Artificial intelligence as a new answer to old challenges in maternal-fetal medicine and obstetrics
Abstract
BACKGROUND:
Following the latest trends in the development of artificial intelligence (AI), the possibility of processing an immense amount of data has created a breakthrough in the medical field. Practitioners can now utilize AI tools to advance diagnostic protocols and improve patient care.
OBJECTIVE:
The aim of this article is to present the importance and modalities of AI in maternal-fetal medicine and obstetrics and its usefulness in daily clinical work and decision-making process.
METHODS:
A comprehensive literature review was performed by searching PubMed for articles published from inception up until August 2023, including the search terms “artificial intelligence in obstetrics”, “maternal-fetal medicine”, and “machine learning” combined through Boolean operators. In addition, references lists of identified articles were further reviewed for inclusion.
RESULTS:
According to recent research, AI has demonstrated remarkable potential in improving the accuracy and timeliness of diagnoses in maternal-fetal medicine and obstetrics, e.g., advancing perinatal ultrasound technique, monitoring fetal heart rate during labor, or predicting mode of delivery. The combination of AI and obstetric ultrasound can help optimize fetal ultrasound assessment by reducing examination time and improving diagnostic accuracy while reducing physician workload.
CONCLUSION:
The integration of AI in maternal-fetal medicine and obstetrics has the potential to significantly improve patient outcomes, enhance healthcare efficiency, and individualized care plans. As technology evolves, AI algorithms are likely to become even more sophisticated. However, the successful implementation of AI in maternal-fetal medicine and obstetrics needs to address challenges related to interpretability and reliability.
1.Introduction
With the latest development of machine learning technology, our society witnessed remarkable progress in the field of computer science [1]. Artificial intelligence (AI) is a technology that simulates human intelligence in machines that are programmed to mimic human cognitive processes [1]. Machine learning (ML), as a subset of AI, deals with the development of algorithms that enable computers to make predictions or decisions without being explicitly programmed to perform such tasks. Finally, Deep learning (DL) is a subfield of ML which utilizes deep neural networks (DNNs), inspired by the function of human brain, to process and learn from data. DNNs are capable of learning high-level features from raw data and this technology has been very successful in different applications such as image and speech recognition, computer vision, and natural language processing [2].
Following the latest trends in the development of AI, the possibility of processing an immense amount of data has created a breakthrough in the medical field [3]. Considering that ultrasound (US) images for almost all obstetric situations are available, AI applications for perinatal medicine are rapidly developing [4]. Practitioners now can utilize AI tools to advance diagnostic protocols and improve patient care.
Even though AI seems to be a very recent trend, its forms have been applied in the medical field for several decades [3]. One of the first AI programs called MYCIN was developed in the 1970s and it was programmed to help with diagnosing bacterial infections and recommending appropriate antibiotic treatments [5]. The development of computer-aided diagnostic systems for US images also started in the 1970s. However, the development of AI in perinatal US is currently still in early stages because fetal US poses several unique challenges, such as the fetal mobility, the developing anatomy of the fetus, and especially the requirement to obtain specific planes which can be limited by changes in fetal positioning and variations in maternal body habitus [6].
Since 2015, there has been a resurgence of interest in maternal well-being research with machine learning. Studies have been published on prenatal diagnosis, fetal heart monitoring, prediction and management of pregnancy-related complications, the prediction of the delivery mode, delivery outcomes, and outcomes after In vitro fertilization (IVF) treatment [7, 8].
The integration of AI into the field of maternal-fetal medicine and obstetrics represents a significant stride towards enhancing healthcare. AI has demonstrated remarkable potential in improving the accuracy and timeliness of diagnoses in obstetrics [9]. Another promising aspect of AI in obstetrics is its capacity to tailor care plans to individual patients. By analyzing a patient’s medical history, genetic factors, and real-time health data, AI systems can formulate personalized treatment strategies [10]. This level of customization not only improves patient outcomes but also reduces the risk of unnecessary medical interventions AI simulations can mimic various case scenarios, from routine prenatal US to high-risk deliveries, which can be used in education and training [11]. AI can also track a learner’s progress and identify areas of weakness, adapting the training content accordingly. These systems can provide immediate feedback and even suggest the best course of action in each scenario.
Looking ahead, the future of AI in obstetrics holds promise for continued innovation. Advancements in wearable devices and remote monitoring may enable real-time data collection, allowing healthcare providers to intervene promptly in response to emerging issues [12]. However, despite the remarkable potential of AI in obstetrics, these tools cannot be a substitute for clinical experience [8].
The aim of this article is to present the importance and modalities of AI in maternal-fetal medicine and obstetrics and its usefulness in daily clinical work and decision-making process.
2.Materials and methods
We conducted a comprehensive literature review searching PubMed for articles published from inception up until August 2023, including the search terms “artificial intelligence in obstetrics”, “maternal-fetal medicine”, and “machine learning” combined through Boolean operators. In addition, references lists of identified articles were further reviewed for inclusion.
3.Perinatal ultrasound
ML was first applied to perinatal US several years ago. Thanks to this, it has become possible to distinguish different body parts of fetus through machine learning [13]. The combination of AI and obstetric US can help optimize fetal US examination by reducing examination time and improving diagnostic accuracy while reducing physician workload that can lead to clinician burnout [14]. Currently, there is a semi-automatic program for fetal US analysis that automatically takes standard biometric measures after the sonographer selects an appropriate image of each body part [15]. AI has been successfully applied to automatic fetal US standard plane detection, measurement of biometric parameters, and disease diagnosis to facilitate conventional imaging approaches [16]. Automatic plane detection a measurement of standard fetal biometric resulting in reduction of repetitive caliper adjustment clicks and operator bias, as well as instant quality control [17].
In recent years, studies have been published that demonstrate the use of ML in the detection of fetal central nervous system (CNS) abnormalities and congenital heart diseases (CHD) which showed potential of AI in fetal anomaly scan [18].
AI-based technology showed reliability in identifying biometric planes, reporting the presence or absence of anatomic malformations, completing a basic US, and classifying a full fetal echocardiogram [19, 20, 21]. However, the main flaw of AI equipment is that its diagnostic enactment was principally based on the biased impression of investigative clinical sonographers [22]. In other words, all available data was non-selectively considered as if all the examinations were performed by experienced examiners [23].
3.1Fetal biometry and gestational age (GA)
Optimal prenatal care relies on accurate gestational age dating. After the first trimester, the accuracy of gestational age estimation from fetal biometrics decreases with increasing gestational age [24]. Automatic measurement of fetal biological parameters using AI can reduce inter- and intra-operator measurement errors and promote clinical efficiency (Fig. 1) [16].
Figure 1.
Automatic estimation of biometric parameters deviation from reference values for gestational age.
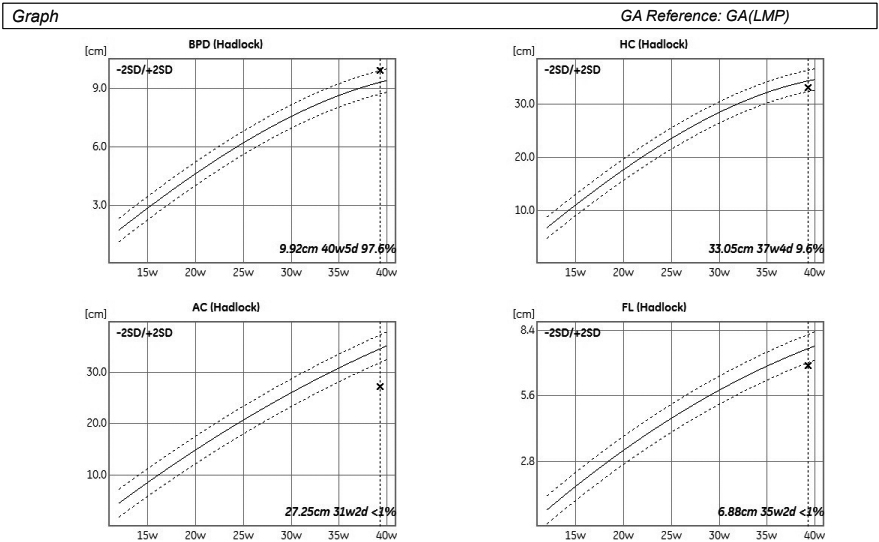
Burgos-Artizz et al. evaluated the performance of an AI method based on automated analysis of fetal brain morphology on standard transthalamic axial plane to estimate the GA in second and third-trimester fetuses compared with the current formulas using standard fetal biometry [25]. The AI method alone yielded somewhat lower gestational age estimation errors than fetal biometric parameters, and errors were even lower when the AI model was combined with biometric parameters [25]. The use of this AI model offers better accuracy in GA estimation; however, there is still deviation when compared with dating by Crown-rump length (CRL) measurement in the first trimester [25].
Grandjean et al. used the Smartplanes® software to identify the transthalamic plane from the 3D volumes and perform biparietal diameter (BPD) and head circumference (HC) measurements automatically [26]. These measurements showed good agreement with those obtained by two experienced sonographers from conventional two-dimensional (2D) images and three-dimensional (3D) volumes. This has the potential advantage of standardizing the measurement technique and therefore minimizing inter-operator variability.
A novel feature-based model that regresses brain development to GA using fetal cranial US images was developed by Namburete et al. [27]. Preliminary analysis identified age-discriminating brain regions observable in fetal US images, i.e. the Sylvian fissure, callosal sulcus, and parieto-occipital fissure, all of which have been reported as undergoing significant change during the gestational period [27]. The estimated GA was close to the value obtained by clinical measurement, with the root mean square error (RMSE) of
Lee et al. used Convolutional Neural Network (CNN) to analyze images from multiple standard US views for GA estimation without any measurement information [28]. The best model estimated GA with a mean absolute error of 3.0 (95% CI, 2.9–3.2) and 4.3 (95% CI, 4.1–4.5) days in the second and third trimesters, respectively, which outperforms current US-based clinical biometry at these gestational ages [28]. Moreover, it applies to both high- and low-risk pregnancies and people in different geographical areas.
Gomes et al. developed mobile-optimized AI models that can help novices in low- and middle-income countries assess GA and fetal malpresentation from freehand US sweeps [29]. AI GA estimates and standard fetal biometry estimates were compared to a previously established ground truth and evaluated for differences in absolute error; fetal malpresentation was compared to sonographer assessment [29]. They showed that the GA estimation accuracy of the AI model is non-inferior to standard fetal biometry estimates, and the fetal malpresentation model has high area under the receiver operating characteristic curve (AUC-ROC) across operators and devices [29]. Software run times on phones for both diagnostic models are less than 3 seconds after completion of a sweep [29].
The nuchal translucency (NT) is the most essential marker for detecting Down syndrome during the first trimester of pregnancy [30]. Moratalla et al. developed an AI algorithm for semi-automatic NT measurements that achieved an inter-operator standard deviation of 0.0149 mm, lower than the manual approach of 0.109 mm, significantly reducing inter- and intra-observer differences [31]. The semi-automated approach, however, involves manual fine-tuning of the NT region, so researchers have developed AI algorithms to achieve fully automatic measurements of NT thickness, with a classification accuracy of 91.7% [32].
3.2Fetal brain assessment
Table 1
Studies on AI aplication in fetal brain assessement
| Study | Technology | Task | Results |
|---|---|---|---|
| Miyagi et al. [35] | CNN | Recognition of fetal expressions | Accuracy of 99.6% for seven categories |
| Lin et al. [36] | MF R-CNN transfer learning | FHSPs detection and US image quality assessment | Accurate quality assessment of an ultrasound plane within 0.5 second |
| Qu et al. [37] | Differential CNN | Automated identification of FHSPs | Accuracy of 92.93% |
| Xi et al. [38] | DCNN | FHSPs classification as normal or abnormal | Accuracy of 96% in classification; precision of 49.7% in lesion localization |
| Xie et al. [19] | CNN based deep learning | FHSPs classification as normal or abnormal | Overall accuracy of 96.3% the sensitivity and specificity of 96.9% and 95.9%; respectively precision of 61.6% in lesion localization |
| Lin et al. [39] | CNN | Recognition of 10 intracranial abnormalities in standard planes | Mean accuracy of 99.2% in the internal validation dataset and 96.3% in the external validation dataset |
CNN – convolution neural network; MF R-CNN – multi-task learning framework using a faster regional convolutional neural network; US – ultrasound; FHSP – fetal head standard plane; AUC – area under receiver operating characteristic curve.
Figure 2.
3D US image of 28 weeks baby, hand-to-face movement.
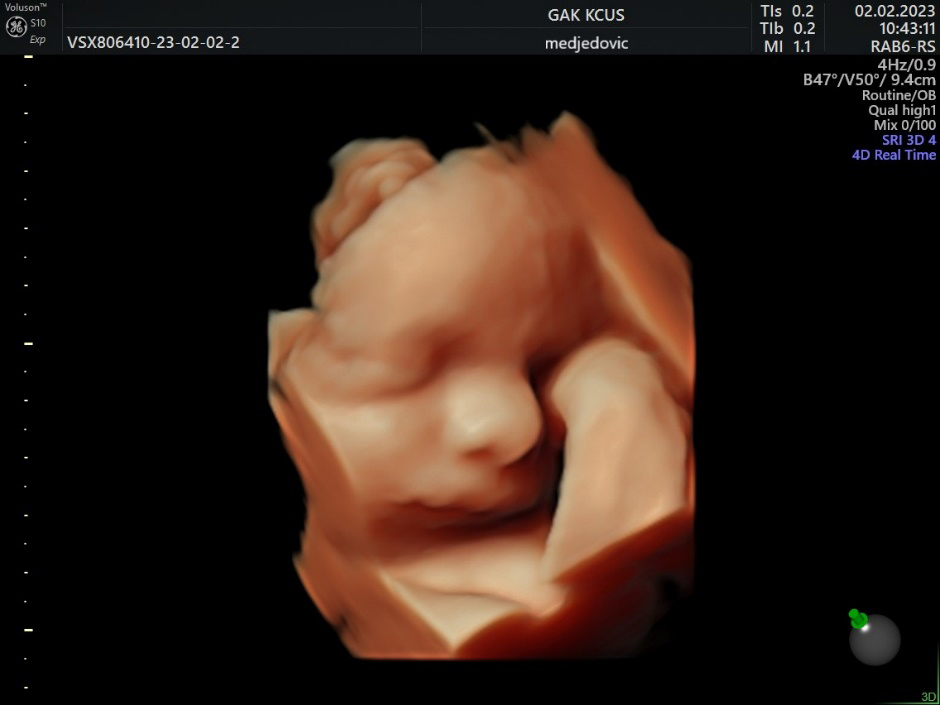
Figure 3.
3D US image of 28 weeks baby, scowling.
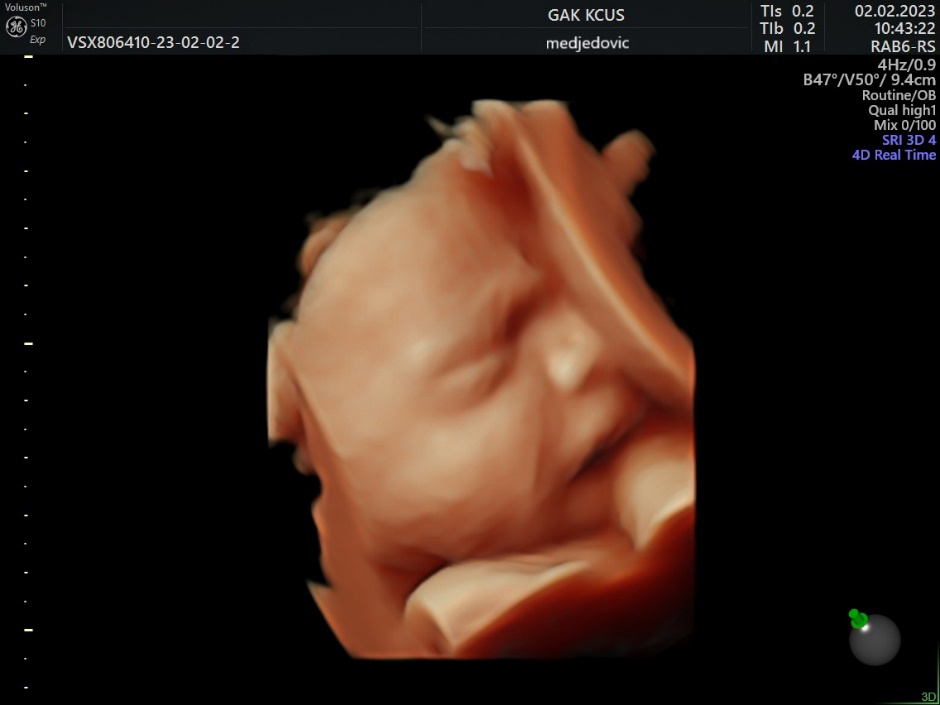
Figure 4.
3D US image of 28 weeks baby, smiling.
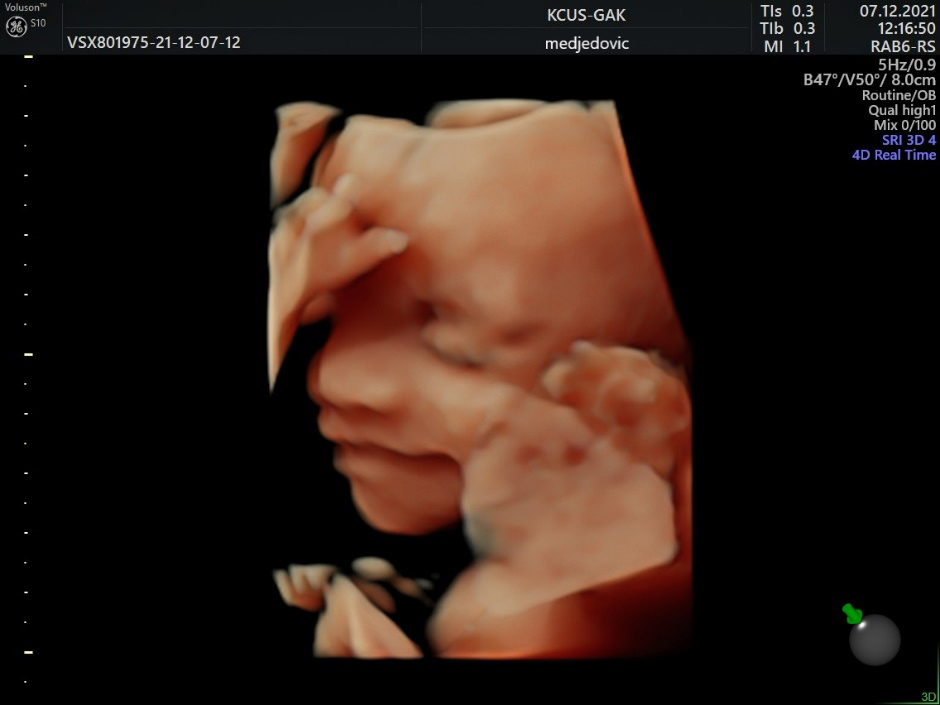
In numerous studies, AI has proven to be useful in fetal brain assessment which requires the observation of several qualitative and quantitative parameters during an ultrasound examination (Table 1). The Kurjak Antenatal Neurodevelopmental Test (KANET) scoring system can assess fetal neurobehavioral development by assessing fetal movements and facial expressions (Figs 2–4) [33, 34]. AI can enable objective assessment of fetal facial expressions. The first reported study on this topic showed an accuracy, reliability score, and ROC curve of AI fetal facial expression analysis of 0.996 for seven categories: eye blinking, mouthing, neutral face, scowling, smiling, tongue expulsion, and yawning [35]. This study also acknowledged its limitations. Observation of the fetal face is time-consuming, there’s a lack of consensus of image classification by examiners, and the perfect classification of fetal facial expressions has not yet been established [35]. Also, the feasibility of recognizing fetal facial expressions using AI depends on data supervised by experienced examiners. Finally, there are still faulty data sets such as for expressions that are rarely seen during screening, such as sucking [35].
The acquisition of a standard plane is highly subjective and depends on the clinical experience of sonographers. A multi-task learning framework proposed by Lin et al. used a faster regional convolutional neural network (MF R-CNN) architecture for standard plane detection and quality assessment [36]. An MF R-CNN can identify the critical anatomical structure of the fetal head, analyze whether the magnification of the US image is appropriate, and then perform a quality assessment of US images based on clinical protocols [36]. By recognizing the key anatomical structure and magnification of the US image, this method can accurately make a quality assessment of an US plane within half a second [36].
The differential CNN proposed by Qu et al. automatically recognized six fetal brain standard planes (the horizontal transverse section of the thalamus, horizontal transverse section of lateral ventricle, transverse section of cerebellum, midsagittal plane, paracentral sagittal section, and coronal section of the anterior horn of the lateral ventricle) with 92.93% accuracy and high computational efficiency [37].
Various algorithms have been developed for the diagnosis of fetal brain abnormalities. Xie et al. presented a model that used a U-Net to segment cranial regions and a VGG-Net network to distinguish between normal and abnormal US images, helping to reduce the false-negative rate of fetal brain abnormalities [38]. Xie et al. used a CNN-based DL model to distinguish between normal and abnormal fetal brains with an overall accuracy of 96.31% [19]. Furthermore, the model was able to visualize the location of the lesion through heat maps and overlapping images, which increased the sensitivity of the essential clinical examination. However, both studies could only distinguish normal from abnormal standard brain planes.
Lin et al. developed an AI system, the Prenatal ultrasound diagnosis Artificial Intelligence Conduct System (PAICS), for the detection of different patterns of fetal intracranial abnormality in standard sonographic reference planes for screening for congenital CNS malformations [39]. In this multicenter, case-control, retrospective diagnostic study 43 890 images from 16 297 pregnancies and 169 videos from 166 pregnancies were used to develop and validate the PAICS [39]. The system achieved excellent performance in identifying 10 types of intracranial image patterns: non-visualization of the cavum septi pellucidi, non-visualization of the septum pellucidum, crescent-shaped single ventricle, mild ventriculomegaly, severe ventriculomegaly, non-intraventricular cyst, intraventricular cyst, open fourth ventricle, mega cisterna magna, and the normal pattern [39]. The performance of the PAICS was comparable to that of expert sonographers but required significantly less time [39]. Both in the image dataset and in the real-time scan setting, the PAICS achieved excellent diagnostic performance for various fetal CNS abnormalities [39]. The PAICS has the potential to be an effective and efficient tool in screening for fetal CNS malformations in clinical practice [39].
3.3AI in fetal echocardiography
Fetal heart US is highly specific and sensitive in experienced hands, however, antenatal detection rates of CHDs remain lower than for most other major structural anomalies. Prenatal detection in some countries is as low as 13% [40].
US imaging of the fetal heart is a challenging and complex task with a high degree of operator dependence. The first step in the diagnosis of CHD is obtaining the standard cardiac imaging planes. AI can be used to automatically retrieve these planes from the US image data stream which is a potential route to improving detection rates. Moreover, it could allow the sonographer to focus on identifying abnormal anatomy, rather than pausing and saving standard planes [41]. Wu et al. proposed a deep convolution neural network (U-Y-net) model which was shown to effectively recognize and classify many important structures and the standard five sections of the fetal heart [42].
Arnaout et al. hypothesized that poor detection results from challenges in obtaining and interpreting diagnostic-quality cardiac views and that deep learning could improve complex CHD detection. Using 107,823 images from 1,326 retrospective echocardiograms and examinations of fetuses aged 18 to 24 weeks, they trained a set of neural networks to identify recommended cardiac views and distinguish normal hearts from complex CHD [20]. In a test set of 4108 fetal studies, the model achieved an AUC of 0.99, 95% sensitivity (95%CI, 84–99), 96% specificity (95%CI, 95–97), and 100% negative predictive value in differentiating normal from abnormal heart [20]. In one study, combining noninvasive fetal electrocardiography with AI for the automatic detection of CHD achieved a detection rate of 63% for all CHD and 75% for critical CHD [43].
The CNN model was used to identify and classify key frames (the aorta, the arches, the atrioventricular flows, and the crossing of the great vessels) of fetal heart echocardiography in the first trimester of pregnancy [44]. A test accuracy of 95% with an F1-score ranging from 90.91% to 99.58% shows the potential in supporting heart scans even from such an early fetal age [44]. Anda et al. proposed the use of learning deep architectures for the interpretation of first-trimester fetal echocardiography (LIFE) to recognize fetal CHD without using 4D sonography [45]. This project’s primary objective is to develop an Intelligent Decision Support System that uses two-dimensional video files of cardiac sweeps obtained during the standard first-trimester fetal echocardiography to signal the presence/absence of previously learned key features [45].
Yeo et al. developed a Fetal Intelligent Navigation Echocardiogram (FINE) that, in conjunction with Virtual Intelligent Sonographer Assistance, automatically generates nine standard fetal echocardiographic views and intelligently identifies surrounding anatomical structures [46]. The FINE model could recognize abnormal cardiac anatomy in four proven cases of CHD (coarctation of the aorta, tetralogy of Fallot, transposition of the great vessels, and pulmonary atresia with an intact ventricular septum) [46]. In the following studies FINE identified d-transposition of the great arteries and double-outlet right ventricle [47, 48]. When performed by non-expert sonographers, FINE has superior performance compared to manual navigation of the normal fetal heart. Specifically, a study showed that FINE obtained appropriate fetal cardiac views in 92–100% of cases [49]. Recently, eight novel and advanced features have been developed for the FINE method with the goal of simplifying FINE further, allowing quantitative measurements to be performed on the cardiac views generated by FINE, and improving the success of obtaining fetal echocardiography views [50].
4.Fetal heart rate monitoring (FHR)
Perinatal asphyxia poses a difficult obstetric challenge which can be better handled by creating a more efficient and reliable way to monitor FHR. Even though cardiotocography (CTG) monitors have been widely used in the last 50 years, and standardized guidelines are available, inter- and intra-observer variability and uncertainty in the classification of unreassuring FHR recordings are still present in clinical practice [51, 52]. Given the clinical relevance of FHR as a marker of fetal well-being and autonomous nervous system development, many different approaches for computerized processing and analysis of FHR patterns have been proposed in the literature [53]. The opportunities offered by AI tools represent the future direction of electronic fetal monitoring (EFM).
The INFANT study is a large trial evaluating the ability of AI interpretation of CTG during labor to assist clinicians in deciding the best management for patients [54]. This technology could aid in making FHR reading more reliable thus helping the practitioner in interpreting and decision-making process. Furthermore, AI technology could be used for out-patient care in the form of home monitors that would provide adequate surveillance of high-risk patients, potentially helping in earlier detection of pregnancy complications.
Another study was conducted to develop an ML model that can identify high-risk fetuses (suspicious as well as pathological states) as accurately as highly trained medical professionals [55]. Ten different ML classification models were trained using CTG data of 2126 pregnant women in the third trimester [55]. The classification model developed using the XGBoost technique had the highest prediction accuracy for predicting a pathological fetal state (
Spairani et al. proposed a hybrid approach based on a neural architecture that receives heterogeneous data in input (a set of quantitative parameters and images) for classifying healthy and pathological fetuses [56]. The quantitative regressors, which represent different aspects of the correct development of the fetus, are combined with features implicitly extracted from various representations of the FHR signal (images) [56]. The neural model with two connected branches, a Multi-Layer Perceptron (MLP) and a CNN, was trained on a huge and balanced set of clinical data (14.000 CTG tracings, 7000 healthy and 7000 pathological) recorded during ambulatory nonstress tests [56]. After hyperparameters tuning and training, the neural network proposed has reached an overall accuracy of 80.1% [56].
Still, when it comes to AI monitoring of the FHR during labor, its application did not show to improve neonatal outcomes [57]. Similar technology is already available in the form of alert automation in monitors for vital sign abnormalities in patients, but its application in everyday practice shows mediocre results [58].
5.Mode of delivery
Mode of delivery is one of the issues that is of great concern in obstetrics. In the last 30 years, the worldwide rate of cesarean section (CS) has increased progressively from 6 to 21% of all births [59], exceeding the limit recommended by the World Health Organization. Besides different medical and paramedical reasons for such an alarming increase of CS, it is a fact that obstetricians generally do lack the necessary technology to help them decide on appropriate mode of delivery based on individual antepartum and intrapartum conditions.
Table 2
Artificial intelligence as a helpful tool in prediction of delivery
| Beksac et al. [63] | Supervised artificial neural network had specificity and sensitivity of 97.5% and 60.9% in |
| prediction of delivery | |
| Wong et al. [64] | Accuracy of predicting vaginal delivery at 4 hours was 87.1% for machine learning model |
| Lipschuetz et al. [65] | Machine-learning algorithm can give personalized risk score for a successful vaginal birth after |
| cesarean delivery and can be helpful in decision-making and reduce cesarean delivery rates |
Using ML, several prediction models were trained to predict the mode of delivery (Table 2), including spontaneous vaginal birth, instrumental delivery, emergency CS, and successful vaginal birth after cesarean (VBAC), with the goal to help the obstetrician in making decisions regarding the appropriate mode of delivery [60, 61].
A study published in 2022 tested the suitability of using three different AI algorithms to develop a clinical decision support system for the mode of delivery prediction, using a large clinical database consisting of 25,038 records of patients with singleton pregnancies [62]. All three implemented algorithms showed a similar performance, reaching an accuracy equal to or above 90% in the classification between vaginal and cesarean deliveries and around 86% for the classification between normal and instrumental vaginal delivery [62].
Beksac et al. developed a supervised artificial neural network (ANN) for predicting the delivery route [63]. During the admission of the patients to the delivery room, at the beginning of the first stage of labor and after pelvic examination and evaluation of the mother and fetus, the input variables for the ANN were obtained: maternal age, gravidity, parity, gestational age at birth, necessity and type of labor induction, presentation of the baby at birth, and maternal disorders and/or risk factors [63]. The outputs of the algorithm were vaginal delivery or CS. This system showed specificity and sensitivity of 97.5% and 60.9%, respectively [63].
Another similar study published in 2022 presented the model that was found to be 87% accurate in predicting a mode of delivery within four hours of patient being admitted to the hospital [64]. It was also reported that the predictor factors that the AI model used mirrored the diagnostic factors that clinicians use to predict a mode of delivery, such as stage of labor and cervix dilation measurement [64].
The algorithm’s ability to predict risks in early labor stages can help reduce adverse birth outcomes and healthcare costs associated with maternal morbidity. This allows preventing a CS from being done if a vaginal delivery is more likely, or preventing a patient from laboring for too long if the model predicts the birth will more likely result in a CS anyway. Lipschuetz et al. developed a machine learning algorithm to assign a personalized risk score for a successful vaginal birth after cesarean delivery. Applying the model to a cohort of parturients who elected a repeat cesarean delivery (
While having a previous vaginal birth strongly predicts VBAC, the delivery outcome in women with only a cesarean delivery is more unpredictable. In another study, three machine-learning methods were used to predict VBAC among women with one previous birth [66]. AUC-ROC ranged from 0.61 to 0.69, with sensitivity (probability of correctly identifying a VBAC for the second delivery) above 91% and specificity (probability of correctly identifying a repeat CS for the second delivery) below 22% for all models [66]. The majority of women with an unplanned repeat cesarean had a predicted probability of vaginal birth after cesarean
5.1Shoulder dystocia
Tsur and al. developed an ML model for the prediction of shoulder dystocia (ShD) and validated the model’s predictive accuracy and potential clinical efficacy in optimizing the use of cesarean delivery in the context of suspected macrosomia [67]. The study included 686 singleton vaginal deliveries in the derivation cohort of which 131 were complicated by ShD, and 2584 deliveries in the validation cohort, of which 31 were complicated by ShD [67]. The model predicted ShD better than estimated fetal weight either alone or combined with maternal diabetes and was able to stratify the risk of ShD and neonatal injury in the context of suspected macrosomia [67].
6.Preterm birth (PTB)
Ultrasonography cervical-length (CL) assessment is considered to be the most effective screening method for preterm birth (PTB), but routine, universal CL screening remains controversial because of its cost [68]. Approximately 30% of preterm births are not correctly predicted due to the complexity of this process and its subjective assessment [69]. In spite of proper screening based on CL measurement, current screening strategies cannot identify a significant percentage of patients who will deliver prematurely. In search of better methods of predicting preterm birth, researchers have investigated the application of artificial intelligence in this area (Table 3). The studies suggest that when we incorporate different uterine cervix assessment methods into the deep learning networks, we may improve the detection rates and contribute additional information to identify women with true spontaneous PTB [69].
Table 3
Artificial intelligence as a helpful tool in prediction of preterm birth
| Zhang et al. [71] | Long short-term memory (LSTM) can be helpful in decision making during obstetric |
| examination (better prediction ability and a lower misdiagnosis rate) | |
| Arabi Belaghi et al. [72] | Artificial neural networks had slightly higher area under the receiver operating |
| characteristic curve than logistic regression in prediction of preterm birth | |
| Goldsztejn and Nehorai [73] | Deep learning method can predict preterm birth for pregnant mothers around their 31st |
| week of gestation | |
| Włodarczyk et al. [74] | Deep neural network with ultrasound data can be helpful in decision making process |
| Włodarczyk et al. [75] | Prediction of preterm birth based on transvaginal ultrasound images compared to state-of- |
| the-art methods |
A systematic review of publications on AI use in the prediction of PTB showed that electrohysterogram images were mostly used, followed by the biological profiles, the metabolic panel in amniotic fluid or maternal blood, and the cervical images on the ultrasound examination. However, the size of the dataset in most studies was a hundred cases and too small for learning [70].
Zhang et al. used continuous EMR data from pregnant women to construct a preterm birth prediction classifier based on long short-term memory (LSTM) networks [71]. The data included more than 25,000 obstetric EMRs from the early trimester to 28 weeks of gestation. Compared with a traditional cross-sectional study, the LSTM model in this time-series study had better overall prediction ability and a lower misdiagnosis rate at the same detection rate. Accuracy, sensitivity, specificity, and AUC were 0.739, 0.407, 0.982, and 0.651, respectively [71]. Important-feature identification indicated that blood pressure, blood glucose, lipids, uric acid, and other metabolic factors were important factors related to PTB [71].
In one study researchers used data during the first and second trimesters to build logistic regression and machine learning models in a “training” sample to predict overall and spontaneous PTB [72]. They identified 13 variables in the first trimester associated with PTB, of which the strongest predictors were diabetes and abnormal pregnancy-associated plasma protein A concentration [72]. In the second trimester, 17 variables were significantly associated with PTB, among which complications during pregnancy had the highest adjusted odds ratio (AOR) [72]. The maximum AUC was 60% (95% CI: 58–62%) and 65% (95% CI: 63–66%) in the first and second trimesters, respectively, with artificial neural networks in the validation sample [72]. Including complications during the pregnancy yielded an AUC of 80% (95% CI: 79–81%). All models yielded 94–97% negative predictive values for spontaneous PTB during the first and second trimesters [72]. They concluded that, although artificial neural networks provided slightly higher AUC than logistic regression, the prediction of preterm birth in the first trimester remains unattainable [72]. Nevertheless, prediction improved to a moderate level by both logistic regression and machine learning approaches including data from the second trimester [72].
Goldsztejn and Nehorai developed a deep-learning method to predict preterm births directly from electrohysterogram (EHG) measurements of pregnant mothers recorded at around 31 weeks of gestation. Their model predicted preterm births with an AUC of 0.78 (95% CI: 0.76–0.80) [73]. Moreover, they found that the spectral patterns of the measurements were more predictive than the temporal patterns, suggesting that preterm births can be predicted from short EHG recordings in an automated process [73].
The first work based on transvaginal ultrasound images that used ML algorithms was the work of Włodarczyk et al. [74]. They used a deep neural network architecture to segment prenatal US images and then automatically extracted two biophysical US markers, cervical length and anterior cervical angle (ACA), from the resulting images [74]. Both biomarkers were used by doctors to manually assess the risk of spontaneous PTB [74]. The results were then classified by classic ML algorithms. The best results were achieved by the Naive Bayesi algorithm: accuracy of 77.5%, precision of 85%, and AUC of 78.13% [74]. The results of predicting spontaneous PTB were better than those presented by gynecologists [74]. Furthermore, they showed that CL and ACA markers, when combined, reduced the false negative rate from 30% to 18% [74].
In another study, Włodarczyk et al. proposed a conceptually simple CNN for cervical segmentation and classification in transvaginal US images while simultaneously predicting PTB based on extracted image features without human supervision [75]. The proposed model was trained and evaluated on a dataset consisting of 354 2D transvaginal US images and achieved segmentation accuracy with a mean Jaccard coefficient index of 0.923
7.Limitations
Obstetrics and gynecology journals report mostly preliminary work (e.g. algorithm or proof-of-concept method) in AI applied to this discipline, and clinical validation remains an unfulfilled prerequisite [9]. AI is primarily well-defined as the study of algorithms. However, an algorithm is only as good as the data it works with. If there is an error in the algorithm, then the potential for negative impact is high compared to a single human error. It is also possible to inadvertently introduce unwanted bias into the algorithm, which would obviously be unacceptable for medical applications [76]. A recent systematic review found that most non-randomized trials in medical imaging are not prospective, have a high risk of bias, and deviate from existing reporting standards [77].
8.Conclusion
The integration of AI in maternal-fetal medicine and obstetrics has the potential to significantly improve patient outcomes, enhance healthcare efficiency, and individualize care plans. As technology evolves, AI algorithms are likely to become even more sophisticated, potentially revolutionizing prenatal care and fetal monitoring. However, the successful implementation of AI in obstetrics necessitates addressing challenges related to interpretability and reliability.
References
[1] | Yazdani A, Costa S, Kroon B. Artificial intelligence: Friend or foe? Australian and New Zealand Journal of Obstetrics and Gynaecology. (2023) ; 63: (2): 127-30. doi: 10.1111/ajo.13661. |
[2] | Egger J, Gsaxner C, Pepe A, Pomykala KL, Jonske F, Kurz M, Li J, Kleesiek J. Medical deep learning-A systematic meta-review. Comput Methods Programs Biomed. (2022) ; 221: : 106874. doi: 10.1016/j.cmpb.2022.106874. |
[3] | Basu K, Sinha R, Ong A, et al. Artificial intelligence: How is it changing medical sciences and its future? Indian J Dermatol. (2020) ; 65: (5): 365-370. doi: 10.4103/ijd.IJD_421_20. |
[4] | Ahn KH, Lee KS. Artificial intelligence in obstetrics. Obstet Gynecol Sci. (2022) ; 65: (2): 113-124. doi: 10.5468/ogs.21234. |
[5] | Shortliffe EH. Mycin: A knowledge-based computer program applied to infectious diseases. Proc Annu Symp Comput Appl Med Care. (1977) ; 5: : 66-9. |
[6] | Horgan R, Nehme L, Abuhamad A. Artificial intelligence in obstetric ultrasound: A scoping review. Prenat Diagn. (2023) ; 43: (9): 1176-1219. doi: 10.1002/pd.6411. |
[7] | Islam MN, Mustafina SN, Mahmud T, et al. Machine learning to predict pregnancy outcomes: A systematic review, synthesizing framework and future research agenda. BMC Pregnancy and Childbirth. (2022) ; 22: (1): 1-9. doi: 10.1186/s12884-022-04594-2. |
[8] | Sarno L, Neola D, Carbone L, et al. Use of artificial intelligence in obstetrics: Not quite ready for prime time. Am J Obstet Gynecol MFM. (2023) ; 5: (2): 100792. doi: 10.1016/j.ajogmf.2022.100792. |
[9] | Dhombres F, Bonnard J, Bailly K, et al. Contributions of artificial intelligence reported in obstetrics and gynecology journals: Systematic review. Journal of Medical Internet Research. (2022) ; 24: (4): e35465. doi: 10.2196/35465. |
[10] | Emin EI, Emin E, Papalois A, et al. Artificial intelligence in obstetrics and gynaecology: Is this the way forward? In Vivo. (2019) ; 33: (5): 1547-1551. doi: 10.21873/invivo.11635. |
[11] | Chen Z, Liu Z, Du M, et al. Artificial intelligence in obstetric ultrasound: An update and future applications. Front Med (Lausanne). (2021) ; 27: (8): 733468. doi: 10.3389/fmed.2021.733468. |
[12] | Escobar GJ, Soltesz L, Schuler A, et al. Prediction of obstetrical and fetal complications using automated electronic health record data. Am J Obstet Gynecol. (2021) ; 224: (2): 137-147.e7. doi: 10.1016/j.ajog.2020.10.030. |
[13] | Kim HY, Cho GJ, Kwon HS. Applications of artificial intelligence in obstetrics. Ultrasonography. (2023) ; 42: (1): 2-9. doi: 10.14366/usg.22063. |
[14] | Gandhi TK, Classen D, Sinsky CA, et al. How can artificial intelligence decrease cognitive and work burden for front line practitioners. JAMIA Open. (2023) ; 6: (3): ooad079. doi: 10.1093/jamiaopen/ooad079. |
[15] | Yaqub M, Kelly B, Papageorghiou AT, et al. A deep learning solution for automatic fetal neurosonographic diagnostic plane verification using clinical standard constraints. Ultrasound Med Biol. (2017) ; 43: (12): 2925-2933. doi: 10.1016/j.ultrasmedbio.2017.07.013. |
[16] | Xiao S, Zhang J, Zhu Y, et al. Application and progress of artificial intelligence in fetal ultrasound. Journal of Clinical Medicine. (2023) ; 12: (9): 3298. doi: 10.3390/jcm12093298. |
[17] | Baumgartner CF, Kamnitsas K, Matthew J, et al. SonoNet: Real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Trans Med Imaging. (2017) Nov; 36: (11): 2204-2215. doi: 10.1109/TMI.2017.2712367. |
[18] | Matsuoka R. Artificial intelligence and obstetric ultrasound. Donald School J Ultrasound Obstet Gynecol. (2021) ; 15: (3): 218-222. doi: 10.5005/jp-journals-10009-1702. |
[19] | Xie HN, Wang N, He M, et al. Using deep-learning algorithms to classify fetal brain ultrasound images as normal or abnormal. Ultrasound Obstet Gynecol. (2020) ; 56: (4): 579-587. doi: 10.1002/uog.21967. |
[20] | Arnaout R, Curran L, Zhao Y, et al. Expert-level prenatal detection of complex congenital heart disease from screening ultrasound using deep learning. medRxiv. 2020: 2020-06. doi: 10.1101/2020.06.22.20137786. |
[21] | Chen Z, Liu Z, Du M, et al. Artificial intelligence in obstetric ultrasound: An update and future applications. Front Med (Lausanne). (2021) ; 8: : 733468. doi: 10.3389/fmed.2021.733468. |
[22] | Malani IVSN, Shrivastava D, Raka MS. A comprehensive review of the role of artificial intelligence in obstetrics and gynecology. Cureus. (2023) ; 15: (2). doi: 10.7759/cureus.34891. |
[23] | Yoldemir T. Artificial intelligence and women’s health. Climacteric. (2020) ; 23: (1): 1-2. doi: 10.1080/13697137.2019.1682804. |
[24] | Skupski DW, Owen J, Kim S, et al. Estimating gestational age from ultrasound fetal biometrics. Obstet Gynecol. (2017) Aug; 130: (2): 433-441. doi: 10.1097/AOG.0000000000002137. |
[25] | Burgos-Artizzu XP, Coronado-Gutiérrez D, Valenzuela-Alcaraz B, et al. Analysis of maturation features in fetal brain ultrasound via artificial intelligence for the estimation of gestational age. Am J Obstet Gynecol MFM. (2021) Nov; 3: (6): 100462. doi: 10.1016/j.ajogmf.2021.100462. |
[26] | Grandjean GA, Hossu G, Bertholdt C, et al. Artificial intelligence assistance for fetal head biometry: Assessment of automated measurement software. Diagn Interv Imaging. (2018) ; 99: (11): 709-716. doi: 10.1016/j.diii.2018.08.001. |
[27] | Namburete AI, Stebbing RV, Kemp B, et al. Learning-based prediction of gestational age from ultrasound images of the fetal brain. Med Image Anal. (2015) Apr; 21: (1): 72-86. doi: 10.1016/j.media.2014.12.006. |
[28] | Lee LH, Bradburn E, Craik R, et al. Machine learning for accurate estimation of fetal gestational age based on ultrasound images. NPJ Digital Medicine. (2023) ; 6: (1): 36. doi: 10.1038/s41746-023-00774-2. |
[29] | Gomes RG, Vwalika B, Lee C, Willis A, Sieniek M, Price JT, et al. A mobile-optimized artificial intelligence system for gestational age and fetal malpresentation assessment. Commun Med (Lond). (2022) ; 11: (2): 128. doi: 10.1038/s43856-022-00194-5. |
[30] | Thomas MC, Arjunan SP. Deep learning measurement model to segment the nuchal translucency region for the early identification of down syndrome. Measurement Science Review. (2022) ; 22: (4): 187-92. doi: 10.2478/msr-2022-0023. |
[31] | Moratalla J, Pintoffl K, Minekawa R, et al. Semi-automated system for measurement of nuchal translucency thickness. Ultrasound Obstet Gynecol. (2010) Oct; 36: (4): 412-6. doi: 10.1002/uog.7737. |
[32] | Thomas MC, Arjunan SP. Deep learning measurement model to segment the nuchal translucency region for the early identification of down syndrome. Measurement Science Review. (2022) ; 22: (4): 187-92. doi: 10.2478/msr-2022-0023. |
[33] | Kurjak A, Miskovic B, Stanojevic M, et al. New scoring system for fetal neurobehavior assessed by three-and four-dimensional sonography. J Perinat Med. (2008) ; 36: (1): 73-81. doi: 10.1515/JPM.2008.007. |
[34] | Stanojevic M, Talic A, Miskovic B, et al. An attempt to standardize Kurjak’s Antenatal Neurodevelopmental Test: Osaka Consensus Statement. Donald School J Ultrasound Obstet Gynecol. (2011) ; 5: (4): 317-29. doi: 10.5005/jp-journals-10009-1209. |
[35] | Miyagi Y, Hata T, Bouno S, et al. Recognition of fetal facial expressions using artificial intelligence deep learning. Donald School J Ultrasound Obstet Gynecol. (2021) ; 15: : 223-8. doi: 10.5005/jp-journals-10009-1710. |
[36] | Lin Z, Li S, Ni D, et al. Multi-task learning for quality assessment of fetal head ultrasound images. Med Image Anal. (2019) ; 58: : 101548. doi: 10.1016/j.media.2019.101548. |
[37] | Qu R, Xu G, Ding C, et al. Standard plane identification in fetal brain ultrasound scans using a differential convolutional neural network. IEEE Access. (2020) ; 8: : 83821-30. doi: 10.1109/ACCESS.2020.2991845. |
[38] | Xie B, Lei T, Wang N, et al. Computer-aided diagnosis for fetal brain ultrasound images using deep convolutional neural networks. Int J Comput Assist Radiol Surg. (2020) ; 15: (8): 1303-12. doi: 10.1007/s11548-020-02182-3. |
[39] | Lin M, He X, Guo H, et al. Use of real-time artificial intelligence in detection of abnormal image patterns in standard sonographic reference planes in screening for fetal intracranial malformations. Ultrasound Obstet Gynecol. (2022) ; 59: (3): 304-316. doi: 10.1002/uog.24843. |
[40] | Bakker MK, Bergman JE, Krikov S, et al. Prenatal diagnosis and prevalence of critical congenital heart defects: An international retrospective cohort study. BMJ Open. (2019) ; 9: (7): e028139. doi: 10.1136/.bmjopen-2018-028139. |
[41] | Day TG, Kainz B, Hajnal J, et al. Artificial intelligence, fetal echocardiography, and congenital heart disease. Prenat Diagn. (2021) ; 41: (6): 733-42. doi: 10.1002/pd.5892. |
[42] | Wu H, Wu B, Lai F, et al. Application of artificial intelligence in anatomical structure recognition of standard section of fetal heart. Comput Math Methods Med. (2023) ; 2023: : 5650378. doi: 10.1155/2023/5650378. |
[43] | de Vries IR, van Laar JO, van der Hout-van der Jagt MB, et al. Fetal electrocardiography and artificial intelligence for prenatal detection of congenital heart disease. Acta Obstet Gynecol Scand. (2023) Aug 10. doi: 10.1111/aogs.14623. |
[44] | Stoean R, Iliescu D, Stoean C, et al. Deep learning for the detection of frames of interest in fetal heart assessment from first trimester ultrasound. In Advances in Computational Intelligence: 16th International Work-Conference on Artificial Neural Networks, IWANN 2021, Virtual Event, June 16–18, 2021, Proceedings, Part I 16 2021 (pp. 3-14). Springer International Publishing. |
[45] | Anda U, Andreea-Sorina M, Laurentiu PC, et al. Learning deep architectures for the interpretation of first-trimester fetal echocardiography (LIFE)-a study protocol for developing an automated intelligent decision support system for early fetal echocardiography. BMC Pregnancy and Childbirth. (2023) ; 23: (1): 20. doi: 10.1186/s12884-023-05825-w. |
[46] | Yeo L, Romero R. Fetal Intelligent Navigation Echocardiography (FINE): A novel method for rapid, simple, and automatic examination of the fetal heart. Ultrasound Obstet Gynecol. (2013) ; 42: (3): 268-84. doi: 10.1002/uog.12563. |
[47] | Huang C, Zhao BW, Chen R, et al. Is fetal intelligent navigation echocardiography helpful in screening for d-transposition of the great arteries. J Ultrasound Med. (2020) ; 39: (4): 775-84. doi: 10.1002/jum.15157. |
[48] | Ma M, Li Y, Chen R, et al. Diagnostic performance of fetal intelligent navigation echocardiography (FINE) in fetuses with double-outlet right ventricle (DORV). Int J Cardiovasc Imaging. (2020) ; 36: (11). doi: 10.1007/s10554-020-01932-3. |
[49] | Swor K, Yeo L, Tarca AL, et al. Fetal intelligent navigation echocardiography (FINE) has superior performance compared to manual navigation of the fetal heart by non-expert sonologists. J Perinat Med. (2023) ; 51: (4): 477-91. doi: 10.1515/jpm-2022-0387. |
[50] | Yeo L, Romero R. New and advanced features of fetal intelligent navigation echocardiography (FINE) or 5D heart. J Matern Fetal Neonatal Med. (2022) ; 35: (8): 1498-516 doi: 10.1080/14767058.2020.1759538. |
[51] | Iftikhar P, Kuijpers MV, Khayyat A, et al. Artificial intelligence: A new paradigm in obstetrics and gynecology research and clinical practice. Cureus. (2020) ; 12: (2). doi: 10.22541/au.158152219.92311429. |
[52] | Amadori R, Vaianella E, Tosi M, et al. Intrapartum cardiotocography: An exploratory analysis of interpretational variation. J Obstet Gynaecol. (2022) ; 42: (7): 2753-7. doi: 10.1080/01443615.2022.2109131. |
[53] | Ponsiglione AM, Cosentino C, Cesarelli G, et al. A comprehensive review of techniques for processing and analyzing fetal heart rate signals. Sensors. (2021) ; 21: (18): 6136. doi: 10.3390/s21186136. |
[54] | INFANT Collaborative Group. Computerised interpretation of fetal heart rate during labour (INFANT): A randomised controlled trial. Lancet. (2017) ; 389: (10080): 1719-1729. doi: 10.1016/S0140-6736(17)30568-8. |
[55] | Hoodbhoy Z, Noman M, Shafique A, et al. Use of machine learning algorithms for prediction of fetal risk using cardiotocographic data. Int J Appl Basic Med Res. (2019) ; 9: (4): 226. doi: 10.4103/ijabmr.IJABMR_370_18. |
[56] | Spairani E, Daniele B, Signorini MG, et al. A deep learning mixed-data type approach for the classification of FHR signals. Front Bioeng Biotechnol. (2022) ; 10: : 887549. doi: 10.3389/fbioe.2022.887549. |
[57] | Balayla J, Shrem G. Use of artificial intelligence (AI) in the interpretation of intrapartum fetal heart rate (FHR) tracings: A systematic review and meta-analysis. Arch Gynecol Obstet. (2019) ; 300: (1): 7-14. doi: 10.1007/s00404-019-05151-7. |
[58] | Lowry AW, Futterman CA, Gazit AZ. Acute vital signs changes are underrepresented by a conventional electronic health record when compared with automatically acquired data in a single-center tertiary pediatric cardiac intensive care unit. J Am Med Inform Assoc. (2022) ; 29: (7): 1183-1190. doi: 10.1093/jamia/ocac033. |
[59] | Betran AP, Ye J, Moller AB, et al. Trends and projections of caesarean section rates: Global and regional estimates. BMJ Glob Health. (2021) ; 6: (6): e005671. doi: 10.1136/bmjgh-2021-005671. |
[60] | Kowsher M, Prottasha NJ, Tahabilder A, et al. Predicting the appropriate mode of childbirth using machine learning algorithm. International Journal of Advanced Computer Science and Applications. (2021) ; 12: (5). doi: 10.14569/IJACSA.2021.0120582. |
[61] | Meyer R, Hendin N, Zamir M, et al. Implementation of machine learning models for the prediction of vaginal birth after cesarean delivery. J Matern Fetal Neonatal Med. (2022) ; 35: (19): 3677-83. doi: 10.1080/14767058.2020.1837769. |
[62] | De Ramón Fernández A, Ruiz Fernández D, Prieto Sánchez MT. Prediction of the mode of delivery using artificial intelligence algorithms. Comput Methods Programs Biomed. (2022) Jun; 219: : 106740. doi: 10.1016/j.cmpb.2022.106740. |
[63] | Beksac MS, Tanacan A, Bacak HO, et al. Computerized prediction system for the route of delivery (vaginal birth versus cesarean section). J Perinat Med. (2018) ; 46: (8): 881-4. doi: 10.1515/jpm-2018-0022. |
[64] | Wong MS, Wells M, Zamanzadeh D, et al. Applying Automated Machine Learning to Predict Mode of Delivery Using Ongoing Intrapartum Data in Laboring Patients. Am J Perinatol. (2022) . doi: 10.1055/a-1885-1697. |
[65] | Lipschuetz M, Guedalia J, Rottenstreich A, et al. Machine learning based algorithm for prediction of vaginal birth after cesarean deliveries. Am J Obstet Gynecol. (2020) ; 222: (1): S214-5. doi: 10.1016/j.ajog.2019.11.334. |
[66] | Lindblad Wollmann C, Hart KD, Liu C, et al. Predicting vaginal birth after previous cesarean: Using machine-learning models and a population-based cohort in Sweden. Acta Obstet Gynecol Scand. (2021) ; 100: (3): 513-20. doi: 10.1111/aogs.14020. |
[67] | Tsur A, Batsry L, Toussia-Cohen S, et al. Development and validation of a machine-learning model for prediction of shoulder dystocia. Ultrasound Obstet Gynecol. (2020) ; 56: (4): 588-96. doi: 10.1002/uog.21878. |
[68] | Rosenbloom JI, Raghuraman N, Temming LA, et al. Predictive value of midtrimester universal cervical length screening based on parity. J Ultrasound Med. (2020) ; 39: (1): 147-54. doi: 10.1002/jum.15091. |
[69] | Włodarczyk T, Płotka S, Szczepałoki T, et al. Machine learning methods for preterm birth prediction: A review. Electronics. (2021) ; 10: (5): 586. doi: 10.1002/jum.15091. |
[70] | Akazawa M, Hashimoto K. Prediction of preterm birth using artificial intelligence: A systematic review. J Obstet Gynaecol. (2022) ; 42: (6): 1662-8. doi: 10.1080/01443615.2022.2056828. |
[71] | Zhang Y, Lu S, Wu Y, et al. The prediction of preterm birth using time-series technology-based machine learning: Retrospective cohort study. JMIR Medical Informatics. (2022) ; 10: (6): e33835. doi: 10.2196/33835. |
[72] | Arabi Belaghi R, Beyene J, McDonald SD. Prediction of preterm birth in nulliparous women using logistic regression and machine learning. PLoS One. (2021) ; 16: (6): e0252025. doi: 10.1371/journal.pone.0252025. |
[73] | Goldsztejn U, Nehorai A. Predicting preterm births from electrohysterogram recordings via deep learning. PLoS One. (2023) ; 18: (5): e0285219. doi: 10.1371/journal.pone.0285219. |
[74] | Włodarczyk T, Płotka S, Trzciński T, et al. Estimation of preterm birth markers with U-Net segmentation network. In Smart Ultrasound Imaging and Perinatal, Preterm and Paediatric Image Analysis: First International Workshop, SUSI 2019, and 4th International Workshop, PIPPI 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13 and 17, 2019, Proceedings 4 2019 (pp. 95-103). Springer International Publishing. doi: 10.48550/arXiv.1908.09148. |
[75] | Włodarczyk T, Płotka S, Rokita P, et al. Spontaneous preterm birth prediction using convolutional neural networks. In Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis: First International Workshop, ASMUS 2020, and 5th International Workshop, PIPPI 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4–8, 2020, Proceedings 1 2020 (pp. 274-283). Springer International Publishing. doi: 10.48550/arXiv.2008.07000. |
[76] | Kelly CJ, Karthikesalingam A, Suleyman M, et al. Key challenges for delivering clinical impact with artificial intelligence. BMC Medicine. (2019) ; 17: : 1-9. doi: 10.1186/s12916-019-1426-2. |
[77] | Nagendran M, Chen Y, Lovejoy CA, Gordon AC, et al. Artificial intelligence versus clinicians: Systematic review of design, reporting standards, and claims of deep learning studies. BMJ. (2020) ; 368. doi: 10.1136/bmj.m689. |




