.0 (46.4, 20.4–68.0) | 45.9 (47.5, 19.8–70.7) | 58.8 (57.6, 25.9–75.5) | |
| HHV-6B | Positive samples | 0 | 0 | 5 (42%) |
| Mean viral DNA load* | 0 | 0 | 1760 | |
| Median viral DNA load* (range) | 0 | 0 | 1030 (59–5220) | |
| HHV-7 | Positive samples | 2 (13%) | 2 (9%) | 7 (58%) |
| Mean viral DNA load* | 81300 | 50 | 15400 | |
| Median viral DNA load* (range) | 81300 (76200–86400) | 50 | 1530 (51–62600) | |
| EBV | Positive samples | 0 | 0 | 3 (25%) |
| Mean viral DNA load* | 0 | 0 | 665 | |
| Median viral DNA load* (range) | 0 | 0 | 419 (63–1520) | |
| Polyomaviridae & Parvoviridae | PA | RPA | CaxPA | |
| Patients | 25 | 21 | 12 | |
| Samples | 25 | 31 | 12 | |
| Gender: female/male | 17/8 | 16/5 | 4/8 | |
| Median age at operation x years (mean x, range x-y) years | 42.0 45.5 (10.8–71.2) | 49.3 46.0 (17.9–70.7) | 58.8 57.6 (25.9–75.5) | |
| MCPyV | Positive samples | 2 (8%) | 2 (6%) | 2 (16%) |
| MWPyV | Positive samples | 1 (4%) | 0 | 0 |
| B19V | Positive samples | 3 (12%) | 0 | 1 (8%) |
| Mean viral DNA load* | 1661 | 0 | 1880 | |
| Median viral DNA load* (range) | 1661 (165–3560) | 0 | 1880 (1880) | |
| Cutavirus | Positive samples | 1 (4%) | 0 | 0 |
| Mean viral DNA load* | 384 | 0 | 0 | |
| Median viral DNA load* (range) | 384 (384) | 0 | 0 | |
*viral load as copies/106 cells. PA, pleomorphic adenoma; RPA recurrent pleomorphic adenoma; CaxPA carcinoma ex pleomorphic adenoma; HHV-6B, human herpesvirus 6B; HHV 7, human herpesvirus 7; EBV, Epstein-Barr virus; MCPyV, Merkel cell virus; MWPyV, Malawi polyomavirus; B19V, parvovirus B19.
3.2Presence of virus DNA in different tumor types
Of the 68 tumor samples, 31 harbored DNA of 7 different viruses (Tables 1 and 2). The viral DNA quantities are shown in Table 1. The human RNase-P qPCR varied between 10e2-10e4 copies/μl among all samples except 2, which were 10e1 copies/μl. Twenty-three viruses were not detected in any sample.
Virus DNA was detected in 19/68 (28%) tumors, more often in CaxPA (7/12, 58%) than in RPA (4/31, 13%, p = 0.0047, Fisher’s exact test) but with no significant difference to that of PA (8/25, 32%, p = 0.1642). Among the different virus families, herpesvirus DNA was detected in 11/50 (22%) tumors, clearly more often in CaxPA (7/12, 58%) than in PA (2/15, 13%, p = 0.0369) or RPA (2/23, 9%, p = 0.0030). Among Herpesviridae, the most common finding was HHV-7, detected significantly more often in CaxPA (7/12, 58%) than in PA (2/15, 13%, p = 0.0369) or in RPA (2/23, 9%, p = 0.0030). In the CaxPA group, 15 herpes virus findings were congregated in 7/12 tumor samples (Table 2 and 3), in which 7 (58%), 5 (42%) and 3 (25%) samples were positive for HHV-7, HHV-6B and EBV, respectively. HHV-6B and EBV DNAs were absent in PAs and RPAs. All samples, in every tumor type, tested DNA negative for HSV-1, HSV-2, VZV, HHV-6A, CMV and HHV-8.
Table 3
Presence of viral DNAs in individual CaxPA samples
| Pat. ID | HHV-7 | HHV-6B | EBV | MCPyV | B19 | Types of viruses/tumor |
| P65 | 1 | 1 | 2 | |||
| P66 | 1 | 1 | ||||
| P67 | 1 | 1 | 1 | 3 | ||
| P68 | 0 | |||||
| P69 | 0 | |||||
| P70 | 1 | 1 | 2 | |||
| P71 | 1 | 1 | 1 | 1 | 4 | |
| P72 | 1 | 1 | 1 | 3 | ||
| P73 | 0 | |||||
| P74 | 0 | |||||
| P75 | 1 | 1 | 1 | 3 | ||
| P78 | 0 | |||||
| Positive no. | 7 | 5 | 3 | 2 | 1 | |
| % | 58 | 42 | 25 | 17 | 8 |
CaxPA carcinoma ex pleomorphic adenoma; HHV-7, human herpesvirus 7; HHV-6B, human herpesvirus 6B; EBV, Epstein-Barr virus; MCPyV, Merkel cell virus; B19, B19 parvovirus.
Polyomaviruses comprised the second most prevalent group with 7 virus findings in 68 samples (10%). The most common viral DNA finding was of MCPyV, which was positive in 2/25 (8%) PAs, in 2/31 (6%) RPAs and in 2/12 (17%) CaxPAs. MWPyV DNA was positive in 1/25 (4%) samples among PAs and negative in the other groups.
Among 68 samples screened for parvoviruses, 3/25 (12%) PAs and 1/12 (8%) CaxPAs were positive for B19V DNA. Of the 3 sequenced B19V amplicons, 1 was of genotype 1 and 2 were of genotype 2, confirming the findings. CuV DNA was detected in a single sample among the 25 (4%) PAs, which was confirmed by cloning and sequencing. Neither BuV, TuV nor bocavirus DNA was found in these samples.
3.3Co-detection of multiple viruses in CaxPA samples
Co-infections with multiple viruses were present in 6/12 (50%) CaxPA samples (Table 3). In the PA group only 1 sample harbored multiple viruses. In the RPA group, there were no multiple virus-DNA findings. In the CaxPA group 3/7 viral DNA-positive samples contained 3, and 1 sample even 4, types of viruses. HHV-7 and HHV-6B DNAs were co-detected in 5/12 (42%) CaxPA samples. Two out of 3 EBV-DNA findings were among the 5 HHV-6B/HHV-7 dual-positive samples, and once EBV DNA was co-detected with HHV-6B and MCPyV.
4Discussion
Our study focused on the potential presence of 30 DNA viruses in PA, RPA or CaxPA. Seven different viruses were detected in these tumors, especially in CaxPA. The most common findings were herpesviruses (altogether 11 HHV-7, 5 HHV-6B, and 3 EBV-DNA findings), which were enriched in CaxPA samples (Table 2). Remarkably, multiple viruses were present in 88% of the virus DNA-positive CaxPA samples (Table 3). In addition, DNAs of altogether 7 polyomaviruses (6 MCPyV, 1 MWPyV) and 5 parvoviruses (4 B19V, 1 CuV) were detected in different tumor types. Only duplicate positive virus DNA findings or sequenced viruses were considered positive for qPCR, whereas single positive findings were accepted for Luminex-based multiplex assay (polyomaviruses).
Due to the lack of previous studies on the presence of viral DNA in CaxPA, our results showing multiple simultaneous herpesviruses in CaxPA samples are noteworthy. Seven out of 12 samples (58%) contained herpesvirus DNA, and 6 samples harbored multiple herpesviruses, which was in striking contrast to PA and RPA, where single herpesvirus DNA was detected only in 2 samples each (13% and 9%, respectively) and all were single detections. Furthermore, in all these 4 cases the virus was HHV-7, which was also detected in 58% of CaxPA samples. EBV DNA was detected only in the CaxPA group, and it was always found together with HHV-6B or HHV-7. Due to the unknown etiology of CaxPA, the potential role of herpesviruses in carcinogenesis is intriguing. Malignant oral tissues such as squamous cell carcinomas exhibit HHV-6 DNA [42], and nasopharyngeal carcinomas may often harbor EBV [43].
Our results on the presence of viral DNA in PA samples are largely in line with previous reports. In our material MCPyV DNA was found in 2/25 (8%) PA. Chen et al. [8] found MCPyV DNA in 10/51 (20%) and EBV DNA in 2/51 (4%) PA samples. In our series, EBV DNA was detected only in CaxPA. Shanehsazzadeh et al. [9] found HHV-7 DNA in 10/23 (43%) pleomorphic or monomorphic adenoma tumors, whereas in our study HHV-7 DNA was present only in 2/15 (13%) PAs. They also reported HHV-6 DNA in 4/23 (17%) PA samples, whereas we detected none.
Regarding RPA, we were unable to shed much light on its etiology as HHV-7 and MCPyV were the only viruses detected, and they were present in only a few samples (2/23, 9% and 2/31, 6%, respectively). However, as there are no previous reports concerning the possible virome of RPA, these findings are novel.
Importantly, for our herpesvirus analyses we collected punch biopsies, which allowed us to minimize the amount of surrounding normal salivary gland tissue. With these samples we were able to demonstrate that benign PA, even when recurrent, seems to lack HHV-6B and EBV. However, these viruses are present in malignant tumors. HHV-6 and -7 are frequently found in the gastrointestinal tract and its organs, as well as in the salivary glands and saliva [44, 45]. In the salivary glands the latent virus infection is assumed to take place in monocytes, macrophages and CD4 + T lymphocytes [44, 46]. HHV-6 and HHV-7 have the unique feature of being able to modify the host-cell immune response and might facilitate the effects of other agents or viruses on these cells [47]. This could be a possible explanation for the presence of HHV-6B and -7 in the tumors that undergo malignant transformation.
Apparently, EBV does not cause epithelial cell transformation alone and additional host gene mutations are needed for malignant transformation of EBV-infected cells [48]. Interestingly, all our 3 EBV-positive CaxPAs had 2 additional concomitant viruses present, HHV-6B and HHV-7 in 2 cases and HHV-7 and MCPyV in 1 case. It is intriguing to speculate that the malignant transformation of these tumors might be related to the possible interplay between the different viruses.
In our study, we found traces of MCPyV in 6 samples and MWPyV in 1. These positive findings were scattered across all tumor groups and were not repeatable with either the PCR-Luminex-based system or with qPCR, suggesting that the virus loads were very low.
CuV DNA was found in one PA tissue slice sample. The presence of CuV in PA has not been reported previously. This is an interesting finding as CuV has been associated with cutaneous T-cell lymphoma and has been detected in other skin cancers, including melanoma [33–36]. The other parvovirus detected in this study, B19V, persists in various tissues without a known clinical significance [28, 49]. It is, however, intriguing that we found B19V only in PA and CaxPA but not in RPA.
The limitations of the study included having only FFPE samples available. The formalin treatment is harmful for the DNA structure and causes DNA degradation. However, all our samples showed RNase-P amplification, indicating that at least some DNA was intact. In addition, most of our qPCRs target short genome regions allowing even fractioned virus DNA sections to be traced. In some samples the tumor material was rather scant. Thus, we were unable to harvest punch biopsies from the whole series but ended up with 50 punch biopsies and slices from 18 samples. A slice sample includes usually both the tumor and the surrounding healthy tissue, and viral DNA positivity can therefore not be considered specific for the tumor alone. Thus, larger FFPE samples or fresh-frozen samples could contribute to more intact DNA structure, higher cell counts and tumor-specific results.
One has to bear in mind that the mere presence of viral DNA in the tumor specimens is not as such an indication of an oncogenic role –the virus can also be oncotropic and oncolytic, or an innocent bystander. Viruses can be found ubiquitously in human normal and neoplastic tissues. It is possible that viruses are just attracted on site by the rich vasculature, rapid cell division, or other factors of the malignancy. The presence of HHV-6B, HHV-7 and EBV in CaxPA, and the lack of the viruses in benign PAs, do require further studies and characterization of their cellular location and activity. This remains to be addressed with additional methods in future studies to define the role of viruses in tumorigenesis and malignant transformation.
Modern molecular biology techniques have enabled rapid progression in virus discovery and research [30]. The potential of viral genomes to integrate into the host genome, or alter the cell function, can cause a diversity of effects yet to be detected and explored.
Acknowledgments
The Helsinki University Hospital research fund, the Sigrid Jusélius Foundation, the Life and Health Medical Support Association, and the Finnish-Norwegian Medical Foundation supported the study.
Author contributions
CONCEPTION: MKJ, TA, KA, AAM, MSV and STS
ANALYSIS AND INTERPRETATION OF DATA: MKJ, MX, LP, AAM, MSV and STS
PREPARATION OF THE MANUSCRIPT: MKJ, AAM and STS
REVISION FOR IMPORTANT INTELLECTUAL CONTENT: MKJ, MX, LP, TA, KA, AM, CH, JH, AAM, MSV and STS
SUPERVISION: AAM, MSV and STS
Conflict of interest
The authors have reported to have no conflicts of interest.
Supplementary material
[1] The supplementary Tables are available in the electronic version of this article: https://dx.doi.org/10.3233/TUB-211519.
References
[1] | Valstar MH , de Ridder M , van den Broek EC , Stuiver MM , van Dijk BAC , van Velthuysen MLF , Balm AJM , Smeele LE . Salivary gland pleomorphic adenoma in the Netherlands: A nationwide observational study of primary tumor incidence, malignant transformation, recurrence, and risk factors for recurrence. Oral Oncol. 2017; doi:S1368-8375(17)30011-8. |
[2] | Colella G , Cannavale R , Chiodini P . Meta-analysis of surgical approaches to the treatment of parotid pleomorphic adenomas and recurrence rates. J Craniomaxillofac Surg. 2015; doi:S1010-5182(15)00071-2. |
[3] | Andreasen S , Therkildsen MH , Bjorndal K , Homoe P . Pleomorphic adenoma of the parotid gland 1985-2010: A Danish nationwide study of incidence, recurrence rate, and malignant transformation. Head Neck. 2016; doi:10.1002/hed.24228. |
[4] | Aro K , Valle J , Tarkkanen J , Makitie A , Atula T . Repeatedly recurring pleomorphic adenoma: a therapeutic challenge. Acta Otorhinolaryngol Ital. 2019; doi:10.14639/0392-100X-2307. |
[5] | Guzzo M , Locati LD , Prott FJ , Gatta G , McGurk M , Licitra L . Major and minor salivary gland tumors. Crit Rev Oncol. (2010) ;74: :134–48; doi: https://doi.org/10.1016/j.critrevonc.2009.10.004. |
[6] | GROSS L . A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc Soc Exp Biol Med. (1953) ;83: :414–21. |
[7] | Morgan GJ . Ludwik Gross, Sarah Stewart, and the 1950s discoveries of Gross murine leukemia virus and polyoma virus. Stud hist philos biol biomed sci. 2014; doi: https://dx.doi.org/10.1016/j.shpsc.2014.07.013. |
[8] | Chen AA , Gheit T , Stellin M , Lupato V , Spinato G , Fuson R , Menegaldo A , Mckay-Chopin S , Dal Cin E , Tirelli G , Da Mosto MC , Tommasino M , Boscolo-Rizzo P . Oncogenic DNA viruses found in salivary gland tumors. Oral Oncol. 2017; doi:S1368-8375(17)30358-5. |
[9] | Shanehsazzadeh M , Rad JS , Pourazar A , Behbahani M . Epidemiology of herpes human virus 6 and 7 infections in salivary gland neoplasms in Isfahan, Iran. Med Arch. 2014; doi:10.5455/medarh.2014.68.276-278. |
[10] | Descamps G , Duray A , Rodriguez A , Chantrain G , Depuydt CE , Delvenne P , Saussez S . Detection and quantification of human papillomavirus in benign and malignant parotid lesions. Anticancer Res. 2012; doi:32/9/3929. |
[11] | Plummer M , de Martel C , Vignat J , Ferlay J , Bray F , Franceschi S . Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016; doi:10.1016/S2214-109X(16)30143-7. |
[12] | Gissmann L , Boshart M , Dürst M , Ikenberg H , Wagner D , Hausen HZ . Presence of human papillomavirus in genital tumors. Journal of Investigative Dermatology. (1984) ;83: :S26–S28. |
[13] | Feng H , Shuda M , Chang Y , Moore PS . Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008; doi:10.1126/science.1152586. |
[14] | Syrjänen K , Syrjänen S , Lamberg M , Pyrhönen S , Nuutinen J . Morphological and immunohistochemical evidence suggesting human papillomavirus (HPV) involvement in oral squamous cell carcinogenesis. International Journal of Oral Surgery. (1983) ;12: :418–24. |
[15] | Beasley RP , Lin C , Hwang L , Chien C . Hepatocellular carcinoma and hepatitis B virus: A Prospective Study of 22 707 Men in Taiwan. The Lancet. (1981) ;318: :1129–33. |
[16] | Branda M , Wands JR . Signal transduction cascades and hepatitis B and C related hepatocellular carcinoma Hepatology. Hepatology. (2006) ;43: :891–902 doi: 10.1002/hep.21196. |
[17] | Okuda K . Hepatocellular carcinoma: Recent progress. Hepatology. (1992) ;15: :948–63. doi: 10.1002/hep.1840150532. |
[18] | Chang Y , Cesarman E , Pessin MS , Lee F , Culpepper J , Knowles DM , Moore PS . Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994. |
[19] | Nüesch JP , Lacroix J , Marchini A , Rommelaere J . Molecular pathways: rodent parvoviruses–mechanisms of oncolysis and prospects for clinical cancer treatment. Clin Cancer Res. 2012; doi:10.1158/1078-0432.CCR-11-2325. |
[20] | Beral V , Peterman TA , Berkelman RL , Jaffe HW . Kaposi’s sarcoma among persons with AIDS: a sexually transmitted infection? Lancet. 1990; doi:0140-6736(90)90001-L. |
[21] | Chen JY , Liu MY , Lynn TC , Yang CS . Antibody to Epstein-Barr virus-specific DNase in patients with nasopharyngeal carcinoma. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi. 1982. |
[22] | Thawaranantha D , Chimabutra K , Balachandra K , Warachit P , Pantuwatana S , Inagi R , Kurata T , Yamanishi K . Prevalences of human herpesvirus 6 and human herpesvirus 7 in normal Thai population. Southeast Asian J Trop Med Public Health. 1999. |
[23] | Bhattacharjee S , Chattaraj S . Entry, infection, replication, and egress of human polyomaviruses: an update. Can J Microbiol. 2017; doi:10.1139/cjm-2016-0519. |
[24] | Sadeghi M , Wang Y , Ramqvist T , Aaltonen LM , Pyöriä L , Toppinen M , Söderlund-Venermo M , Hedman K . Multiplex detection in tonsillar tissue of all known human polyomaviruses. BMC Infect Dis. 2017; doi:10.1186/s12879-017-2479-5. |
[25] | Poluschkin L , Rautava J , Turunen A , Wang Y , Hedman K , Syrjanen K , Grenman R , Syrjanen S . Polyomaviruses detectable in head and neck carcinomas. Oncotarget. 2018; doi:10.18632/oncotarget.25202. |
[26] | Dickinson A , Xu M , Silén S , Wang Y , Fu Y , Sadeghi M , Toppinen M , Carpén T , Hedman K , Mäkitie A , Söderlund-Venermo M . Newly detected DNA viruses in juvenile nasopharyngeal angiofibroma (JNA) and oral and oropharyngeal squamous cell carcinoma (OSCC/OPSCC). Eur Arch Otorhinolaryngol. 2019; doi:10.1007/s00405-018-5250-7. |
[27] | Qiu J , Söderlund-Venermo M , Young NS . Human parvoviruses. Clin Microbiol Rev. 2017; doi:30/1/43. |
[28] | Hokynar K , Norja P , Hedman K , Söderlund-Venermo M . Tissue persistance and prevalence of B19 virus types 1-3. Future Medicine. (2007) ;2: :377–388. |
[29] | Pyöriä L , Toppinen M , Mäntylä E , Hedman L , Aaltonen LM , Vihinen-Ranta M , Ilmarinen T , Söderlund-Venermo M , Hedman K , Perdomo MF . Extinct type of human parvovirus B19 persists in tonsillar B cells. Nat Commun. 2017;doi:10.1038/ncomms14930. |
[30] | Söderlund-Venermo M . Emerging human parvoviruses: The rocky road to fame. Annu Rev Virol. 2019;doi:10.1146/annurev-virology-092818-015803. |
[31] | Phan TG , Vo NP , Bonkoungou IJ , Kapoor A , Barro N , O’Ryan M , Kapusinszky B , Wang C , Delwart E . Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J Virol. 2012; doi:JVI.01427-12. |
[32] | Phan TG , Sdiri-Loulizi K , Aouni M , Ambert-Balay K , Pothier P , Deng X , Delwart E . New parvovirus in child with unexplained diarrhea, Tunisia. Emerg Infect Dis. 2014; doi:10.3201/eid2011.140428. |
[33] | Phan TG , Dreno B , da Costa AC , Li L , Orlandi P , Deng X , Kapusinszky B , Siqueira J , Knol AC , Halary F , Dantal J , Alexander KA , Pesavento PA , Delwart E . A new protoparvovirus in human fecal samples and cutaneous T cell lymphomas (mycosis fungoides). Virology. 2016; doi: S0042-6822(16)30153-2. |
[34] | Mollerup S , Fridholm H , Vinner L , Kjartansdóttir KR , Friis-Nielsen J , Asplund M , Herrera JA , Steiniche T , Mourier T , Brunak S , Willerslev E , Izarzugaza JM , Hansen AJ , Nielsen LP . Cutavirus in cutaneous malignant melanoma. Emerg Infect Dis. 2017; doi:10.3201/eid2302.161564. |
[35] | Kreuter A , Nasserani N , Tigges C , Oellig F , Silling S , Akgül B , Wieland U . Cutavirus infection in primary cutaneous B- and T-cell lymphoma. JAMA Dermatol. 2018; doi:10.1001/jamadermatol.2018.1628. |
[36] | Väisänen E , Fu Y , Koskenmies S , Fyhrquist N , Wang Y , Keinonen A , Mäkisalo H , Väkevä L , Pitkänen S , Ranki A , Hedman K , Söderlund-Venermo M . Cutavirus DNA in malignant and nonmalignant skin of cutaneous T-cell lymphoma and organ transplant patients but not of healthy adults. Clin Infect Dis. 2019; doi:10.1093/cid/ciy806. |
[37] | Toppinen M , Norja P , Aaltonen LM , Wessberg S , Hedman L , Söderlund-Venermo M , Hedman K . A new quantitative PCR for human parvovirus B19 genotypes. J Virol Methods. 2015; doi: S0166-0934(15)00082-8. |
[38] | Pyöriä L , Jokinen M , Toppinen M , Salminen H , Vuorinen T , Hukkanen V , Schmotz C , Elbasani E , Ojala PM , Hedman K , Välimaa H , Perdomo MF . HERQ-9 Is a New multiplex PCR for differentiation and quantification of all nine human herpesviruses. mSphere. 2020; doi:10.1128/mSphere.00265-20. |
[39] | Wang Y , Strassl R , Helanterä I , Aberle SW , Bond G , Hedman K , Weseslindtner L . Multiplex analysis of human polyomavirus diversity in kidney transplant recipients with BK virus replication. J Clin Virol. 2019; doi: S1386-6532(19)30195-7. |
[40] | Kantola K , Sadeghi M , Antikainen J , Kirveskari J , Delwart E , Hedman K , Söderlund-Venermo M . Real-time quantitative PCR detection of four human bocaviruses. J Clin Microbiol. 2010; doi:10.1128/JCM.00686-10. |
[41] | Väisänen E , Lahtinen A , Eis-Hübinger AM , Lappalainen M , Hedman K , Söderlund-Venermo M . A two-step real-time PCR assay for quantitation and genotyping of human parvovirus 4. JVirol Methods. 2014; doi: S0166-0934(13)00415-1. |
[42] | Yadav M , Arivananthan M , Chandrashekran A , Tan BS , Hashim BY . Human herpesvirus-6 (HHV-6) DNA and virusencoded antigen in oral lesions. J Oral Pathol Med. 1997; doi:10.1111/j.1600-0714.1997.tb00238.x. |
[43] | Ruuskanen M , Irjala H , Minn H , Vahlberg T , Randen-Brady R , Hagström J , Syrjänen S , Leivo I . Epstein-Barr virus and human papillomaviruses as favorable prognostic factors in nasopharyngeal carcinoma: A nationwide study in Finland. Head Neck. 2019; doi:10.1002/hed.25450. |
[44] | Yamanishi K , Mori Y , Pellett P . Chapter 64 - Human Herpesviruses 6 and 7. In: Knipe DM, Howley PM, editors. Fields Virology, 6th Edition. Philadelphia, PA: Wolters Kluwer/LippincottWilliams &Wilkins,; (2013) . pp. 2058–2079. |
[45] | Wyatt LS , Frenkel N . Human herpesvirus 7 is a constitutive inhabitant of adult human saliva. J Virol. 1992;doi:10.1128/JVI.66.5.3206-3209.1992. |
[46] | Agut H , Bonnafous P , Gautheret-Dejean A . Human Herpesviruses 6A, 6B, and 7. Microbiol Spectr. 2016; doi:10.1128/microbiolspec.DMIH2-0007-2015. |
[47] | Eliassen E , Lum E , Pritchett J , Ongradi J , Krueger G , Crawford JR , Phan TL , Ablashi D , Hudnall SD . Human herpesvirus 6 and malignancy: A review. Front Oncol. 2018;doi:10.3389/fonc.2018.00512. |
[48] | Tsao SW , Tsang CM , To KF , Lo KW . The role of Epstein-Barr virus in epithelial malignancies. J Pathol. 2015;doi:10.1002/path.4448. |
[49] | Adamson-Small LA , Ignatovich IV , Laemmerhirt MG , Hobbs JA . Persistent parvovirus B19 infection in non-erythroid tissues: possible role in the inflammatory and disease process. Virus Res. 2014; doi: S0168-1702(14)00265-2. |




