Applications of deep learning models in precision prediction of survival rates for heart failure patients
Abstract
BACKGROUND:
Heart failure poses a significant challenge in the global health domain, and accurate prediction of mortality is crucial for devising effective treatment plans. In this study, we employed a Seq2Seq model from deep learning, integrating 12 patient features. By finely modeling continuous medical records, we successfully enhanced the accuracy of mortality prediction.
OBJECTIVE:
The objective of this research was to leverage the Seq2Seq model in conjunction with patient features for precise mortality prediction in heart failure cases, surpassing the performance of traditional machine learning methods.
METHODS:
The study utilized a Seq2Seq model in deep learning, incorporating 12 patient features, to intricately model continuous medical records. The experimental design aimed to compare the performance of Seq2Seq with traditional machine learning methods in predicting mortality rates.
RESULTS:
The experimental results demonstrated that the Seq2Seq model outperformed conventional machine learning methods in terms of predictive accuracy. Feature importance analysis provided critical patient risk factors, offering robust support for formulating personalized treatment plans.
CONCLUSIONS:
This research sheds light on the significant applications of deep learning, specifically the Seq2Seq model, in enhancing the precision of mortality prediction in heart failure cases. The findings present a valuable direction for the application of deep learning in the medical field and provide crucial insights for future research and clinical practices.
1.Introduction
Heart failure is a severe disease that significantly impacts global health, and its associated mortality rates remain alarmingly high [1]. Precise prediction of mortality risk in heart failure patients is crucial for formulating personalized treatment plans and enhancing patient survival. This study addresses this challenging issue by introducing the Seq2Seq model from deep learning, aiming to effectively and accurately predict the mortality rates of heart failure patients [2].
Accurate prediction of survival rates in heart failure patients has consistently posed a formidable challenge in the medical field. Despite the continuous development of medical technology and the accumulation of extensive patient health data, accurately capturing and predicting patient survival within these data remain a daunting task. This challenge stems from the diverse and nonlinear nature of factors influencing the physiological status of heart failure patients [3].
Primarily, the progression of heart failure is often influenced by various chronic diseases such as hypertension, diabetes, and coronary artery disease. These underlying diseases manifest differently among different patients, leading to individual differences in survival rates. Additionally, the clinical manifestations of heart failure itself exhibit high heterogeneity, including but not limited to decreased cardiac contractile function, fluid retention, and fatigue [4]. This complexity introduces temporal and dynamic features that increase the difficulty of the prediction task.
Moreover, modern medicine increasingly emphasizes personalized healthcare, requiring the comprehensive consideration of various factors such as patients’ basic physiological characteristics, medical history, and medication usage for predicting survival rates [5]. This necessitates prediction models capable of extracting key information from extensive and complex temporal data [6], aligning more closely with individual patient characteristics to enhance prediction accuracy.
In the face of these challenges, traditional machine learning methods may face limitations in handling temporal data and complex relationships. These methods typically rely on manually extracted features, overlooking potential patterns in the data and struggling to capture dynamic changes between different features [7]. Therefore, the introduction of deep learning models becomes a potent approach to improve the accuracy of survival rate predictions [8]. In addressing the aforementioned difficulties and challenges, this study leverages the Seq2Seq model in deep learning. Through an end-to-end approach to model complex temporal data, we strive to more accurately predict the survival rates of heart failure patients. By comprehensively considering patient features and utilizing advanced neural network structures, our commitment is to find a better balance between performance and interpretability, providing new perspectives and methods for addressing this challenging problem.
2.Related work
In the domain of predicting mortality rates in heart failure, researchers have extensively explored various modeling approaches, including traditional machine learning techniques and recently emerged deep learning methods. The following outlines some relevant prior work.
2.1Traditional machine learning methods
Previous studies have seen many scholars employ traditional machine learning algorithms to forecast heart failure mortality rates. These include K-Nearest Neighbors (KNN), Support Vector Machines (SVM), Decision Trees, Random Forests, XGBoost, and Gradient Boosting. Typically, these methods utilize manually extracted features such as patient age, biochemical indicators, etc. The models then undergo learning and prediction processes. However, traditional machine learning methods might face limitations in handling temporal data and complex relationships, making it challenging to capture long-term dependencies within the data [9].
2.2Deep learning methods
With the rise of deep learning, an increasing number of studies have focused on applying neural networks to predict heart failure mortality rates [10]. Recurrent Neural Networks (RNN) have been a common choice for better handling temporal data. However, traditional RNNs face challenges related to the vanishing gradient problem, leading to suboptimal performance in learning long sequences [11]. To address this, some studies have started adopting structures like Gated Recurrent Units (GRU) and Long Short-Term Memory Networks (LSTM) to enhance their modeling capability for long-term dependencies [12]. While these deep learning methods have improved predictive performance to some extent, they still face issues related to insufficient data and poor interpretability.
2.3Sequence-to-Sequence (Seq2Seq) models
Initially designed for natural language processing tasks, Seq2Seq models have gained popularity in the medical field in recent years [13]. Utilizing an encoder-decoder structure, these models effectively capture key information in input sequences and generate corresponding output sequences. In predicting heart failure mortality rates, Seq2Seq models offer flexibility in handling continuous medical records, modeling temporal relationships, and thereby improving prediction accuracy.
2.4Feature importance analysis
To comprehensively understand the workings of predictive models, some studies have conducted analyses of feature importance. Models like Random Forest inherently possess mechanisms for assessing feature importance, revealing which features are most critical for mortality rate prediction. These analyses provide insights for clinical practice, assisting healthcare professionals in better understanding patient risk factors [14].
In summary, prior research has made progress, but there still exist shortcomings in the modeling capability and interpretability of temporal data [15]. This study aims to address these issues by introducing the Seq2Seq model, combined with the GRU structure, seeking a better balance between performance and interpretability, and providing new perspectives and methods for accurately predicting mortality rates in heart failure patients.
3.Method
We utilized 12 features relevant to heart failure mortality (age, red blood cell levels, CPK enzyme levels, diabetes status, percentage of blood leaving the heart during systole, hypertension status, platelet count, creatinine levels, serum sodium levels, gender, smoking status, follow-up period) to predict whether patients would experience mortality.
For prediction, we employed a deep learning approach. As the patient’s medical condition features constitute a sequence, traditional Convolutional Neural Network (CNN) methods do not leverage their advantages effectively [16]. Therefore, we adopted a neural network model based on the Sequence-to-Sequence (Seq2Seq) structure.
The Seq2Seq model comprises an encoder and a decoder. The encoder is responsible for encoding the input sequence into an intermediate vector of fixed dimensions to capture essential information from the input sequence. The decoder is responsible for decoding the intermediate vector output by the encoder into the output sequence.
To allow the network to flexibly learn dependencies in long sequences and mitigate the vanishing gradient problem, we employed Gated Recurrent Units (GRU). GRU is an improved Recurrent Neural Network (RNN) structure with fewer parameters and better performance [17].
GRU is primarily based on gate control, including a reset gate and an update gate. The reset gate determines whether to consider the previous memory at the current time step. The update gate decides how the previous hidden state from the last time step combines with the current temporary memory state.
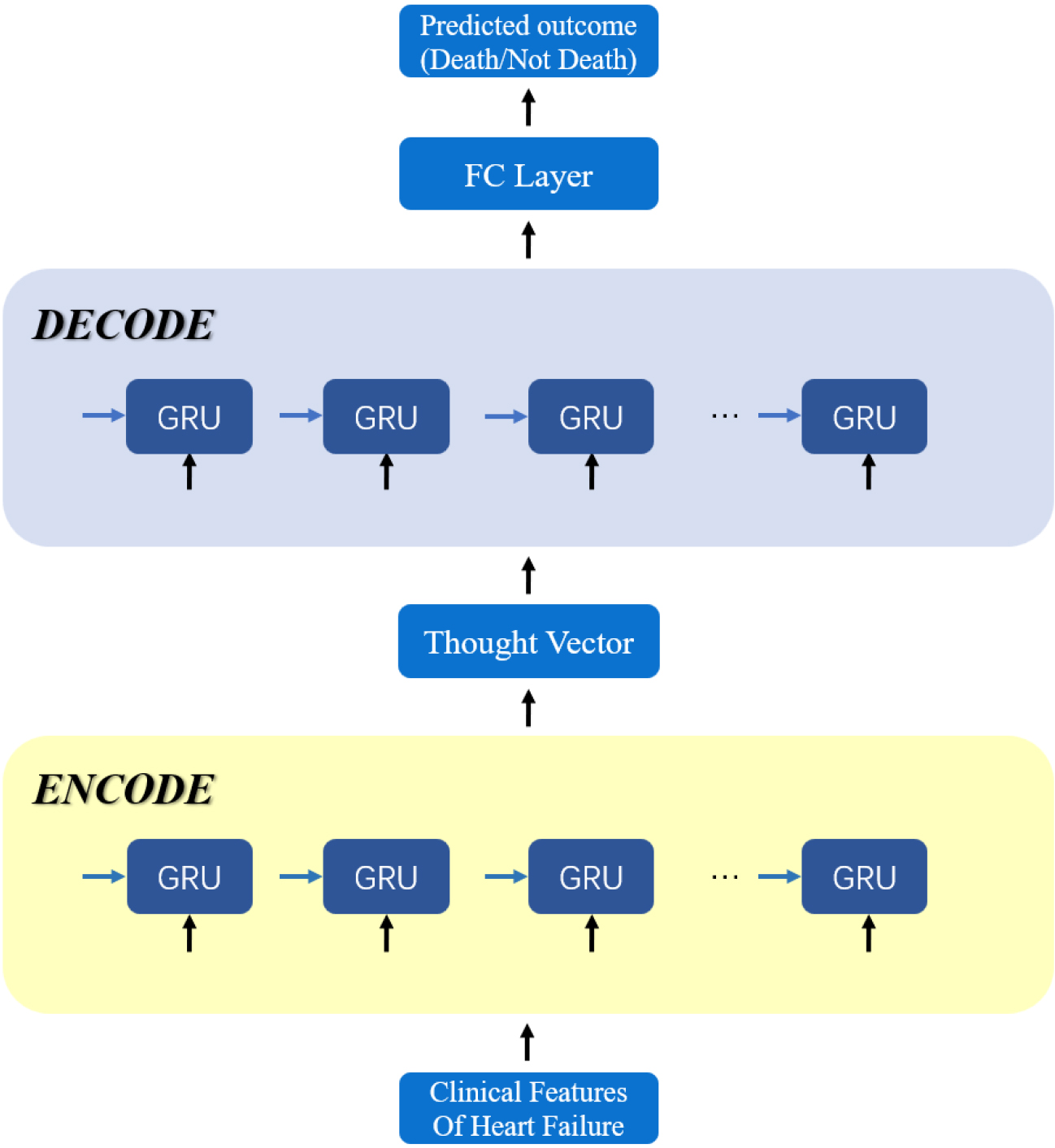
The Seq2Seq model combines the encoder and decoder to form an end-to-end network. At each time step, the encoder produces a hidden state, which is then passed to the decoder. The decoder generates the output for the next time step based on the previous time step’s output and the current time step’s hidden state. This process continues until the entire feature sequence is processed. To adapt to heart failure prediction, we utilized a fully connected layer in the model’s final layer, and the output results underwent probabilization through the Softmax function to obtain the final results. The model architecture is depicted in Fig. 1.
Figure 1.
The overall model diagram for heart failure prediction using the Seq2Seq model.
4.Experiment
We divided the data into training and validation sets in an 8:2 ratio. The validation set data did not undergo any data processing or model training stages and was solely used for the final model performance validation. When evaluating the model, the performance of the validation set represents the model’s generalization ability. Before training, we normalized the patient feature data to ensure that the mean and variance of the input data were within a reasonable range.
In this study, we employed cross-entropy loss as our loss function. We used the Adam optimizer for parameter updates. To prevent overfitting, we employed a series of strategies. Firstly, we introduced a Dropout layer before the model’s fully connected layer. Dropout randomly deactivates neurons during training, aiding in improving the model’s generalization ability. Secondly, we employed the Early Stopping strategy to monitor model performance on the validation set, halting training once performance ceased to improve.
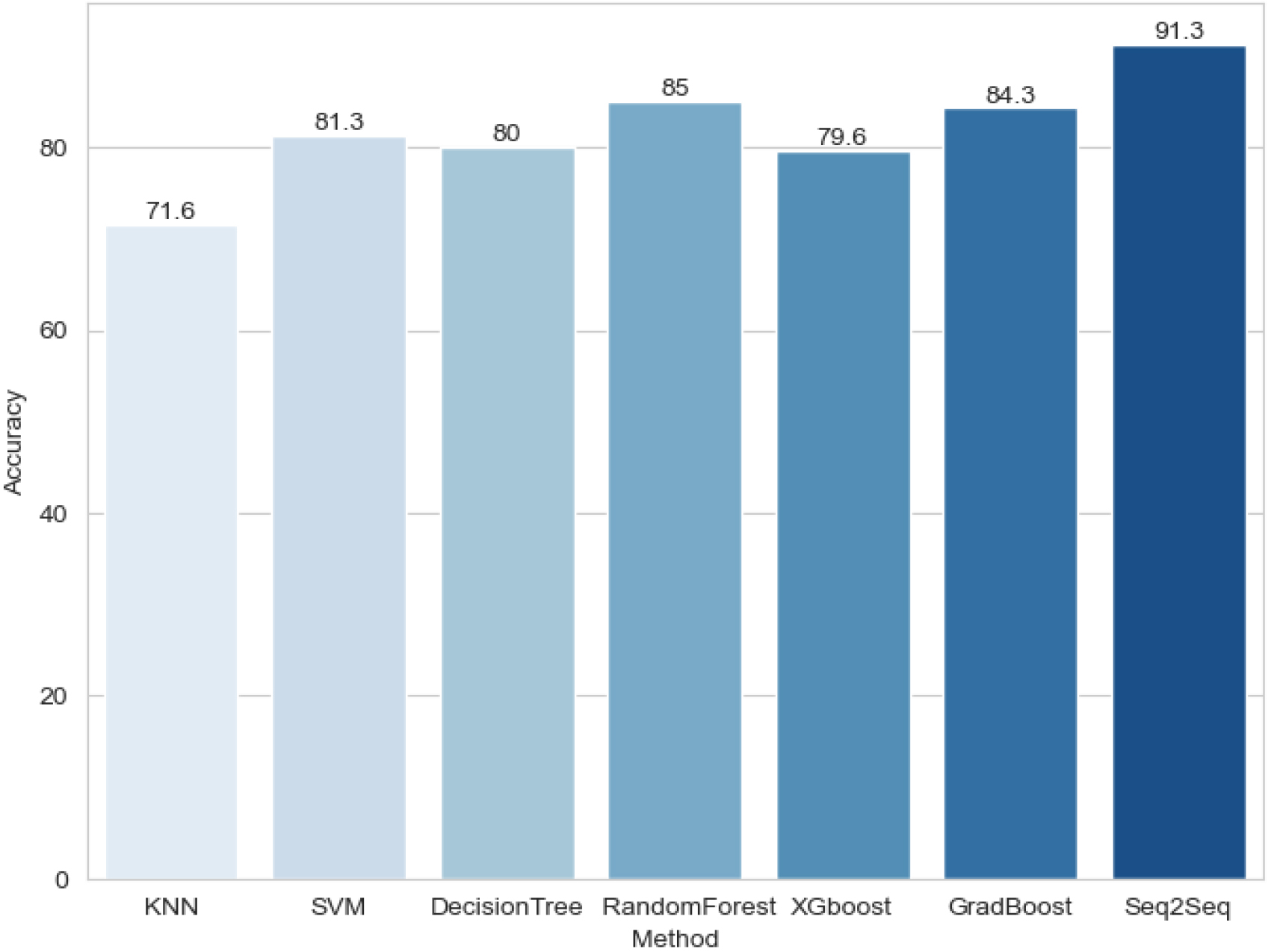
To demonstrate the effectiveness of the model, we conducted a comparative study, contrasting it with traditional machine learning methods (including K-Nearest Neighbors (KNN), Support Vector Machine (SVM), Decision Tree, Random Forest, XGBoost, and Gradient Boosting). Encouragingly, our model exhibited excellent performance, showcasing the highest accuracy and generalization ability. The accuracy comparison chart for different models is presented in Fig. 2.
To better reflect the performance of the model, we also calculated the “Precision,” “Recall,” and “Accuracy” for different models, and the results are presented in Table 1.
Table 1
Model results
| Model | Recall | Precision | Accuracy |
|---|---|---|---|
| KNN | 78.5% | 39.2% | 71.6% |
| SVM | 82% | 50% | 81.3% |
| Decision tree | 67.9% | 56% | 80% |
| Random Forest | 71.4% | 83.3% | 85% |
| XGBoost | 86% | 44% | 79.6% |
| GradBoost | 75% | 75% | 84.3% |
| Seq2Seq | 86.9% | 90.9% | 91.3% |
Figure 2.
Comparison chart of accuracy between our model and traditional machine learning methods.
Accuracy: It summarizes the performance of the classification task, measuring the number of instances correctly predicted among all data instances.
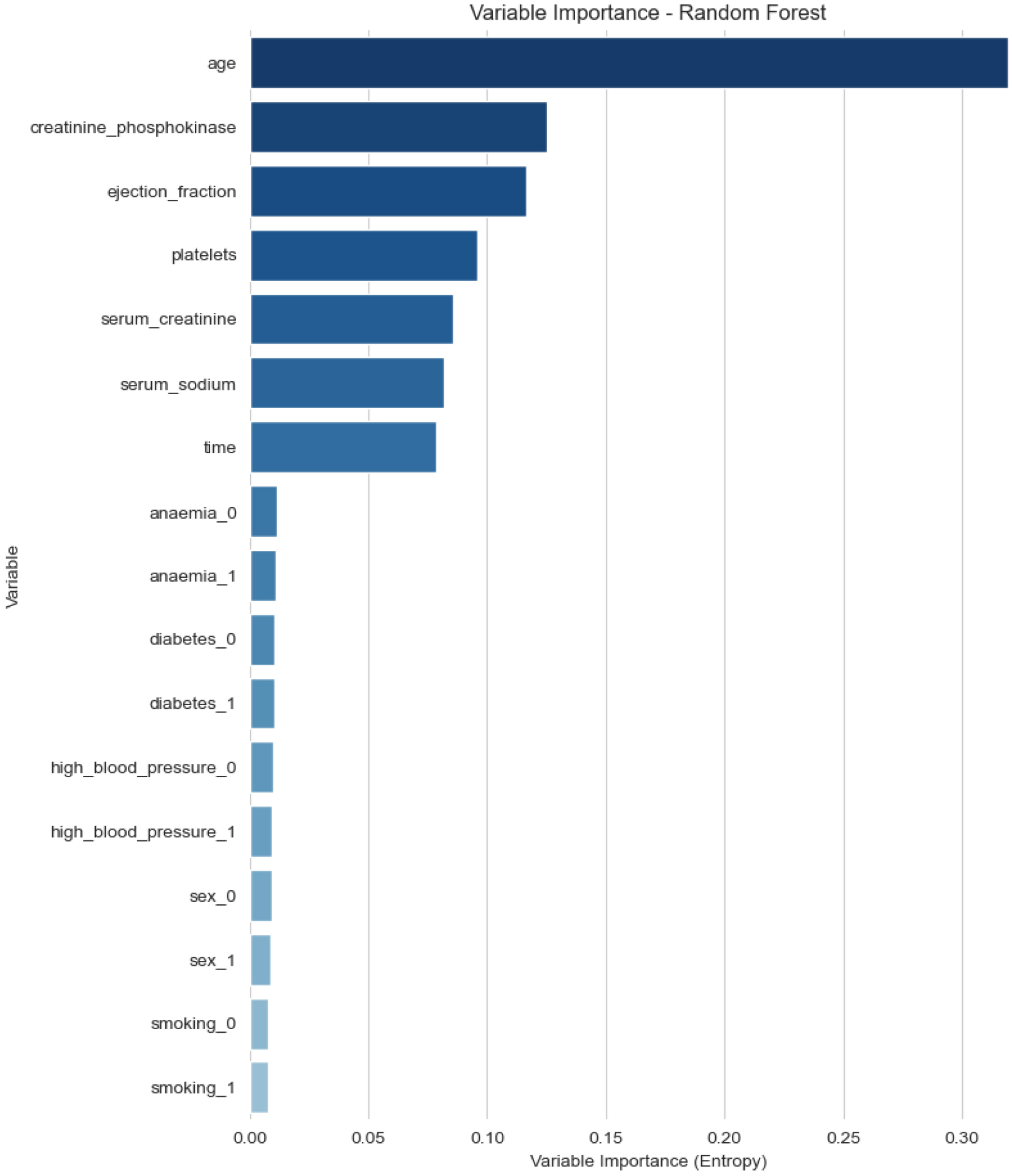
Figure 3.
Visualization of feature importance in heart failure prediction.
Recall: It represents the proportion of participants who experienced an ulcer event and were correctly classified among all participants.
Precision: It indicates how many of those identified as having an ulcer event actually belong to that category.
The formulas for calculating Recall, Precision, and Accuracy are as follows:
Recall
Precision
Accuracy
Where TP is true positive, TN is true negative, FP is false positive, and FN is false negative.
In addition, to better analyze the importance of different factors in causing death due to heart failure, we conducted feature importance analysis on the well-trained random forest model. The visualized feature importance output is presented in Fig. 3.
The experimental results of this study demonstrate the significant effectiveness of the Seq2Seq model in predicting heart failure mortality. The model not only surpasses traditional machine learning methods such as KNN, SVM, and decision trees in overall accuracy but also exhibits stronger performance in handling medical data with time-series characteristics. Specifically, the model can effectively capture and integrate long-term health records, providing accurate predictions of the patient’s future condition.
Further analysis reveals that certain features, such as ejection fraction, serum sodium levels, and CPK enzyme levels, have higher importance in the predictive model. This finding aligns with clinical experience, emphasizing the importance of continuous monitoring and management of these physiological parameters in heart failure treatment. Additionally, the model’s performance evaluation indicates that the introduction of the Dropout layer and Early Stopping strategy effectively prevents overfitting, ensuring the model’s generalization ability.
Overall, these experimental results not only validate the potential application of deep learning in the medical prediction field but also provide valuable insights for future clinical decision-making. Especially in the context of increasing emphasis on personalized medicine and precision treatment, the outcomes of this study will contribute to the development of more accurate and personalized treatment plans to improve the treatment outcomes and quality of life for heart failure patients.
5.Discussion
In this study, we utilized the Seq2Seq model for predicting heart failure mortality and achieved promising performance. Firstly, compared to traditional machine learning methods, the Seq2Seq model demonstrated higher accuracy and generalization ability, especially excelling in handling time-series data and long-term dependencies. Secondly, by introducing the GRU structure, the model could more effectively capture continuous medical records of patients, enhancing its ability to model potential risk factors.
However, it is essential to acknowledge the black-box nature of deep learning models and their dependency on data [18]. Although the Seq2Seq model exhibited superior predictive performance, challenges still exist in interpreting the decision-making process of the model. In future research, further exploration can be conducted on how to enhance the interpretability of the model, allowing physicians and patients to better understand the prediction results [19]. Additionally, the model’s robustness and handling of imbalanced data should also be areas of focus [20]. Overall, this study provides new insights into the application of deep learning in predicting heart failure mortality, but there is still room for further improvement.
6.Conclusion
In this study, we employed the Seq2Seq model from deep learning to address the challenges in predicting the survival rate of heart failure patients, achieving significant outcomes. By comprehensively considering the temporal features of patients and introducing advanced neural network structures, we successfully improved the accuracy of survival rate predictions, providing robust support for individualized medical care and treatment planning in the field of medicine. This offers a more reliable and precise tool for predicting the survival rates of future heart failure patients.
Our research achievements not only demonstrate technological advancements but also bring positive impacts to clinical practice. By accurately predicting the survival rates of heart failure patients, we offer healthcare professionals more detailed information, aiding in the formulation of personalized treatment plans and enhancing treatment effectiveness. This holds significant implications for achieving personalized healthcare and precision treatment, positively influencing the survival and quality of life of heart failure patients. In the future, we anticipate widespread applications of this research in medical practice, bringing positive impacts to the health of a broader patient population.
Acknowledgments
This work was supported by the Foundation of Xiamen Municipal Bureau of Science and Technology (3502Z20199039) and Xiamen City Key Medical and Health Project (3502Z20234018).
Conflict of interest
None to report.
References
[1] | Sajja GS. A Comprehensive Review of Various Machine Learning Techniques for Heart Disease Prediction. International Journal of Computer Applications. (2021) ; 183: (37): 53-56. |
[2] | Saha T, Chopra S, Saha S, et al. A large-scale dataset for motivational dialogue system: An application of natural language generation to mental health//2021 International Joint Conference on Neural Networks (IJCNN). IEEE, (2021) : 1-8. |
[3] | Choi E, Schuetz A, Stewart WF, et al. Using recurrent neural network models for early detection of heart failure onset. Journal of the American Medical Informatics Association. (2017) ; 24: (2): 361-370. |
[4] | Lee J, Woo J, Kang AR, et al. Comparative analysis on machine learning and deep learning to predict post-induction hypotension. Sensors. (2020) ; 20: (16): 4575. |
[5] | Roger VL. Epidemiology of heart failure: a contemporary perspective. Circulation Research. (2021) ; 128: (10): 1421-1434. |
[6] | Franssen FME, Alter P, Bar N, et al. Personalized medicine for patients with COPD: where are we? International Journal of Chronic Obstructive Pulmonary Disease. (2019) : 1465-1484. |
[7] | Gottdiener JS, Fohner AE. Risk prediction in heart failure: new methods, old problems. JACC: Heart Failure. (2020) ; 8: (1): 22-24. |
[8] | Guo A, Pasque M, Loh F, et al. Heart failure diagnosis, readmission, and mortality prediction using machine learning and artificial intelligence models. Current Epidemiology Reports. (2020) ; 7: : 212-219. |
[9] | Verduijn M, Sacchi L, Peek N, et al. Temporal abstraction for feature extraction: A comparative case study in prediction from intensive care monitoring data. Artificial Intelligence in Medicine. (2007) ; 41: (1): 1-12. |
[10] | Maragatham G, Devi S. LSTM model for prediction of heart failure in big data. Journal of Medical Systems. (2019) ; 43: (5). |
[11] | Silva Ferreira BW. In-hospital mortality prediction for patients with congestive heart failure using long short-term memory and gated recurrent units. Wichita State University, (2021) . |
[12] | Gong J, Bai X, Li D, et al. Prognosis analysis of heart failure based on recurrent attention model. Irbm. (2020) ; 41: (2): 71-79. |
[13] | Pourjafari E, Ziaei N, Rezaei MR, et al. Survival seq2seq: A survival model based on sequence to sequence architecture//Machine Learning for Healthcare Conference. PMLR. (2022) : 79-100. |
[14] | Ke J, Chen Y, Wang X, et al. Machine learning-based in-hospital mortality prediction models for patients with acute coronary syndrome. The American Journal of Emergency Medicine. (2022) ; 53: : 127-134. |
[15] | Ng K, Steinhubl SR, DeFilippi C, et al. Early detection of heart failure using electronic health records: practical implications for time before diagnosis, data diversity, data quantity, and data density. Circulation: Cardiovascular Quality and Outcomes. (2016) ; 9: (6): 649-658. |
[16] | Porumb M, Iadanza E, Massaro S, et al. A convolutional neural network approach to detect congestive heart failure. Biomedical Signal Processing and Control. (2020) ; 55: : 101597. |
[17] | Gao S, Zheng Y, Guo X. Gated recurrent unit-based heart sound analysis for heart failure screening. Biomedical Engineering Online. (2020) ; 19: : 1-17. |
[18] | Stiglic G, Kocbek P, Fijacko N, et al. Interpretability of machine learning-based prediction models in healthcare. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery. (2020) ; 10: (5): e1379. |
[19] | Chen P, Dong W, Wang J, et al. Interpretable clinical prediction via attention-based neural network. BMC Medical Informatics and Decision Making. (2020) ; 20: (3): 1-9. |
[20] | Ishaq A, Sadiq S, Umer M, et al. Improving the prediction of heart failure patients’ survival using SMOTE and effective data mining techniques. IEEE Access. (2021) ; 9: : 39707-39716. |




