Deep learning-based anatomical position recognition for gastroscopic examination
Abstract
BACKGROUND:
The gastroscopic examination is a preferred method for the detection of upper gastrointestinal lesions. However, gastroscopic examination has high requirements for doctors, especially for the strict position and quantity of the archived images. These requirements are challenging for the education and training of junior doctors.
OBJECTIVE:
The purpose of this study is to use deep learning to develop automatic position recognition technology for gastroscopic examination.
METHODS:
A total of 17182 gastroscopic images in eight anatomical position categories are collected. Convolutional neural network model MogaNet is used to identify all the anatomical positions of the stomach for gastroscopic examination The performance of four models is evaluated by sensitivity, precision, and F1 score.
RESULTS:
The average sensitivity of the method proposed is 0.963, which is 0.074, 0.066 and 0.065 higher than ResNet, GoogleNet and SqueezeNet, respectively. The average precision of the method proposed is 0.964, which is 0.072, 0.067 and 0.068 higher than ResNet, GoogleNet, and SqueezeNet, respectively. And the average F1-Score of the method proposed is 0.964, which is 0.074, 0.067 and 0.067 higher than ResNet, GoogleNet, and SqueezeNet, respectively. The results of the
CONCLUSION:
The method proposed exhibits the best performance for anatomical positions recognition. And the method proposed can help junior doctors meet the requirements of completeness of gastroscopic examination and the number and position of archived images quickly.
1.Introduction
Gastroscopic examination is a pivotal diagnostic tool for the detection of upper gastrointestinal diseases, with the identification of anatomical positions within gastric images being a primary goal to enhance the quality and efficacy of such examinations. Stomach disease is globally regarded as one of the most common disorders with an alarmingly high incidence rate [1]. Specifically, gastric cancer presents a stark statistic, accounting for one million new diagnoses and approximately 800,000 fatalities annually [2]. The significance of early detection and treatment is particularly critical for gastrointestinal tumours, as it substantially improves patient survival rates. Despite the importance of gastroscopy, the procedure is intricate and highly dependent on the proficiency of the operator due to various factors such as object heterogeneity and individual skill levels [3, 4]. Operator variability can lead to inconsistent examination quality [5, 6], and manual scope manipulation may cause motion blur that detracts from image clarity, potentially impacting lesion detection rates [7, 8]. Ensuring high-quality, effective gastroscopic examinations is, therefore, imperative, given the considerable risk of misdiagnosis in cancer detection through endoscopy [8]. This study revolves around augmenting the skillset of less experienced doctors to enhance the detection accuracy of gastric cancer, aiming to provide an effective method for identification of gastric anatomical positions.
A fundamental prerequisite for a comprehensive gastroscopic examination is the thorough inspection of the entire gastric mucosa to ensure no pathological regions are overlooked. New practitioners often encounter challenges in achieving this comprehensive coverage due to the complex anatomy of the stomach. Implementing an automated system that can identify and label the anatomical positions within gastroscopic images could significantly aid novices. Such a system would provide real-time feedback on whether all gastric regions have been visualized and if the acquired images are sufficient for diagnostic purposes, ensuring the detection of potential neoplastic lesions. Moreover, clear visualization of key anatomical areas is crucial for early detection of gastric cancer, as precise localization of lesions is essential for accurate assessment and subsequent treatment. The integration of this technology would enhance both the educational and clinical aspects of gastric endoscopy, particularly for those newly adopting this technique. The processing methods of gastroscopic image have been extensively studied, which focus on image noise reduction, enhancement, and multimodal fusion [9, 10, 11]. With the rapid development of deep learning technology [12, 13, 14, 15], it has made great progress in image segmentation, classification, and recognition [16, 17, 18, 19]. Gastric cancer and polyps have been widely studied as their high potential risk [20, 21, 22, 23, 24, 25, 26]. Some researchers have studied the image quality of endoscopy to ensure the reliability of gastroscopic examination [27, 28].
Few studies have been conducted on the recognition of anatomical parts in gastroscopic images. Takyama et al. have reported that GoogLeNet was successfully performed to identify six anatomical positions of the esophagogastroduodenoscopy images [29]. It demonstrates that deep learning technology is efficient to solve the problem of recognition of anatomical parts of the endoscope which is consistent with existing research indicating that deep learning methods are more effective than traditional methods in analysing and classifying medical images [30]. However, the anatomical parts of the stomach identified by this study remain unclear. In fact, the image covering the whole stomach is required during gastroscopic examination. This requires that all stomach positions be automatically recognized, which is not available in the studies currently reported.
In this paper, the automatic recognition method of anatomical positions for gastroscopic images is reported. To our knowledge, there is no research applying deep learning to identify entire the anatomical positions of the stomach for gastroscopic examination. The innovation of this paper is that, for the first time, a deep learning model based on convolutional neural network is used to identify entire the anatomical positions of the stomach for gastroscopic examination. The performance of the method proposed using MogaNet is compared with some existing methods, and it is superior to existing methods. The method proposed is helpful for junior doctors to quickly master the operation of gastroscopic examination to ensure completeness. We believe that this work can apply artificial intelligence technology to help doctors ensure the integrity of gastroscopic examination, which is significant for the development of more intelligent and standardized automatic gastroscopic examination in the future.
2.Materials and methods
2.1Image data
The images saved by gastroscopic examination are required to cover entire areas of the stomach. Since the anatomical structure of the stomach is relatively complex, eight anatomical positions were selected for identification based on the characteristics of the anatomical structure of the stomach. The entire stomach is covered by anatomical parts, as shown in Fig. 1. Due to the circular structure of the stomach, there are different views on the image of each part. For instance, corpus ventriculi includes images of the front wall, back wall, small angles, and large angles.
Data of gastroscopic images was collected from Weihai Municipal Hospital from October 2021 to March 2022 with Olympus CV-290 endoscopic imaging system (Olympus, Japan). The study was approved by the Hospital Ethics Committee and was in accordance with the principles of the Declaration of Helsinki. All patients had signed an informed consent. The registration number of this research at chictrorg.cn is ChiCTR2100052273.
Table 1
Image number of each anatomical position
| Position | Total | Training | Test |
|---|---|---|---|
| Cardia | 1898 | 1518 | 380 |
| Incisura angularis | 2870 | 2296 | 574 |
| Pylorus | 1843 | 1474 | 369 |
| Proximal duodenum | 1976 | 1580 | 396 |
| Descending duodenum | 1864 | 1491 | 373 |
| Pyloric antrum | 2034 | 1627 | 407 |
| Corpus ventriculi | 2849 | 2279 | 570 |
| Fundus ventriculi | 1848 | 1478 | 370 |
Figure 1.
Anatomical position to recognize for the gastroscopic examination.
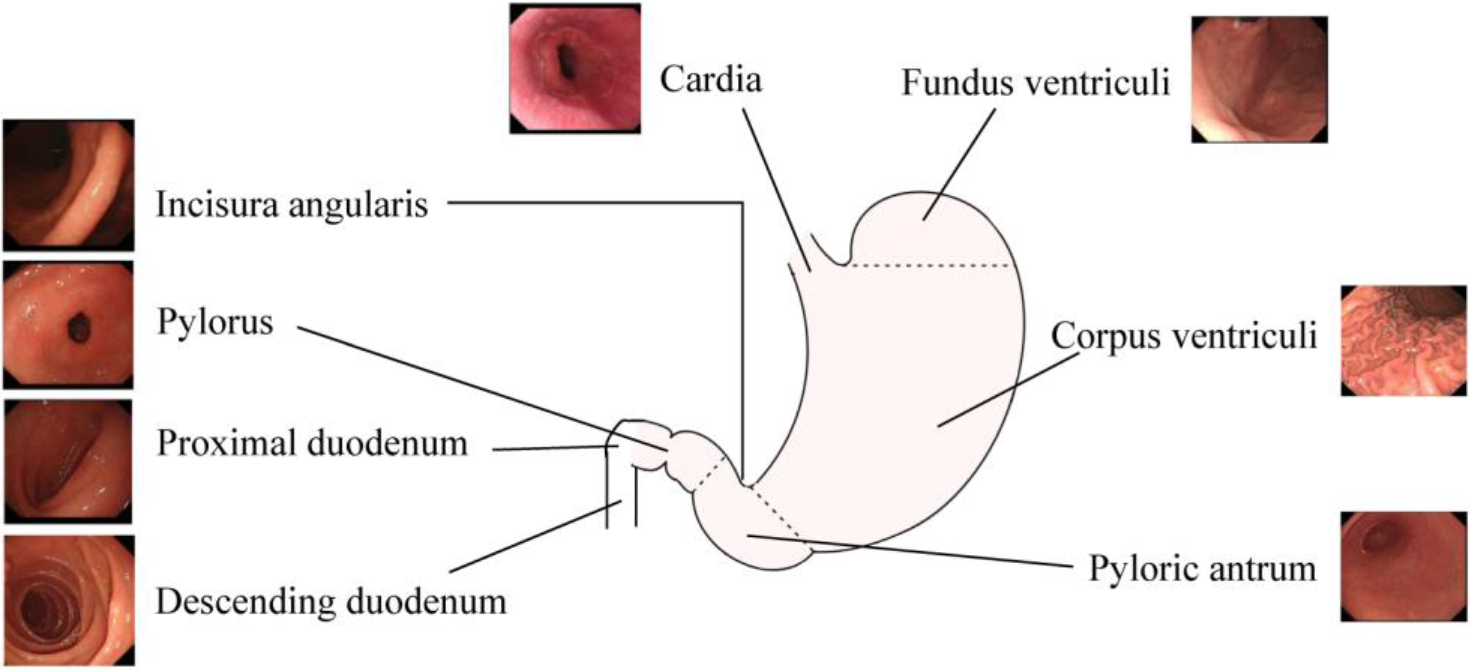
The quality of each image from the clinical examinations was strictly checked by three professional physicians. Images with blurry focus, large bubbles, and other coverings were considered as unqualified images, which should be excluded. The classification of these images was based on anatomical location by three professional physicians. When the judgment conclusions of the three doctors were consistent, the anatomical location was used as the label of the image. A total of 17182 gastroscopic images in eight categories were collected. Each data class was divided into training set and test set according to the ratio of 8:2 as shown in Table 1.
2.2Convolutional neural network architecture of MogaNet
Anatomical positions recognition is an important task for gastroscopic examination, which can be achieved by using deep learning methods based on convolutional neural networks (CNNs). CNNs can learn feature representations from images by applying convolution and pooling operations, which can capture the spatial and structural information of the images. However, commonly used CNNs have some limitations, such as losing the original resolution of the images, being sensitive to noise and illumination, and being weak in extracting texture features, which are crucial for distinguishing different anatomical parts. To address these issues, we employ a modified CNN model, named MogaNet, which incorporates the gated aggregation module to aggregate the local perception information in the middle layer, to enhance the performance of anatomical parts recognition [31]. The lightweight MogaNet-T network is used for anatomical position identification in this study. This architecture of MogaNet is illustrated in Fig. 2.
Figure 2.
Architecture of MogaNet.
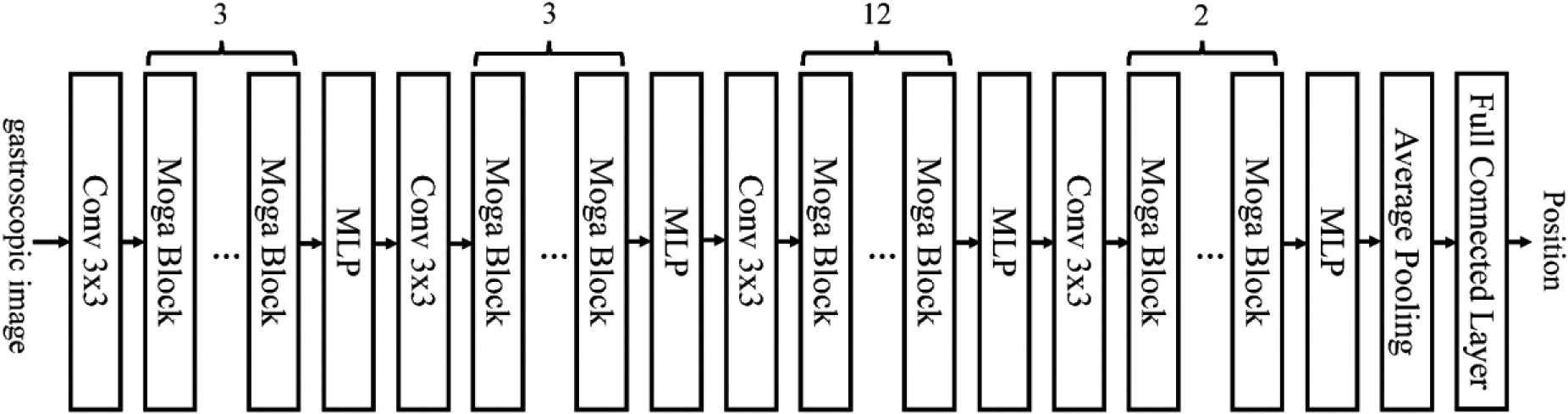
The backbone of MogaNet is composed of four stages with the same architecture. The number of Moga blocks in each stage is different. Each Moga block contains two parts: spatial aggregation and channel aggregation. In the spatial aggregation, multi-stage features with static and adaptive region awareness are extracted. With the help of this feature extraction, the network focuses on multi-level interaction. It makes up for the problem that traditional convolution only focuses on low-order and high-order [31]. In channel aggregation, unlike most networks, which increase a large number of parameters to ensure feature diversity, lightweight channel aggregation module is used to reweight high-dimensional hidden space. Previous studies have shown that by using the spatial and channel focus strategies to improve the global relevance, this model can achieve better classification performance than typical networks such as ResNet [31].
2.3Training and optimization
Due to the limited amount of data in the research, the transfer learning method is adopted. Transfer learning refers to the application of knowledge learned in one source domain to another target domain using models as a medium [32]. Convolutional neural network automatically learns image classification features through hierarchical method. The shallow neural network can extract the basic and general features of the image, such as points and lines, while the deep neural network can extract the special image features most relevant to the classification. Different image classification tasks have different special classification features, but the general image features of these classification methods are similar. When the weights of convolutional neural network obtained from image classification task on large datasets are applied to a new classification task, it is helpful to solve this image classification task. The transfer learning is realized through the pre training model, which is conducive to narrowing the search scope of the model and speeding up the training speed [32, 33]. With the transfer learning, the model can be fine-tuned to learn the feature distribution of the new task. In this application, the ImageNet data set is the source domain, the gastroscopic image is the target domain, and the image classification task is the target task. In this study, MogaNets trained weights on ImageNet are used as initial weights of the same layer in the proposed convolutional neural network. Then the network is trained on gastroscopic images.
Python 3.7 and PyTorch are used for the implementation of the method proposed. In this study, momentum acts as an optimizer, which is an improved method of the stochastic gradient descent method, and the loss function is cross-entropy loss. The learning rate is set to 0.002 at the beginning. From half of the maximum training times to the end of training, the learning rate decays exponentially to 0 with a training time of 1000 and a batch size of 32. The proposed network is trained on a PC with NVIDIA GeForce GTX 2080Ti GPU and Windows 10.
2.4Performance evaluation
The method proposed is compared with other convolutional neural network methods, including SqueezeNet [21], GooLeNet [29], and ResNet [22]. These methods have been widely used for various image recognition tasks, such as anatomical location classification [29] and cancer detection [21, 22]. To ensure a fair comparison, we adopt the transfer learning method for all the methods, and use the pre-trained weights based on ImageNet as the initial parameters. The performance is evaluated by sensitivity, accuracy, specificity, and F1 score. All methods are fine-tuned on the basis of the corresponding literature of the same data set. In the following sections, we present the experimental results and analysis of these methods on our dataset.
3.Results and discussion
The confusion matrix can provide detailed information on the classification of the test set. The confusion matrices of the four different methods are shown in Tables 2–5. The image number and percentage value of the classification result are all provided in the same figure. The diagonal of the matrix is the number and percentage of the correct classification of the corresponding category. The proposed method stands out as the singular approach that consistently achieves a sensitivity, precision, and F1-Score exceeding 90% across all categories. Upon meticulous examination of the accurate classification results by category, it is evident that the proposed method furnishes the precise assessment of anatomical positions for each distinct category.
Table 2
Confusion matrix of the GooLeNet method
| Real position | Cardia | Incisura andularis | Pylorus | Proximal duodenum | Descending duodenum | Pyloric antrum | Corpus ventriculi | Fundus ventriculi |
|---|---|---|---|---|---|---|---|---|
| Cardia | 339 | 9 | 19 | 7 | 0 | 6 | 0 | 0 |
| Incisura andularis | 0 | 531 | 12 | 7 | 9 | 4 | 0 | 11 |
| Pylorus | 24 | 3 | 314 | 4 | 6 | 13 | 5 | 0 |
| Proximal duodenum | 16 | 3 | 1 | 346 | 0 | 14 | 11 | 5 |
| Descending duodenum | 0 | 11 | 14 | 0 | 335 | 0 | 13 | 0 |
| Pyloric antrum | 3 | 8 | 4 | 11 | 7 | 362 | 8 | 4 |
| Corpus ventriculi | 5 | 0 | 12 | 0 | 7 | 8 | 538 | 0 |
| Fundus ventriculi | 4 | 0 | 11 | 0 | 15 | 7 | 0 | 333 |
Table 3
Confusion matrix of the ResNet method
| Real position | Cardia | Incisura andularis | Pylorus | Proximal duodenum | Descending duodenum | Pyloric antrum | Corpus ventriculi | Fundus ventriculi |
|---|---|---|---|---|---|---|---|---|
| Cardia | 338 | 12 | 14 | 5 | 3 | 8 | 0 | 0 |
| Incisura andularis | 0 | 530 | 8 | 9 | 11 | 6 | 3 | 7 |
| Pylorus | 18 | 6 | 309 | 9 | 4 | 14 | 9 | 0 |
| Proximal duodenum | 14 | 8 | 0 | 345 | 0 | 12 | 9 | 8 |
| Descending duodenum | 0 | 13 | 12 | 0 | 332 | 0 | 16 | 0 |
| Pyloric antrum | 1 | 9 | 7 | 6 | 9 | 357 | 13 | 5 |
| Corpus ventriculi | 6 | 0 | 14 | 2 | 9 | 6 | 533 | 0 |
| Fundus ventriculi | 0 | 2 | 16 | 0 | 17 | 5 | 0 | 330 |
Table 4
Confusion matrix of the SqeezeNet method
| Real position | Cardia | Incisura andularis | Pylorus | Proximal duodenum | Descending duodenum | Pyloric antrum | Corpus ventriculi | Fundus ventriculi |
|---|---|---|---|---|---|---|---|---|
| Cardia | 348 | 0 | 23 | 6 | 0 | 3 | 0 | 0 |
| Incisura andularis | 0 | 525 | 8 | 14 | 13 | 9 | 0 | 5 |
| Pylorus | 26 | 7 | 307 | 9 | 0 | 12 | 8 | 0 |
| Proximal duodenum | 13 | 0 | 6 | 343 | 0 | 19 | 7 | 8 |
| Descending duodenum | 0 | 8 | 12 | 0 | 340 | 4 | 9 | 0 |
| Pyloric antrum | 3 | 12 | 0 | 12 | 9 | 363 | 6 | 2 |
| Corpus ventriculi | 2 | 0 | 9 | 0 | 11 | 2 | 539 | 7 |
| Fundus ventriculi | 4 | 0 | 8 | 0 | 17 | 5 | 2 | 334 |
Table 5
Confusion matrix of the MogaNet method
| Real position | Cardia | Incisura andularis | Pylorus | Proximal duodenum | Descending duodenum | Pyloric antrum | Corpus ventriculi | Fundus ventriculi |
|---|---|---|---|---|---|---|---|---|
| Cardia | 370 | 0 | 6 | 4 | 0 | 0 | 0 | 0 |
| Incisura andularis | 0 | 562 | 2 | 4 | 3 | 3 | 0 | 0 |
| Pylorus | 12 | 3 | 334 | 5 | 4 | 11 | 0 | 0 |
| Proximal duodenum | 3 | 0 | 0 | 387 | 0 | 4 | 2 | 0 |
| Descending duodenum | 0 | 2 | 2 | 0 | 368 | 0 | 1 | 0 |
| Pyloric antrum | 6 | 4 | 0 | 5 | 4 | 381 | 7 | 0 |
| Corpus ventriculi | 0 | 4 | 3 | 0 | 1 | 0 | 558 | 4 |
| Fundus ventriculi | 0 | 0 | 2 | 2 | 3 | 0 | 4 | 359 |
Table 6
Average performance comparison of different methods
| Methods | Sensitivity | Precision | Accuracy | Specificity | F1-Score |
|---|---|---|---|---|---|
| ResNet | 0.889 | 0.892 | 0.973 | 0.985 | 0.890 |
| GoogLeNet | 0.897 | 0.897 | 0.975 | 0.986 | 0.897 |
| SqueezeNet | 0.898 | 0.896 | 0.975 | 0.986 | 0.897 |
| MogaNet | 0.963 | 0.964 | 0.991 | 0.995 | 0.964 |
Sensitivity, precision, accuracy, specificity, and F1 score are calculated to assess the performance of different methods for anatomical position classification. The average performance indicators of the four methods are listed in Table 6 for comparison. The performances of GoogLeNet and SqueezeNet are very similar, and both of them are better than the ResNet method. The average sensitivity of the MogaNet method proposed is 0.963, which is 0.074, 0.066 and 0.065 higher than ResNet, GoogleNet and SqueezeNet, respectively. The average precision of the MogaNet method proposed is 0.964, which is 0.072, 0.067 and 0.068 higher than ResNet, GoogleNet, and SqueezeNet, respectively. And the average F1-Score of the MogaNet method proposed is 0.964, which is 0.074, 0.067 and 0.067 higher than ResNet, GoogleNet, and SqueezeNet, respectively.
Based on the
The Sensitivity score for MogaNet is statistically higher compared to the other methods, with a t-statistic value of 23.993 and an extremely small
For Precision, the superiority of MogaNet is clear, as reflected by a t-statistic of 45.171 and a
In terms of Accuracy, again MogaNet outperforms the other approaches, shown by a t-statistic of 25.000 and a
When evaluating Specificity, MogaNet leads with a t-statistic of 28.000, with the
Finally, the F1-Score test also highlights MogaNet’s better performance with a t-statistic of 29.714 and a
These statistical metrics consistently demonstrate MogaNet’s improved performance across all tested indicators when compared to ResNet, GoogLeNet, and SqueezeNet. All tests reveal
Figure 3.
Performance comparison of each anatomical position category for different methods.
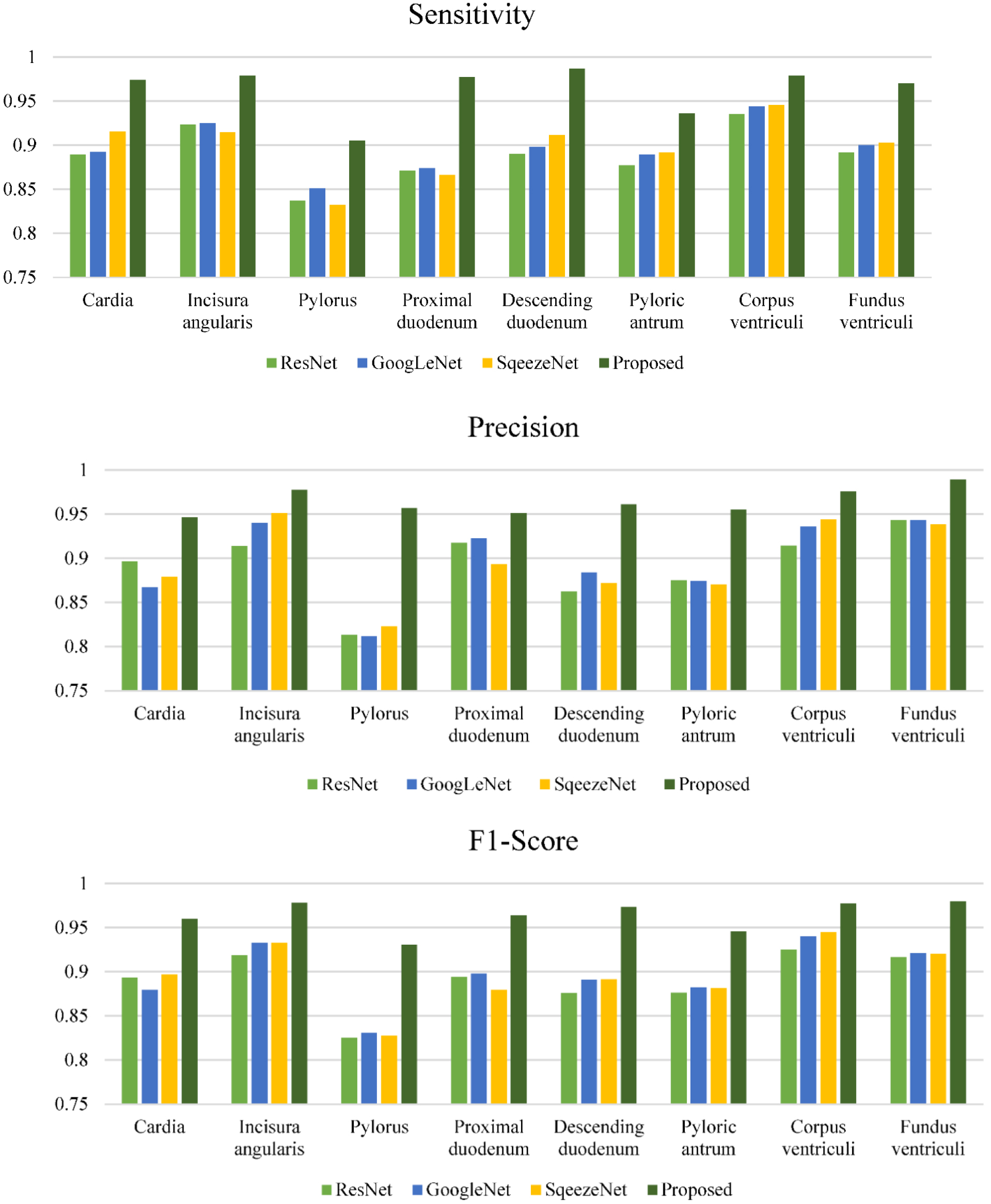
The indicators of each anatomical position category of different methods are also calculated and analysed. These indicators are illustrated in Fig. 3 for comparison. The accuracy and specificity of all methods for all anatomical positions are better than 90%. Different methods have great differences for different types of sensitivity and precision indicators. From these two indicators, among the four methods, ResNet has the lowest indicators, the MogaNet method proposed has the highest ones, and GoogLeNet and SqueezeNet are equivalent. In terms of different anatomical positions, the pylorus is the hardest to classify. Due to the introduction of spatial aggregation and channel aggregation, the MogaNet method in this paper exhibits obvious advantages in the recognition of all the anatomical positions.
GoogLeNet has been reported to recognize a few anatomical positions with a sensitivity and specificity of 98.9% and 93.0% [27]. But when it was used to identify more anatomical locations, the performance decreases significantly. When less training sample data is used, ResNet is not as effective as SqueezeNet. SqueezeNet is a lightweight network, which is not prone to overfitting. The MogaNet method proposed achieves better performance than SqueezeNet by introducing spatial aggregation and channel aggregation, and it is also the method with the best performance among the four methods, sensitivity, precision, accuracy, specificity and F1 score are increased by 0.065, 0.068, 0.016, 0.009 and 0.067 respectively.
Our proposed method for automatic recognition of anatomical positions of gastroscopic images can also facilitate the training of junior doctors. To perform a complete gastroscopic examination of the stomach, 40 images of different positions must be captured and saved. For beginners, it is a challenging task to ensure that the archived images are neither repeated nor omitted. Usually, they need two weeks of practice to meet the requirements. The method proposed is applied to the training of primary physicians in Weihai Municipal Hospital. The archived gastroscopic examination image was immediately transmitted to the recognition software according to the examination sequence. The recognition software displayed the completed positions and the number of image archives, as well as the missing image positions and the number of required images. The classification output of the convolutional neural network was probability, which helps users get more accurate information. The classification results of gastroscopic examination images and their probabilities were displayed. When the probability of the first classification result of a gastroscopic examination image was less than 90%, the software also showed the second most likely classification result and its probability. By using these TOP2 classification results, the accuracy of the recognition reached 100%. The information was very helpful for beginners, as they can save enough images in time according to the prompts. With the help of this software, all two primary trained doctors could ensure the completeness of gastroscopic examination in one week and meet the requirements of the position and the number of archived images. This is a preliminary result based on the trial of two junior doctors. We will conduct more detailed experiments and report the results in the future.
4.Conclusions
In comparison with various methods outlined in existing literature, the MogaNet method introduced in this study demonstrates promising potential in the recognition of gastroscopic positions. Performance metrics such as sensitivity, precision, and F1-score suggest that MogaNet may offer improvements over established approaches. Its application could potentially aid novice physicians in fulfilling the requirements for comprehensive gastroscopic examinations and the systematic archiving of images, both in terms of quantity and position.
Acknowledgments
This work was supported by the Scientific and Technological Development Plan Project of Shandong Province Medical and Health (2018WS104) and Shandong Provincial Natural Science Foundation (ZR2022MH047).
Conflict of interest
None to report.
References
[1] | Qin Y, Tong X, Fan J, Liu Z, Zhao R, Zhang T, et al. Global burden and trends in incidence, mortality, and disability of stomach cancer from 1990 to 2017. Clinical and Translational Gastroenterology. (2021) ; 12: (10): e00406. |
[2] | Park J, Herrero R. Recent progress in gastric cancer prevention. Best Practice & Research Clinical Gastroenterology. (2021) ; 50: : 101733. |
[3] | Gulati S, Patel M, Emmanuel A, Haji A, Hayee B, Neumann H. The future of endoscopy: Advances in endoscopic image innovations. Digestive Endoscopy. (2020) ; 32: (4): 512-522. |
[4] | East J, Vleugele J, Roelandt P, Bhandari P, Bisschops R, Dekker E, et al. Advanced endoscopic imaging: European society of gastrointestinal endoscopy (ESGE) technology review. Endoscopy. (2016) ; 48: (11): 1029-1045. |
[5] | Ponsky J, Strong A. A history of flexible gastrointestinal endoscopy. Surgical Clinics of North America. (2020) ; 100: (6): 971. |
[6] | Luo X, Mori K, Peters T. Advanced endoscopic navigation: Surgical big data, methodology, and applications. Annual Review of Biomedical Engineering. (2018) ; 20: : 221-251. |
[7] | He Z, Wang P, Liang Y, Fu Z, Ye Z. Clinically available optical imaging technologies in endoscopic lesion detection: Current status and future perspective. Journal of Healthcare Engineering. (2021) ; 2021: : 7954513. |
[8] | Namasivayam V, Uddo N. Quality indicators in the endoscopic detection of gastric cancer. DEN Open. (2023) ; 3: (1): e221. |
[9] | Perperidis A, Dhaliwal K, Mclaughlin S, Vercauteren T. Image computing for fibre-bundle endomicroscopy: A review. Medical Image Analysis. (2020) ; 62: : 101620. |
[10] | Fu Z, Jin Z, Zhang C, He Z, Zha Z, Hu C. The future of endoscopic navigation: A review of advanced endoscopic vision technology. IEEE Access. (2021) ; 9: : 41144-411167. |
[11] | Ali H, Yasmin M, Sharif M, Rehmani M. Computer assisted gastric abnormalities detection using hybrid texture descriptors for chromoendoscopy images. Computer Methods and Programs in Biomedicine. (2018) ; 157: : 39-47. |
[12] | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. (2015) ; 521: (7553): 436-444. |
[13] | Puttagunta M. Medical image analysis based on deep learning approach. Multimedia Tools and Applications. (2021) ; 80: (16): 24365-98. |
[14] | Ma L, Ma C, Liu Y, Wang X. Thyroid diagnosis from SPECT images using convolutional neural network with optimization. Computational Intelligence and Neuroscience. (2019) ; 2019: : 6212759. |
[15] | Zhou S, Greenspan H, Davatzikous C, Duncan J, Van Ginneken B, Madabhushi A, et al. A review of deep learning in medical imaging: Imaging traits, technology trends, case studies with progress highlights, and future promises. Proceedings of IEEE. (2021) ; 109: (5): 820-838. |
[16] | Du W, Rao N, Liu D, Jiang H, Luo C, Li Z, et al. Review on the applications of deep learning in the analysis of gastrointestinal endoscopy images. IEEE Access. (2019) ; 7: : 142053-1402069. |
[17] | Le Berre C, Sandborn W, Aridhi S, Devignes M, Fournier L, Smail-Tabbone M, et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology. (2020) ; 158: (1): 76-79. |
[18] | Min J, Kwak M, Cha J. Overview of deep learning in gastrointestinal endoscopy. Gut Liver. (2019) ; 13: (4): 388-393. |
[19] | Ali H, Sharif M, Yasmin M. A survey of feature extraction and fusion of deep learning for detection of abnormalities in video endoscopy of gastrointestinal-tract. Artificial Intelligence Review. (2020) ; 53: (4): 2635-707. |
[20] | Gholami E, Tabbakh S, Kheirabadi M. Increasing the accuracy in the diagnosis of stomach cancer based on color and lint features of tongue. Biomedical Signalal Processing and Control. (2021) ; 69: : 102782. |
[21] | Zhang X, Hu W, Chen F, Liu J, Yang Y, Wang L, et al. Gastric precancerous diseases classification using CNN with a concise model. Plos One. (2017) ; 12: (9): e0185508. |
[22] | Zhu Y, Wang Q, Xu M, Zhang Z, Cheng J, Zhong Y, et al. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointestical Endoscopy. (2019) ; 89: (4): 806-9. |
[23] | Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, et al. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. (2020) ; 23: (1): 126-32. |
[24] | Sun M, Zhang X, Qu G, Zou M, Du H, Ma L, et al. Automatic polyp detection in colonoscopy images: Convolutional neural network, dataset and transfer learning. Journal of Medical Imaging and Health Informatics. (2019) ; 9: (1): 126-133. |
[25] | Majid A, Khan M, Yasmin A, Rehman A, Yousafzai A, Tariq U. Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection. Microscopy Research and Technique. (2020) ; 83: (5): 562-576. |
[26] | Wimmer G, Hafner M, Uhl A. Improving CNN training on endoscopic image data by extracting additionally training data from endoscopic videos. Computerized Medical Imaging and Graphics. (2020) ; 86: : 101798. |
[27] | Wang L, Lin H, Xin L, Qian W, Wang T, Zhang J. Establishing a model to measure and predict the quality of gastrointestinal endoscopy. World Journal of Gastroenterology. (2019) ; 25: (8): 1024-30. |
[28] | Vinsard D, Mori Y, Misawa M, Kudo S, Rastogi A, Bagci U, et al. Quality assurance of computer-aided detection and diagnosis in colonoscopy. Gastrointestinal Endoscopy. (2019) ; 90: (1): 55-63. |
[29] | Takyama H, Ozawa T, Ishihara S, Fujishiro M, Ahichijo S, Nomura S, et al. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Scientific Reports. (2018) ; 8: : 7494. |
[30] | Iqbal S, Qureshi A, Li J, Mahmood T. On the analyses of medical images using traditional machine learning techniques and convolutional neural networks. Archives of Computational Methods in Engineering. (2023) ; 30: : 3173-3233. |
[31] | Li S, Wang Z, Liu Z, Tan C, Lin H, Wu D, et al. Moganet: multi-order gated aggregation network. arXiv, doi: 10.48550/arXiv.2211.03295. |
[32] | Shin H, Roth H, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Transactions on Medical Imaging. (2016) ; 35: (5): 1285-1298. |
[33] | Kim H, Cosa-Linan A, Santhanam N, Jannesari M, Maros M, Ganslandt T. Transfer learning for medical image classification: A literature review. BMC Medical Imaging. (2022) ; 22: : 69. |




