HPC+ in the medical field: Overview and current examples
Abstract
BACKGROUND:
To say data is revolutionising the medical sector would be a vast understatement. The amount of medical data available today is unprecedented and has the potential to enable to date unseen forms of healthcare. To process this huge amount of data, an equally huge amount of computing power is required, which cannot be provided by regular desktop computers. These areas can be (and already are) supported by High-Performance-Computing (HPC), High-Performance Data Analytics (HPDA), and AI (together “HPC
OBJECTIVE:
This overview article aims to show state-of-the-art examples of studies supported by the National Competence Centres (NCCs) in HPC
METHOD:
The included studies on different applications of HPC in the medical sector were sourced from the National Competence Centres in HPC and compiled into an overview article. Methods include the application of HPC
RESULTS:
This article showcases state-of-the-art applications and large-scale data analytics in the medical sector employing HPC
CONCLUSION:
HPC
1.Introduction
“Data is the unifying force driving our predictions for 2025; enabling more proactive, preventative, personalised and participatory care, changing care delivery models, and revolutionising the discovery of new medical interventions, whether drugs or devices or services.” – That’s what Deloitte concludes in its extensive report in 2020, titled “Predicting the future of healthcare and life sciences in 2025” [1].
However, to say data is revolutionising the medical sector would be a vast understatement. Wearables are collecting individual health related data, digital twins enable virtual testing of treatments, algorithms help with diagnostics, and these are just three among at least thousands of application possibilities. The amount of medical data available today is unprecedented and has the potential to enable totally new forms of healthcare.
This potential is where we come to High-Performance Computing (HPC), also known as supercomputing. To process this huge amount of data and tame it, an equally huge amount of computing power is required, which cannot be provided by regular desktop computers. Although HPC has been around since the 1960s, it is still relatively new to clinical research and practice. Supercomputing supports scientific fields from research to industrial applications, enabling researchers to run calculations and data analyses that would take years on desktop computers.
As diverse as the European Countries are, as diverse are their skills and applications when it comes to Supercomputing. To level the playing field and strengthen HPC in Europe, the EuroHPC Joint Undertaking, a legal and funding entity, with both public (European Union and Member States and Associated Countries) and private members, has funded the EuroCC project [35] in 2020, which establishes National Competence Centres for High Performance Computing in every participating country. These National Competence Centres (NCCs) consist of universities, research institutes and supercomputing centres with decades of experience in every field, more recently also including healthcare. The EuroCC project will continue to provide support and act as an information hub for all HPC
HPC
In the following sections, different use cases of supercomputing in medicine are each presented by an NCC from the EuroCC project. From the important aspects of surgical pre-planning, to aortic valve replacements, surgery is only one of the fields covered. Other examples include medical image processing in diagnostics, nutritional support of patients in hospitals, treating congenital heart diseases in children, and basic research. Moreover, the examples described here are just the beginning of what is yet to come. As our world becomes more digital, so will healthcare. More data opens up new horizons when it comes to AIassisted diagnostics and preventive care. Digital Twins will become a standard practice and help in treatment as well as in development of new pharmaceutical drugs. Simulations of the spread of infectious diseases will help manage future pandemics. Applications will equally affect physical, mental, and environmental healthcare. Seeing how disruptive HPC has been in other fields, these applications will only be a starting point.
Our first example from the National Competence Centre of the Czech Republic introduces medical image processing, which can help to understand irregularities in the human body, and to detect and predict diseases. We have developed a software concept that uses the computing power of a High-Performance Computing cluster and provides a deep learning-based approach for specific tissue segmentation. It works as a service using state-of-the-art algorithms to segment desired tissues automatically. It can also collect the data after being validated by a professional and facilitates improvement of the segmentation quality over time. The image data can be sent, and results received by a remote computer.
Highperformance computing is also omnipresent in Belgian life sciences and the medical field. It is an important driver of innovation as illustrated here by two further exemplary use cases. The first case presented by the NCC Belgium, concerns VIPUN Medical, a company that develops innovations to feed critically ill and other vulnerable patients. Malnutrition is associated with increased morbidity and mortality rates. To combat this problem, VIPUN Medical develops a software tool to automatically quantify gastric motility from the measurement of intragastric pressure. An HPC expert was consulted to develop an efficient signal analysis and to optimize the parametrization of the method. The second case demonstrates the use and impact of HPC in pharmaceutics, as the dynamics of protein drug targets can be studied in detail using molecular dynamics simulations. Both case studies show the importance of having dedicated HPC staff and infrastructure.
Our third study presented by the NCC Switzerland, covers mechanical and biological aortic valve prostheses. Such prostheses suffer from limited durability and biocompatibility. In part, this is related to unphysiological turbulent blood flow patterns downstream of the valves. The onset and intensity of this turbulent flow is directly related to the prosthesis design which has not undergone any major design changes over the past decades. We use high-fidelity, GPU-accelerated solvers for fluid-structure interaction (FSI) to study this flow system on GPU-based supercomputing systems at the Swiss Supercomputing Center CSCS. We investigate the mechanisms of laminar-turbulent transition in valve prostheses and explore new valve designs to reduce or to delay the onset of turbulence. Results indicate that even small design changes may lead to significant reduction of turbulence.
Pediatric radiation dosimetry is of great interest in modern medicine, as children provide a higher risk of developing cancer compared to adults, when exposed in equivalent radiation doses. The aim of our fourth article, by the NCC Greece in collaboration with an experiment enabled by the FF4EuroHPC project [36], is to present the innovative online application “PediDose” for internal dosimetry prediction, which acts as a decision support tool for clinicians, to estimate absorbed doses per organ, prior to a pediatric nuclear medical procedure. Monte Carlo simulations were applied on a pediatric computational population to create a realistic dosimetry database for 5 commonly used radiopharmaceuticals. This simulated database was used as ground truth to develop an internal dosimetry prediction toolkit for nuclear medical pediatric applications by exploiting Artificial Intelligence techniques. The “PediDose” software has been developed with a user-friendly Graphical User Interface. The online application’s interface requires the input of the patient’s physical characteristics (e.g., age, gender, weight, height) by the clinician and exports predictions of the absorbed doses for 30 different organs of interest. Advanced computational techniques were combined (MC simulations, Machine Learning, anthropomorphic phantoms) to predict internal doses taking into consideration the patients’ specific anatomical characteristics. The developed models are integrated in a novel dosimetry software, assisting clinicians towards personalized pediatric dosimetry.
The fifth and last study, a cloud-based HPC platform to support systemic pulmonary shunting procedures application is presented by the NCC Italy in collaboration with an experiment enabled by the FF4EuroHPC project [36]. The cloud-based HPC platform will use patient-specific computationally intensive models running in a reliable and cost-efficient way on a cloud-based HPC environment. The experiment aims at building up an affordable decision support web application that, thanks to the medical digital twin (MDT), allows surgeons to approach the mBTS medical intervention at best. The tool generates the ROM of a patient-specific vascular district in which the shunt implantation is geometrically parameterized. The ROM generates pre-defined CFD results of different configurations of the vasculature with shunt implantation. With a dedicated User Interface, the medical staff will be able to inspect the MDT of the patient in an interactive way, to finalize the decisions on surgical intervention. The geometry implementation and model setup is carried out with the adoption of the well-established commercial software provided thanks to the cooperation with ANSYS.
Section 1. National Competence Centre Czech Republic: Medical image processing as a service
Petra Svobodova, Khyati Sethia and Petr Strakos
IT4Innovations, VSB – Technical University of Ostrava, Ostrava-Poruba, Czech Republic
Method
The visualised concept for remote automatic tissue segmentation consists of two main sections as depicted in Fig. 1. The first part runs on site at a medical doctor’s office in a hospital, and the second part operates at the supercomputing centre. The part based at the hospital is represented by a frontend that mediates the interaction between the doctor and the data. An open-source 3D Slicer [2] was chosen to provide this functionality and allow for the development of the required interconnection to a backend part. The backend part provides the computational power of an HPC cluster and other necessary features. It allows the training of deep learning models for automatic segmentation and model inference of incoming image data as an AI-based annotation service. NVIDIA’s Clara Train SDK [3] was used to create the backend part.
Figure 1.
The main concept of the tool for medical image processing and analysis.
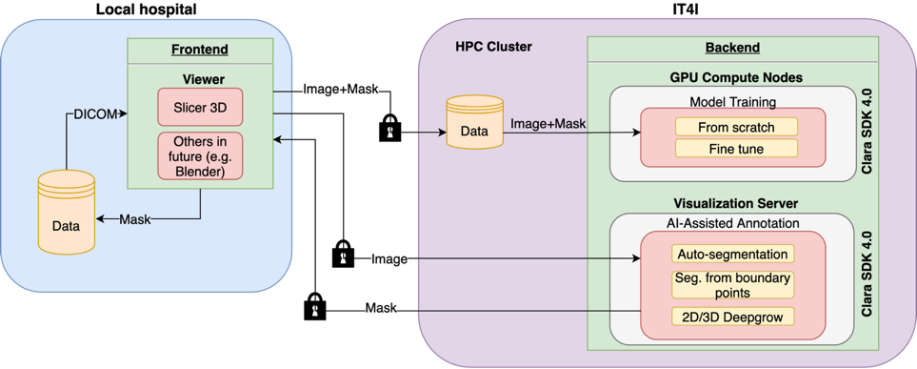
Only anonymised image data in the NIFTI format [4] and binary label masks are exchanged between the hospital and the supercomputing centre. Additionally, all the connections are secured by secure encrypted communication (SSH).
Conclusion
The created toolset offers medical doctors the possibility to establish or refine the patient’s diagnosis from image data. When detailed data analysis such as image segmentation is required, e.g., to prepare a patient for surgery, the process is lengthy and highly manual therefore supporting tools that enable the efficient and accurate automation of this kind of radiological examination whose quality evolves with each additional case are very valuable and in high demand.
Section 2. NCC Belgium: HPC support in innovative medtech
Pierre Beaujean
Belgium has strong roots in research and innovation. This is very clear in the life sciences industry, with an exceptionally diverse and innovative biotech, medtech, pharma and healthcare industry. NCC Belgium has several actions aimed at supporting this sector by providing expert services and state-of-the art high performance computing infrastructure, illustrated by two interesting cases of a Flemish SME and fundamental research in the Walloon region.
Development of an algorithm to automatically determine gastric motility from intragastric pressure
Belgian life sciences industry is diverse; from young start-ups to multinationals. VIPUN
Several factors, such as feeding tube positioning problems or gastrointestinal intolerance, contribute to the overall incidence of malnutrition which could be as high as 50% in Intensive Care Units. The VIPUN Gastric Monitoring System aims to make it easy for medical staff to make a well-informed and faster nutrition therapy decision thus reducing malnutrition and feeding-related complications. One aspect of this assessment is the measurement of intragastric pressure, dictated by several non-related phenomena such as breathing and abdominal pressure. Moreover the quantification of gastric contractility is difficult as the stomach contractions are variable in duration and amplitude [9]. VIPUN Medical is developing a software tool called the “VIPUN Motility Explorer” to automatically quantify gastric motility from the measurement of intragastric pressure, assessed with an intragastric balloon [8]. To develop this software, VIPUN Medical required expert support to process the clinical data, to develop an efficient signal analysis and to computationally optimize the parametrization of the method. To this end, VIPUN Medical was able to partner with the Flemish Supercomputing Center (VSC; partner of NCC Belgium) and PRACE. An easy-to-use tool with advanced graphical user interaction features was developed in Python/Matplotlib by the VSC that enables easy and fast manual scoring of existing pressure recordings from the clinical studies. The output file of the VIPUN MI Recorder matches the proposed output of the VIPUN Motility Explorer and as such it allows to iterate the settings of the VIPUN Motility Explorer to optimize the VIPUN Motility Explorer output to the expert opinion.
Once a working methodology was programmed in the VIPUN Motility Explorer, the parametrisation of that methodology had to be computationally optimized. The parametrization of the application was based on the least square difference method, minimizing the difference between a reference output and the target output (generated by the VIPUN Motility Explorer). Computational speed-up of the parameter optimization was accomplished by the HPC expert by parallelizing the code with MPI, improving performance by as much as 50%. This example clearly demonstrates the impact the support of HPC staff can have for innovative SMEs which in this case was provided by the VSC [1], a consortium of the Flemish universities in Belgium that provides supercomputer infrastructure and services to academia and industry.
Using HPC to unravel the dynamic behaviour of hiDO1, a drug target for immunotherapy
The second use case is academic research that nevertheless has important applications in pharmaceutics. Manon Mirgaux (University of Namur) studies the dynamic behavior of the human protein Indoleamine 2,3-dioxygenase 1 (hIDO1) [11]. This protein is involved in the thryptophane-kynuranine pathway and when suractivated, immune escape is induced, increasing resistance to immune therapy [11]. Hence, a better understanding of the dynamics of this protein may lead to the development of inhibitors that would improve the success rate of immune therapy for, e.g., cancer.
The 3D structure of hIDO1 was determined using X-ray crystallography [11]. However, this only provides static information, while a dynamic loop in this protein is involved in the substrate positioning, and hence influences the response of hIDO1 to inhibitors. Using the crystallographic structure as a starting point, molecular dynamics simulations were performed to study the motion of this dynamic loop during substrate positioning.
These simulations of the dynamic behavior of hIDO1 were performed on the HPC infrastructure of CECI [12], a consortium of the Walloon universities in Belgium to provide such infrastructure and related services to their researchers. Manon Mirgaux stresses the importance of her interaction with the support team of CECI to help her complete her simulations efficiently.
Both use cases illustrate that the support of the HPC staff of CECI and VSC is crucial for success. Researchers are obviously specialists in their respective fields but require guidance on how to run their computations efficiently on HPC infrastructure. SMEs often lack the resources to invest in fine-tuning algorithms so they can scale to larger data sets or more involved workflows. CECI and VSC are member organizations of NCC Belgium [13].
Section 3. NCC Switzerland: Improved prosthesis design for aortic valve replacement with HPC
Lukas Drescher
Aortic valve replacement (AVR) is a commonly performed treatment for patients with moderate to severe aortic stenosis which may lead to heart failure if left untreated. Despite its clinical success, AVR still suffers from severe restrictions related to limited durability of valve prostheses and to blood trauma caused by unphysiological blood flow through the prosthesis.
The design of the bi-leaflet mechanical heart valve (BMHV), which is one of most widely used heart valve prostheses, reaches back to the 1970’s and submits patients to life-long anticoagulation therapy because of its thrombogenicity. Valve prostheses made from biological tissue (biological heart valves, BHV) were first implanted only a few years after the first BMHV. Although their introduction mostly resolved the problem of valve thrombogenicity, they durability is limited (
Under the light of these problems, it seems difficult to understand why the basic design of aortic valve prostheses has barely seen any major design changes over the past decades. So far, development efforts went mostly in improved chemical fixation of the prosthetic tissue to mitigate BHV calcification and in catheter-based delivery systems to allow for minimally invasive implantation techniques, although there is strong evidence that the quality blood flow through valve prostheses is an important factor in valve performance, biocompatibility and durability. Therefore, we use high-performance computing (HPC) to simulate blood flow for different valve designs aiming order to better understand the relation between valve design and blood flow.
Reducing turbulence in bileaflet mechanical heart valves
BMHV are associated with vigorous turbulent blood flow. Next to increased viscous losses and unphysiological wall shear stresses in the ascending aorta, turbulence may also lead to blood platelet activation which is the first step toward valvular thrombus formation. To find ways to reduce turbulence behind BMHV, we used high-fidelity blood flow simulations which resolved flow structures down to 50
Although we used a highly optimized flow solver which was designed for massively-parallel computing using the message passing interface MPI [15], modifications of the leaflet design could not be tested because the requirements on spatial resolution prohibited three-dimensional simulations even on large supercomputing systems. Porting of compute-intensive kernels to GPUs [16] (using a Fortran-C-CUDA interface to enable implementation in our legacy flow solver) and the replacement of the pressure Poisson solver (originally using distributed multigrid) by a GPU-accelerated Jacobi solver with successive over-relaxation [17] yielded an implementation which was about 150 times faster than the original CPU-based implementation. This allowed running a simulation with a three-dimensional model of a modified BMHV design with approximately 337 million grid points which was arguably the largest simulation ever of a heart valve. This simulation was completed on the Piz Daint system (Cray XC-40/50) at the Swiss Supercomputing Centre CSCS in three days using 20 nodes (whereas the CPU-based implementation would have taken 1.5 years to complete). The computational results confirmed that the modified leaflet geometry significantly delayed the onset of turbulence behind the BMHV.
Laminar-turbulent transition in bioprosthetic heart valves
In contrast to BMHV, complex fluid-structure interaction (FSI) between blood flow and soft tissue governs the biomechanics of BHV, which creates additional computational challenges and further increases the computational cost. We developed the high-performance FSI solver AV-FLOW [18] which uses an immersed boundary scheme [19] to couple our high-order Navier-Stokes solver with a finite-element solver for soft tissue mechanics using hyperelastic material models. The solver has been partially accelerated for GPUs and employs a parallel library for efficient variational transfer operations between the fluid grid and the structure mesh [20].
Simulations with a generic BHV model [21] exhibited transition from laminar to turbulent flow which was governed by a FSI instability leading leaflet fluttering and shedding of vortex rings which rapidly break down to turbulent flow. Based on these results, modifications of the leaflet material and the leaflet geometry were tested with the aim to eliminate leaflet fluttering and to suppress or delay the laminar-turbulent transition behind BHV. Whereas changes in the bulk material properties did not yield the desired effect, modified leaflet designs with reduced belly curves were shown to stabilize the leaflet fluttering [22].
Where do we go from here?
HPC is an enabling technology for improving the design of heart valve prostheses. Although the Reynolds number of valvular flow is moderate compared to classical technical applications (e.g., in aerospace engineering) the computational cost of the simulations is high and efficient turn-around requires the use of modern supercomputing platforms including GPUs.
The use of such accelerated codes allows us to go beyond classical forward simulations and to employ, e.g., adjoint-based methods of automatic design optimization for heart valves [23] or data assimilation to enhance clinical diagnostic imaging [25]. In particular, the second example presents an application which can connect daily clinical work in a hospital to HPC. This was explored by the authors in the HPC-PREDICT project in which a computational workflow for the analysis of blood flow in the ascending aorta was developed. This end-to-end workflow includes elements for fast data acquisition with 4D-Flow-MRI [24] automatic segmentation algorithms a Kalman filter for data assimilation of 4D-Flow-MRI images [25], and a deep learning network for anomaly detection in the enhanced MRI data [26]. The vision of HPC-PREDICT is to enable hospitals to improve their diagnostic imaging by providing fast, high-resolution, low-noise and automatically annotated 4D-Flow-MRI images using HPC.
Clinical implementation of such a workflow presents challenges that still need to be addressed. This includes the efficient data transfer between hospital and supercomputing center which respects regulatory requirements and all aspects of data privacy. First steps were taken in this direction by establishing a data store using an encrypted file system encfs and data version control DVC.
As modern imaging modalities yield more and more data (higher resolution, 4D imaging, functional imaging), the need for efficient tools increases as the meaningful interpretation of this data is limited by the availability of trained personnel. This bottleneck could be resolved by HPC which can take over some of these tasks in an efficient manner. It leads the way toward computer-augmented diagnostic data and implants with optimized patient-specific designs to provide better diagnosis and treatment in the spirit of personalized medicine.
Section 4. NCC Greece and FF4EuroHPC: PediDose: An innovative tool for personalised pediatric dosimetry in nuclear medicine using advanced computational techniques
Panagiotis Papadimitroulas
Introduction
Radiation dose calculations in humans from radiotracers have been a challenge to healthcare society for more than 50 years. Exposure to ionizing radiation in children can cause radiation-induced complications that may include cancer [27]. Current advances in computer science, i.e., high-performance computing (HPC) and artificial intelligence (AI) allow for the development of precise medicine and personalization of clinical protocols.
Figure 2.
Functionality of PediDose backend.
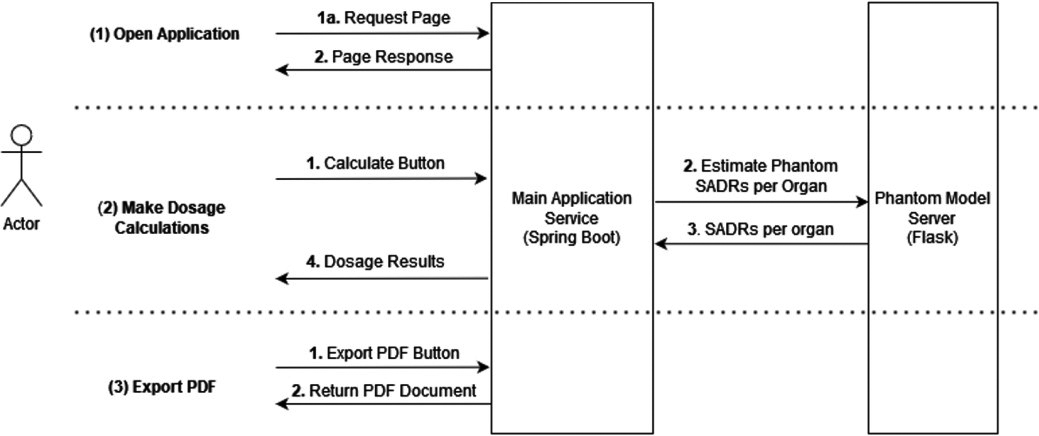
Materials and methods
PediDose utilizes: i) MC simulations using GATE MC toolkit [28], ii) a population of 28 pediatric computational models [29] and iii) ensemble learning by combining predictions of ML regression models, trained and evaluated on the simulated dataset with LOOCV [30]. The dosimetry calculations via MC simulations have been based on the SADR method [31].
Results and conclusions
The platform has been built in a modular way, designed to separate the main application server functionality from the model predictions. This design allows multiple instances to be active simultaneously
Figure 3.
PediDose software implementation. Left: GUI for clinicians to import parameters. Middle: Horizontal diagram of the predicted absorbed doses per organ. Right: Exported report in pdf format.
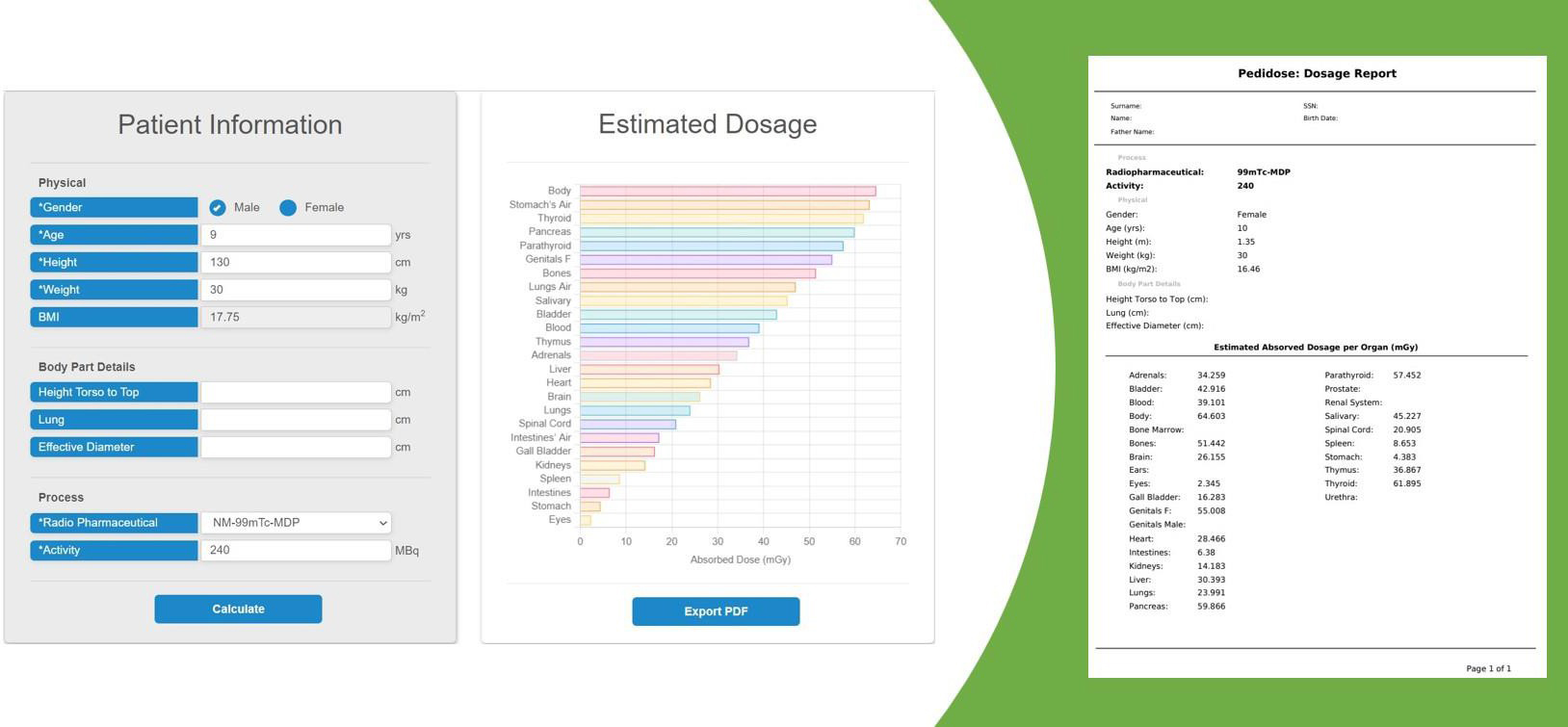
for the process heavy tasks and balances the load (i.e., model predictions) for performance improvement. The provided functionality of the backend is demonstrated in Fig. 1. PediDose is developed as an online tool with a graphical user interface (GUI). Figure 2 depicts the software implementation flow. Initially, the clinician imports the specific characteristics of the patient, selects the appropriate radiopharmaceutical and defines the injected activity according to the corresponding protocol. Consequently, the software provides a prediction of the doses per organ that the patient is going to receive and finally, the absorbed doses, along with the patient’s anatomical information can be exported in pdf format. The proposed methodology of combining the predictive power of AI utilizing MC ground truth for dosimetry assessment, can be further extended to other populations (adult, obese, pregnant) and medical applications (radioimmunotherapy), where fast and personalized absorbed dose determination is critical.
Figure 4.
Workflow for MDT application development.
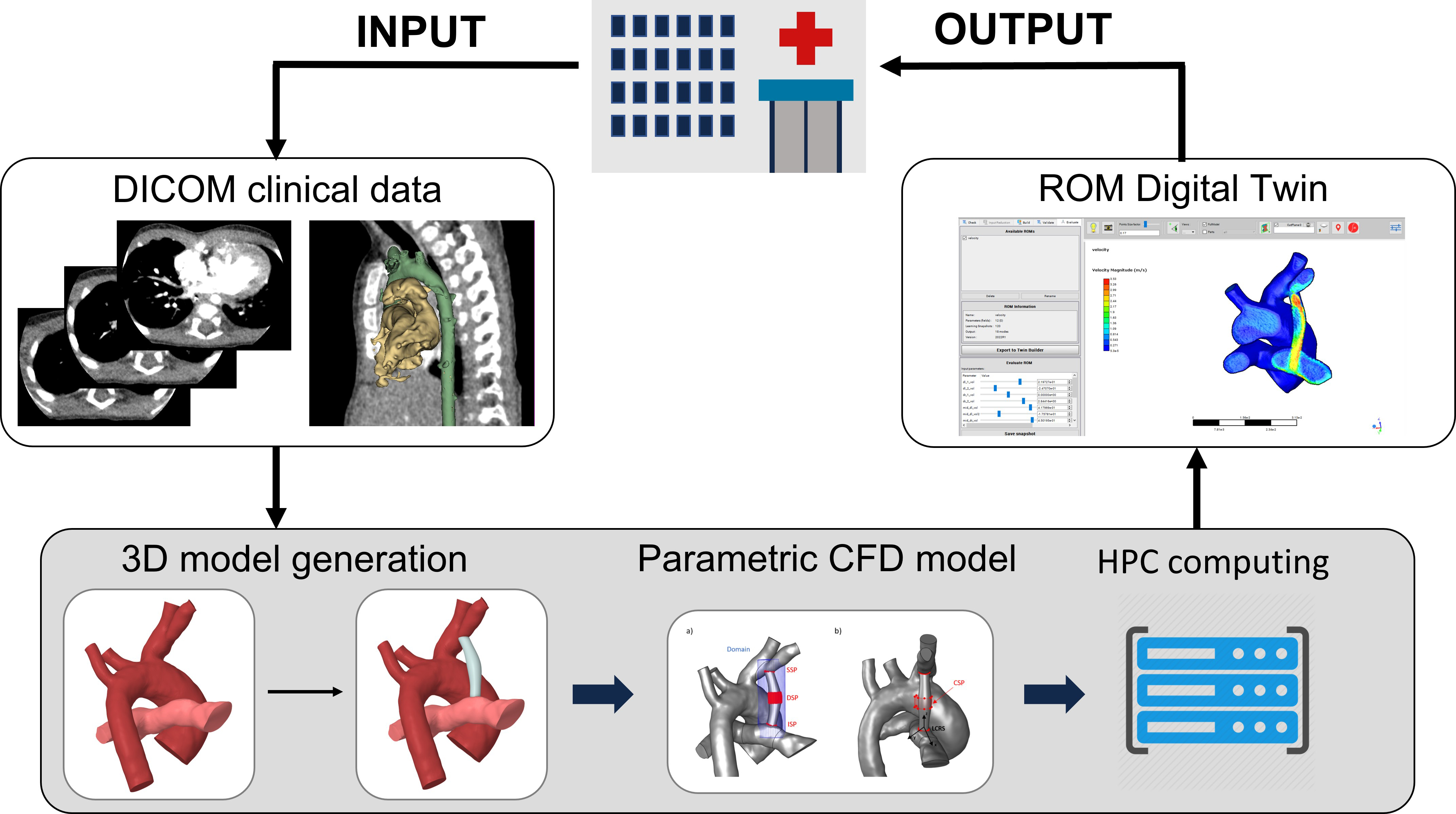
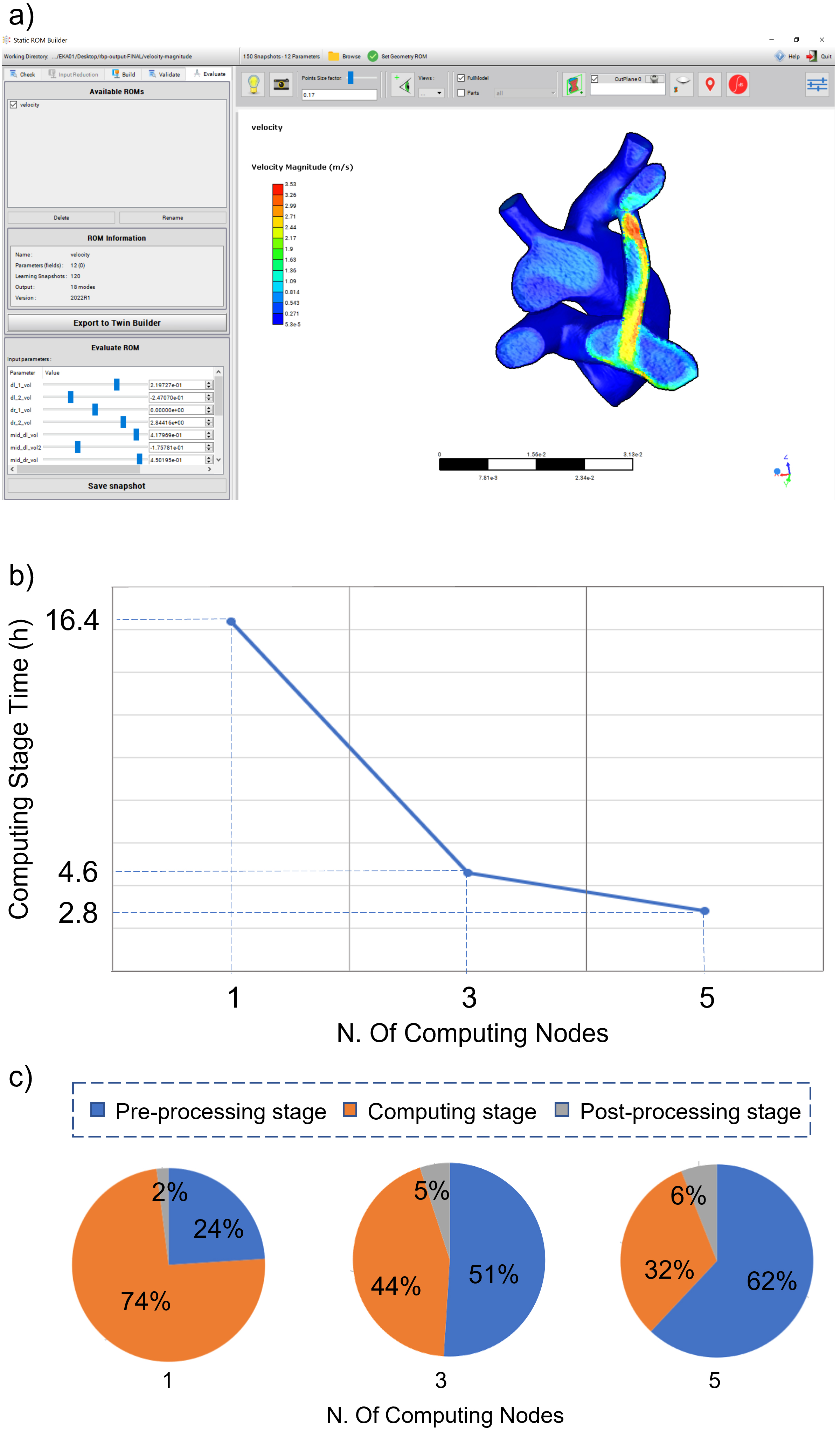
Figure 5.
Example of interface for MDT of MBTS (a), computational times (b) and relative times percentages for the different application stages (c) as a function of number of HPC nodes.
Section 5. NCC Italy and FF4EuroHPC: Cloud-based HPC platform to support systemic pulmonary shunting procedures
Emanuele Vignali
Congenital heart diseases (CHDs) are the most common type of birth defects and the leading cause of death in children with congenital malformations. The Modified Blalock Taussig Shunt (MBTS) is a widely used surgical technique to restore the connection between systemic and pulmonary circulation in patients with critical CHD [32]. The procedure is, however, associated with significant morbidity and mortality. Surgical planning in terms of MBTS sizing and positioning is pivotal for the management of the disease. In this context, an affordable decision support application was built to allow surgeons approaching the MBTS medical intervention at best [33]. The application output is a high-fidelity medical digital twin (MDT) of a patient-specific MBTS procedure, modelled starting from patient’s clinical images. The MDT is obtained from a Reduced Order Model (ROM) that represents the Computational Fluid Dynamics (CFD) behaviour of the shunt (Fig. 1) [34]. Usage of HPC resources accelerates the time-demanding ROM generation step allowing to obtain the MDT within a time scale compatible with clinical needs.
The MDT procedure foresaw a pre-processing, a computation and a post-processing stage. For the pre-processing stage, a 480 multi-slice CT dataset was segmented and the corresponding CAD model of MBTS was generated. The model was meshed and a baseline CFD case was setup. RBF morphing shape modifiers were introduced to allow for shunt modifications. The 12 shape modifiers were defined according to clinical needs to cover all the relevant MBTS positions.
For the computing stage, a combination of 150 simulation cases was designed for the ROM population by parametrizing the shape modifiers. As this stage is the most computationally onerous, different HPC strategies were tested. A HPC cluster (2 x CPU Intel CascadeLake 8260, 48 cores, 2.4 GHz, 384 GB RAM for each computing node) was adopted. To assess the HPC performances and allow for fast analysis results, the computation was carried out both by using a single computing node and by using multiple nodes in parallel (i.e. 3 and 5 nodes). For the post-processing stage, ROM consumption was carried out from the generated scenarios. The resulting MDT was provided to clinical staff for evaluation and computational times were analysed.
An example of the generated MDT interface is reported in Fig. 2a. The computing stage time as a function of the number of HPC nodes is reported in Fig. 2b. Performing the computation on 5 HPC nodes reduces this stage time down to 2.8 h. In this case, the MDT generation time, including the pre- and post-processing stages, was
Acknowledgments
The project EuroCC (including all NCCs’ work presented here) has received funding from the European High-Performance Computing Joint Undertaking (JU) under grant agreement no 951732. The JU received support from the European Union’s Horizon 2020 Research and Innovation Programme and Germany, Bulgaria, Austria, Croatia, Cyprus, Czech Republic, Denmark, Estonia, Finland, Greece, Hungary, Ireland, Italy, Lithuania, Latvia, Poland, Portugal, Romania, Slovenia, Spain, Sweden, United Kingdom, France, The Netherlands, Belgium, Luxembourg, Slovakia, Norway, Switzerland, Turkey, Republic of North Macedonia, Iceland, and Montenegro.
Section 2: This work was also financially supported by the PRACE project funded in part by the EU’s Horizon 2020 Research and Innovation Programme (2014–2020) under grant agreement no. 823767.
Section 3: The work described for the NCC Switzerland has also been supported by the Platform for Advanced Scientific Computing PASC.
Section 4: The experiment “PediDose” received funding from the European High-Performance Computing Joint Undertaking (JU) through the FF4EuroHPC project under grant agreement no 951745. The JU received support from the European Union’s Horizon 2020 Research and Innovation Programme and Germany, Italy, Slovenia, France, and Spain.
Section 5: This project received funding from the experiment number 1006 of the FF4EuroHPC project no. 951745 and the Marie Skłodowska-Curie grant agreement MeDiTATe project no 859836.
Conflict of interest
None to report.
References
[1] | |
[2] | Fedorov A, et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. (2012) Nov; 30: (9): 1323-41. PMID: 22770690. PMCID: PMC3466397. |
[3] | NVIDIA Clara Imaging. https://developer.nvidia.com/clara-medical-imaging ((2020) ). |
[4] | NIfTI: Neuroimaging Informatics Technology Initiative. https://nifti.nimh.nih.gov/ ((2021) ). |
[5] | VIPUN Medical, 2022, accessed 20/10/2022, https://www.vipunmedical.com/#about. |
[6] | Kim H, Stotts NA, Froelicher ES, Engler MM, Porter C. Why patients in critical care do not receive adequate enteral nutrition? A review of the literature. Journal of Critical Care. (2012) ; 27: (6): 702-713. doi: 10.1016/j.jcrc.2012.07.019. |
[7] | Saunders J, Smith T. Malnutrition: causes and consequences. Clinical Medicine. (2010) ; 10: (6): 624-627. doi: 10.7861/clinmedicine.10-6-624. |
[8] | Goelen N, Hoon J, Morales JF, Varon C, van Huffel S, Augustijns P, Mols R, Herbots M, Verbeke K, Vanuytsel T, Tack J, Janssen P. Codeine delays gastric emptying through inhibition of gastric motility as assessed with a novel diagnostic intragastric balloon catheter. Neurogastroenterology & Motility. (2020) ; 32: (1). doi: 10.1111/nmo.13733. |
[9] | Phillips RS, Iradukunda EC, Hughes T, Bowen JP. Modulation of enzyme activity in the kynurenine pathway by kynurenine monooxygenase inhibition. Frontiers in Molecular Biosciences. (2019) ; 6: (FEB). doi: 10.3389/fmolb.2019.00003. |
[10] | Vlaams Supercomputing Centrum (VSC), https://vscentrum.be/. |
[11] | Mirgaux M, Leherte L, Wouters J. Influence of the presence of the heme cofactor on the JK-loop structure in indoleamine 2,3-dioxygenase 1. Acta Crystallographica Section D Structural Biology. (2020) ; 76: (12): 1211-1221. doi: 10.1107/S2059798320013510. |
[12] | Consortium des Équipements de Calcul Intensif (CECI), https://www.ceci-hpc.be/. |
[13] | National Competence Center Belgium (NCC Belgium), EuroCC, https://www.enccb.be/. |
[14] | Zolfaghari H, Obrist D. Absolute instability of impinging leading edge vortices in a submodel of a bileaflet mechanical heart valve. Phys. Rev. fluids, (2019) ; 4: (12): 123901. |
[15] | Henniger R, Obrist D, Kleiser L. High-order accurate solution of the incompressible Navier-Stokes equations on massively parallel computers. J. Comput. Phys. (2010) ; 229: (10). doi: 10.1016/j.jcp.2010.01.015. |
[16] | Zolfaghari H, Becsek B, Nestola MGC, Sawyer WB, Krause R, Obrist D. High-order accurate simulation of incompressible turbulent flows on many parallel GPUs of a hybrid-node supercomputer. Comput. Phys. Commun. (2019) ; 244: . doi: 10.1016/j.cpc.2019.06.012. |
[17] | Zolfaghari H, Obrist D. A high-throughput hybrid task and data parallel Poisson solver for large-scale simulations of incompressible turbulent flows on distributed GPUs. J. Comput. Phys. (2021) ; 437: : 110329. |
[18] | Nestola MGC, et al. An immersed boundary method for fluid-structure interaction based on variational transfer. J. Comput. Phys. (2019) ; 398: . doi: 10.1016/j.jcp.2019.108884. |
[19] | Peskin CS. The immersed boundary method. Acta Numer. (2002) ; 11: : 479-517. |
[20] | Krause R, Zulian P. A parallel approach to the variational transfer of discrete fields between arbitrarily distributed unstructured finite element meshes. SIAM Journal on Scientific Computing. (2016) ; 38: (3): C307-C333. |
[21] | Becsek B, Pietrasanta L, Obrist D. Turbulent Systolic Flow Downstream of a Bioprosthetic Aortic Valve: Velocity Spectra, Wall Shear Stresses, and Turbulent Dissipation Rates. Front. Physiol. (2020) ; 11: . doi: 10.3389/fphys.2020.577188. |
[22] | Corso P, Jahren SE, Vennemann B, Obrist D. Leaflet fluttering and turbulent systolic blood flow after bioprosthetic heart valves, CMBBE – International Symposium on Computer Methods in Biomechanics and Biomedical Engineering, 7–9 September 2021, https://youtu.be/ug_-I_94opc, 2021. |
[23] | Zolfaghari H, Kerswell RR, Obrist D, Schmid PJ. Sensitivity and downstream influence of the impinging leading-edge vortex instability in a bileaflet mechanical heart valve. J. Fluid Mech. (2022) ; 936: . |
[24] | Vishnevskiy V, Walheim J, Kozerke S. Deep variational network for rapid 4D flow MRI reconstruction. Nature Machine Intelligence. (2020) ; 2: (4): 228-235. |
[25] | De Marinis D, Obrist D. Data assimilation by stochastic ensemble kalman filtering to enhance turbulent cardiovascular flow data from under-resolved observations. Front. Cardiovasc. Med. (2021) ; 8: : 742110. |
[26] | You S, Tezcan KC, Chen X, Konukoglu E. Unsupervised lesion detection via image restoration with a normative prior. In International Conference on Medical Imaging with Deep Learning. PMLR. (2019) May. pp. 540-556. |
[27] | Papadimitroulas P, et al. A Review on Personalized Pediatric Dosimetry Applications Using Advanced Computational Tools IEEE trans. Radiat. Plasma Med. Sci. (2019) ; 3: (6): 607-620. |
[28] | Sarrut D, et al. The OpenGATE ecosystem for Monte Carlo simulation in medical physics. Phys Med Biol. (2022) ; 67: (18): 184001. |
[29] | Segars WP, et al. The development of a population of 4D pediatric XCAT phantoms for imaging research and optimization Med. Phys. (2015) ; 42: (8): 4719-26. |
[30] | Sammut C, Webb GI (eds). Encyclopedia of Machine Learning Boston, MA Springer Leave-One-Out Cross-Validation, (2011) . |
[31] | Papadimitroulas P, et al. A personalized, Monte Carlo-based method for internal dosimetric evaluation of radiopharmaceuticals in children Med. Phys. (2018) ; 45: : 3939-3949. |
[32] | Bentham JR, Zava NK, Harrison WJ, Shauq A, Kalantre A, Derrick G, et al. Duct stenting versus modified Blalock-Taussig shunt in neonates with duct-dependent pulmonary blood flow: Associations with clinical outcomes in a multicenter national study. Circulation. (2018) ; 137: (6): 581-8. |
[33] | Cloud-based HPC platform to support systemic-pulmonary shunting procedures; Available from: https://www.ff4eurohpc.eu/en/success-stories/2022111921555173/cloudbased_hpc_platform_to_support_systemicpulmonary_shunting_procedures. |
[34] | Kardampiki E, Vignali E, Haxhiademi D, Federici D, Ferrante E, Porziani S, et al. The hemodynamic effect of modified blalock-taussig shunt mor-phologies: A computational analysis based on reduced order modeling. Electronics. (2022) ; 11: (13): 1930. |
[35] | EuroCC, https://www.eurocc-access.eu. |
[36] | FF4EuroHPC, https://www.ff4eurohpc.eu. |
[37] | Medical Solution Center – CASE4Med, https://case4med.de/. |
[38] | Computational Immediate Response Center for Emergencies, CIRCE, https://www.hlrs.de/projects/detail/circe. |
[39] | https://www.hlrs.de/news/detail/hlrs-and-bib-win-hpc-innovation-award-for-pandemic-monitoring-tool. |



