Evidence of Increased Hemangioblastic and Early Hematopoietic Potential in Chronic Myeloid Leukemia (CML)-derived Induced Pluripotent Stem Cells (iPSC)
Abstract
Hemangioblasts derived from mesodermal lineage are the earliest precursors of hematopoietic stem cells and endothelial cells. Embryonic stem cells (ESC) and induced pluripotent stem cells (iPSC) are the only experimental systems in which these cells can be assayed and quantified. We show here using CML-derived iPSC and blast-cell colony forming (Bl-CFC) assays that hemangioblasts are highly expanded in CML derived iPSC as compared to human H1-ESC-derived hemangioblasts. BCR-ABL signaling pathway is intact in these cells with evidence of CRK-L phosphorylation which is reduced by the use of Imatinib. Hematopoietic progenitor assays generated using blast-CFC demonstrates also a highly increased hematopoietic progenitor potential of these cells as compared to H1-ESC. The same results were also obtained using hematopoietic progenitor assays via embryoid body formation. In CML iPSC, we have also found a significant reduction of Aryl Hydrocarbon Receptor (AHR) expression. Further inhibition of AHR using StemRegenin (SR1), an AHR antagonist, led to an increase of blast-cell colonies in CML iPSC whereas the use of an AHR agonist inhibited blast cell colonies. Thus, our results show for the first time, the possibility of establishment of a myeloproliferative phenotype using patient-derived iPSC and the presence of a major expansion of the hemangioblast compartment and hemangioblast-derived hematopoietic progenitors in this context. They also suggest that the AHR signaling pathway could represent a novel druggable target in CML.
INTRODUCTION
Chronic myeloid leukemia is the prototype of hematopoietic stem cell malignancy involving a highly primitive hematopoietic stem cell (HSC). The disease presents essentially by the expansion and proliferation of hematopoietic progenitors of myeloid lineage but lymphoid B cells lineage can also be involved [1]. There is a controversy with regard to the involvement of endothelial lineage in CML which would suggest that a stem cell with both hematopoietic and endothelial potential would be the target of transformation by BCR-ABL [2, 3].
Patient-derived induced pluripotent stem cells (iPSC) represent a powerful tool to model several types of diseases [4] and especially chronic myeloid leukemia [5–8]. We report here for the first time that the earliest common hematopoietic and endothelial precursor designed as hemangioblast is highly expanded in stem cells assays using iPS cell lines derived from a patient with CML. We show that CML-derived iPSCs generate high numbers hemangioblasts detected by blast-cell colony forming cell (Bl-CFC) assays. Likewise, hematopoietic progenitors derived from either Bl-CFC or embryoid bodies are also highly increased in CML-derived iPSC. Finally we show the potential of manipulating this expansion by using AHR pathway.
MATERIALS AND METHODS
Patient-derived iPSC and characterization
Generation of CML iPSC has been previously described [9]. Briefly, peripheral blood cells were obtained at diagnosis from a 14-year-old CML patient with informed consents according to the declaration of Helsinki. This patient was treated with Imatinib mesylate as a first line therapy. At +12 months post-therapy, there was no molecular response and no other targeted therapy was available in 2004. An allogenic bone marrow transplant was performed. The patient remains in complete remission since the transplant performed in 2004. To generate iPSC, leukemic CD34+ isolated and cryopreserved at diagnosis were used as previously described [9]. Briefly, CD34+ cells were cultured for 4 days in the presence of growth factors hSCF (100 ng/ml), hFLT-3 (100 ng/ml), hIL-6 (20 ng/ml), hIL-3 (20 ng/ml), and hIL7 (20 ng/ml) (all from Peprotech). At day + 4, 2×105 CD34 + cells were transduced overnight with Sendai viruses containing Oct3/4, Sox2, Klf4, and c-Myc (CytoTune®-iPS Sendai Reprogramming Kit, Life technologies), at multiplicity of infection (MOI) of 15.
48 hours after the transduction, cells were placed in Mitomycin-C-treated murine embryonic fibroblast (MEF) feeders in the presence of bFGF, until the appearance of iPSC colonies as previously described [9]. The control iPSC cell line (PB33) was obtained using hematopoietic cells of a normal donor using the same protocol. All analyses were performed using a polyclonal stock of iPSC. Both cell lines were characterized using in vitro and in vivo pluripotency tests [9].
iPSC and ESC cultures
hIPS cell lines were maintained on the mitomycin-C-inactivated mouse embryonic fibroblast feeder cells with DMEMF12 supplemented with 0.1 mg/ml of bFGF. Human embryonic stem cell line H1 was obtained from WiCell (Wisconsin, USA) and cultured as previously described, after the authorization obtained from Agence de Biomedicine [10]. (Authorization for A.G.Turhan and A. Bennaceur-Griscelli).
Passaging was performed every 7 days using 1 mg/ml collagenase IV applied for 20 minutes to detach cells prior to transfer to new MEF layers. In some experiments cells were cultured in feeder-free conditions in Essential 8 medium. Passaging was done every 3-4 days, using DPBS containing 0.5 mM EDTA, Life Technologies) and 1.8 mg/L NaCl (Sigma). Cell lines were regularly tested for mycoplasma infection which always remained negative. Authenticity of cell lines was tested using short tandem repeat (STR) analyses. Karyotype analyses were performed before the experiments and regularly, showing a normal karyotype for H1 and PB33 and the presence of the Ph1 chromosome in 20/20 mitoses in PB32 cell line. To determine the potential effect of Imatinib on the iPSC, cells were cultured for 5 days in the presence of this inhibitor at 1μM final concentration, before protein extraction for Western blots, to analyze the expression of BCR-ABL.
Embryoid body assays
For Embryoid Body (EB) formation, ES and iPS cells, at day 6–7 after cell passage, were treated with 1 mg/ml Collagenase type IV for 20 minutes (Gibco Life Technologies). Clumps were cultured in Iscove’s modified Dulbecco’s medium (IMDM, Invitrogen) supplemented with 1% penicillin/streptomycin, 1 mM L-glutamine, 15% fetal calf serum (FCS, Invitrogen), 450μM monothioglycerol, 50μg/mL ascorbic acid (Sigma Aldrich) and 200μg/L transferrin (Sigma Aldrich) and supplemented with hematopoietic cytokines: 100 ng/mL stem cell factor (SCF), 100 ng/mL fms-like tyrosine kinase 3 ligand (Flt-3 L) and 50 ng/mL thrombopoietin (TPO) (all from Peprotech). ESC and IPSC-derived-EBs were cultured in ultralow attachment 6-well plates (Costar) for 16 days. Media was changed two or three times depending on EB proliferation. All cultures were incubated at 37°C in 5% CO2. For cultures with Imatinib mesylate, cells were cultured for 16 days at 1μM before protein and RNA extraction.
Blast-colony forming assays
To obtain Bl-CFC, iPSC pellets were resuspended in Stemline II Hematopoietic Stem Cell Expansion medium (Sigma-Aldrich S0192) supplemented with 1% peni-streptomycin and L-glutamine, 50 ng/ml rhVEGF (Peprotech) and 50 ng/ml rhBMP4 (Peprotech). After 48 h, we added 20 ng/ml of each of the following growth factors including rh-SCF (Peprotech), rh-TPO (Miltenyi Biotec) and rh-Flt-3 L (Peprotech). The embryoid bodies obtained at day 3.5 were collected, dissociated with pre-heated stable trypsin replacement enzyme TrypLE Express (Gibco by life technologies ref 12605-10) and filtered using 40μm Nylon Mesh sterile cell strainer (Fisher Scientific). Cells were suspended in Stemline II Hematopoietic Stem Cell Expansion medium (Sigma-Aldrich S0192) supplemented with 1% peni/streptomycin and L-glutamine, 50 ng/ml of rh-VEGF, rh-BMP4, rh-FLT3 L and rh-TPO, 20 ng/ml of bFGF (Peprotech) and 5 Units/ml of EPO (Peprotech) and transferred in Methocult SF H4436 (Stem Cell Technologies) for 5 to 7 days.
Hematopoietic differentiation
CFC assays from ESC and iPSC-derived EB or Bl-CFC were performed by plating 104 cells/mL into MethoCult GF (H4435 StemCell Technologies) in triplicate. For testing the effects of tyrosine kinase inhibitors (TKI), the MethoCult GF were supplemented with Imatinib mesylate (1μM) or Dasatinib (5nM). Hematopoietic colonies (BFU-E, CFU-M, CFU-G, CFU-GM and CFU-Mix) were counted at day+14 according to standard morphological criteria. CFC assays were performed by the same manipulator during all clonogenic differentiation experiments.
Culture of K562 and UT7 cells
K562 cells were cultured in the RPMI medium containing 10% Fetal Calf Serum (FCS). Parental UT7 cells were also cultured in RPMI containing 10% FCS but in the presence of 10 ng/ml of rh-GM-CSF, whereas BCR-ABL-expressing UT7-11 cells were cultured without GM-CSF as they were growth-factor-independent. Cells were split 2-3 times / week, at the concentration of 105 cells / ml.
RNA and Q-RT PCR analyses
Total RNA was isolated using TRIzol (Ambion from Life Technologies) extraction protocol and treated with DNAse I (Invitrogen). cDNA was synthesized using affinity script multi temperature cDNA synthesis kit. PCR amplification was conducted with Fast start DNA polymerase (Roche) or Platinium taq (Thermofisher) using the following conditions:
Fast Start Mix 2.5μl, MgCl2 2.5 μl, Forward Primer 0.5μl,Reverse Primer 0.5μl, dNPTs 0.5μl, Taq 0.1 μl, cDNA 2μl + qsp H20 to 20 μl.
Amplification protocol included a denaturation step for 4 min at 95°C, followed by PCR 40 cycles of PCR (30 sec at 95°C, 30 sec at 65°C, 2 min at 72°C) terminated by an elongation step of 10 minutes 72°C (Agilent Technologies, Stratagene Mx3005P).
For RT-PCR-mediated BCR-ABL detection, we used the European Leukemia Network primers (BCR-F: TGACCAACTCGTGTGTGAAACTC; ABLK-F: CGCAACAAGCCCACTGTCT; ABLK-R: TCCACTTCGTCTGAGATACTGGATT) The other primers used are listed in Table 2.
Table 2
List of primers used for PCR analyses
| PU.1 | (TGCCCTATGACACGGATCTATAC/TCCAACAGGAACTGGTACAGG) |
| c-MYB | (ATGGGCAGAAATCGCAAAGC/GGTAGCTGCATGTGTGGTTC) |
| GFI.1 | (AAGTGCCACCTGGTCTCCT/CTCAAACTCCGAGCTCCGTT) |
| GFI.1b | (CAACAGCCCTGTCCTTAGCA/GGGGTGGAGAAGACCTTGTT) |
| LMO2 | (TGCAGGGCTTGCTAAGGAAT/CCTTTTTCCTGCCTCTCCACTA) |
| GATA3 | (CAGCACAGAAGGCAGGGAG/CGAGCTGTTCTTGGGGAAGT) |
| IKAROS | (GAATGGGGAAGAATGTGCGG/GGCAGCGCTCTTTATGTTCC) |
| CXCR4 | (GTGACTTTGAAACCCTCAGCG/GAGCCCATTTCCTCGGTGTA) |
Amplification products were run in a 1% or 0,8% agarose gels and detected with the GenoView UV Transilluminator. An Unpaired t.test with Welch’s correction was performed from Prism software.
Phenotypic analysis of hematopoietic progenitors
Hematopoietic cells were obtained using both via EBs and Bl-CFC generation. To analyze hematopoietic cells generated via EB differentiation, cells were dissociated with pre-warmed stable trypsin replacement enzyme TrypLE Express (Gibco Life Technologies) and filtered with 70μm Nylon Mesh sterile cell strainer (Fisher Scientific). Living cells were counted using trypan blue and stained.
To analyze the hematopoietic potential of Bl-CFC, we used colonies obtained at day +5 of differentiation. Day +5 Bl-CFC on methylcellulose were washed 3 times with PBS. The pellet was resuspended in Iscove’s MDM medium and filtered with 70μm Nylon Mesh sterile cell strainer (Fisher Scientific). Living cells were counted in trypan blue and stained with the following antibodies in PBS 4% BSA at 4°C for 45 min: CD45Pe-Vio770 (Miltenyi), CD34Vioblue (Miltenyi), CD41aPE (BD), CD43FITC (Miltenyi), CD309APC (BD), CD31FITC (Beckman Coulter), CD235aAPC (BD), CD144 FITC (Miltenyi), BB9PE (BD). After 1 h, cells were washed and resuspended in PBS 4% BSA with viability staining reagent 7-aminoactinomycin D (7-AAD, 1μg/ml, Sigma Aldrich). Stained cells were analyzed with a MACSQuant 10 (Miltenyi Biotec) flow cytometer and Flowjo analysis software.
Western Blots
Cells were lysed in ice with RIPA buffer containing NaCl (200 mM), Tris pH 8 (50 mM), Nonidet P40 (1%), acide deoxycholate (0.5%), SDS (0.05%), EDTA (2 mM) and supplemented with 100μM phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium fluoride (NaF) and 1 mM orthovanadate (Na3VO4). Separation of proteins was performed by electrophoretic migration on a 3–8% polyacrylamide gel under denaturing conditions. Proteins were then transferred in a semi-liquid condition on PVDF membrane pre-activated in methanol. After saturation with TBS-Tween 5% BSA for 1 h, membranes were and hybridized with primary antibodies (BCR-ABL Ab3 Calbiochem, pCrkL Tyr207 Cell Signaling, pTYR 4G10 Millipore) and secondary antibodies coupled to HRP. The signal was revealed by chemiluminescence with SuperSignal West Dura or Femto reagents and acquired with G:BOX-Chemi Chemiluminescence Image Capture system.
AHR experiments
They were performed with the goal of detecting the effect of SR1 addition during the generation of Bl-CFC and during the hematopoietic progenitor generation (Fig. 9). Bl-CFC assays were performed for iPSC cultures in the presence or absence of SR1 1μM.
Fig. 9
Modulation of hematopoietic potential by AHR signaling in CML iPSC-derived Bl-CFC. (A) AHR gene expression levels analyzed by qRT-PCR in CML IPSC as compared to iPSC from normal donor (PB33, CTL IPSC) and human ESC line H1. (B) Schematic representation of the experimental approach. Upper panel: iPSC were used to generate Bl-CFC in the presence or in the absence of AHR antagonist SR1 for 5 days. Bl-CFCs generated in either way were then used to generate hematopoietic cells in the presence or in the absence of SR1. The same experiments were then performed by the use of FICZ, which is the agonist of AHR. Lower panel: Evaluation of the effect of AHR antagonist SR1 or AHR agonist FICZ in the generation of CML Bl-CFC. (C) Impact of the AHR antagonist SR1 on the generation of CD45+ hematopoietic cells according to the presence or absence of SR1 during Bl-CFC cultures (see text).
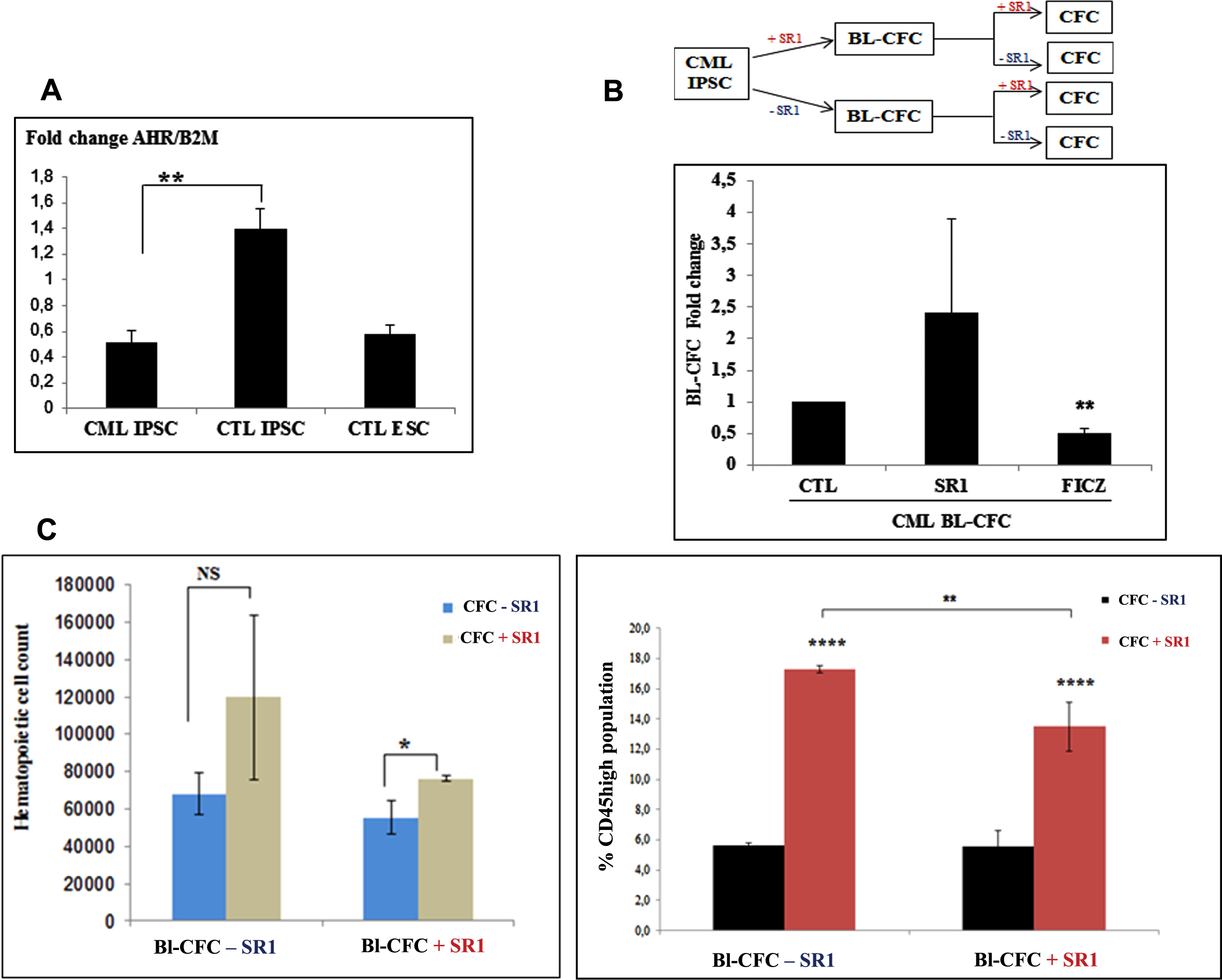
At day + 3.5 of Bl-CFC differentiation, cells were dissociated and divided in 3 separate conditions for CFC assays in MethoCult, including control, sample supplemented with 1μM SR1 (Sigma Aldrich) or with 100nM FICZ (Sigma Aldrich) which is an Aryl Hydrocarbon Receptor agonist, as opposed to SR1 which is the antagonist of AHR. After 14 days of culture, methylcellulose was collected from the culture dishes, washed by centrifugation and cells obtained were counted using Trypan blue.
Statistical analyses
Unpaired t.test and Two way ANOVA statistical analysis were performed with PRISM software from at least 3 experiments.
RESULTS
BCR-ABL expression in CML iPSC
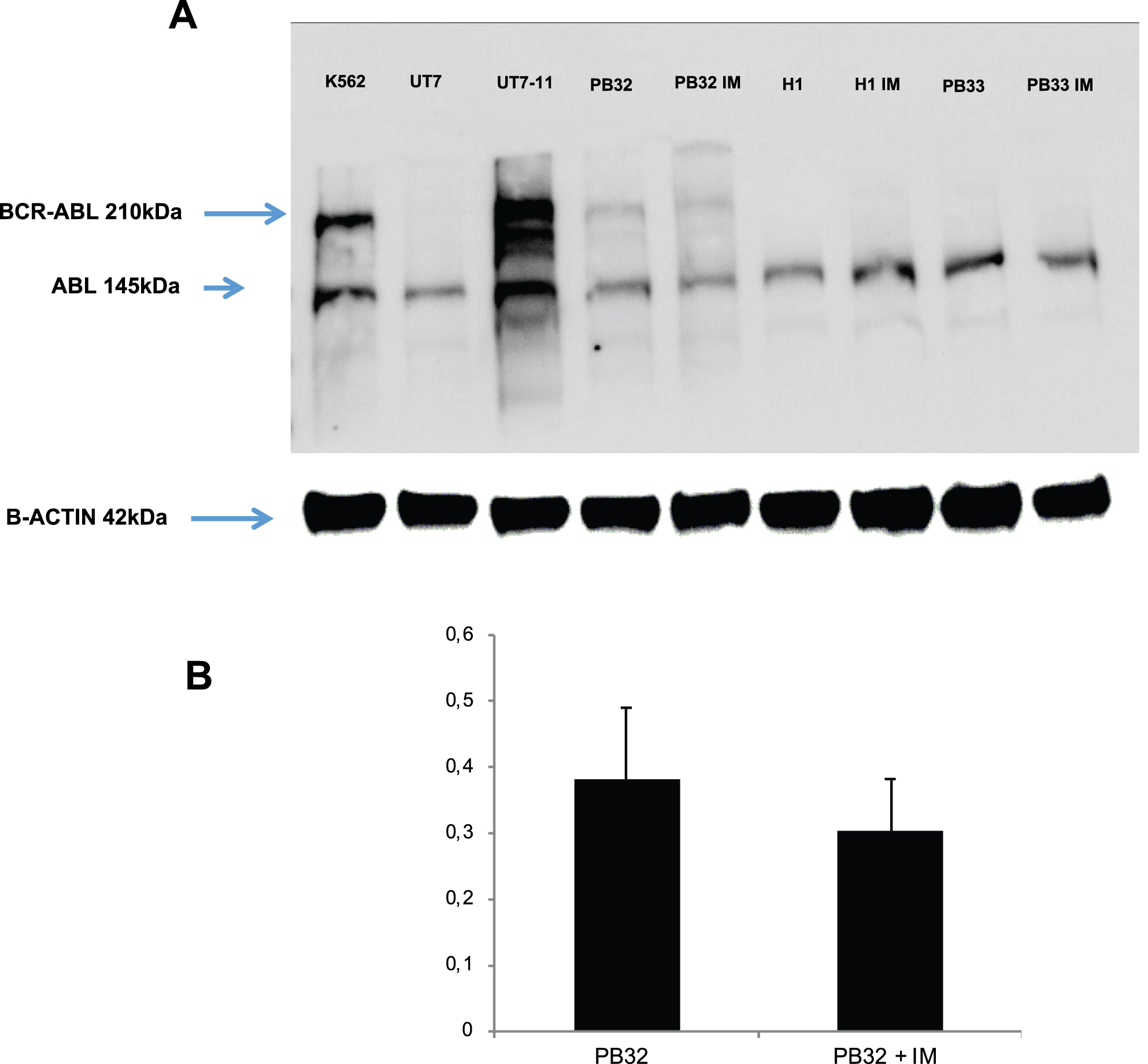
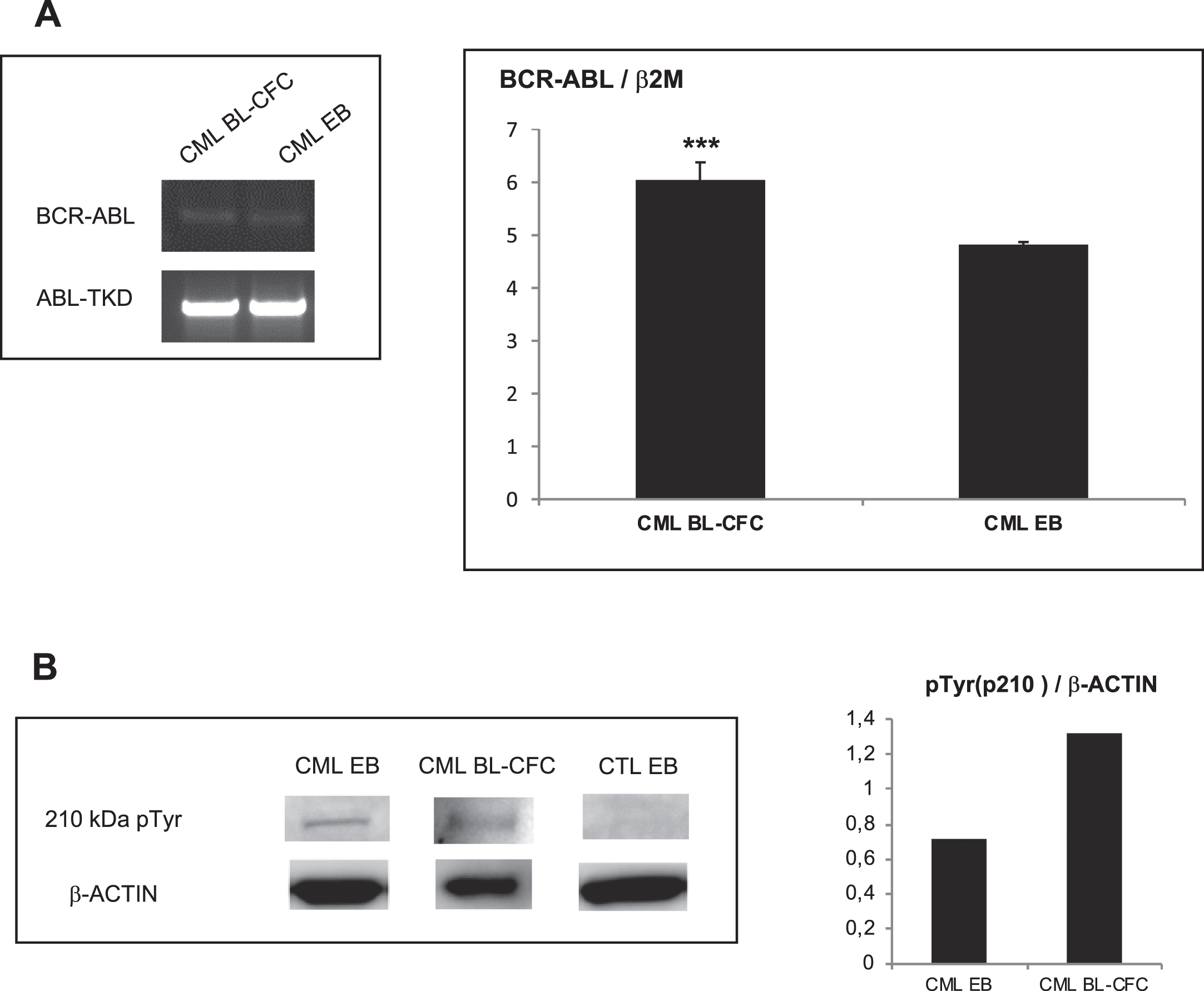
BCR-ABL expression was analyzed in CML IPSC at undifferentiated pluripotent stage by first with Western blots in the presence of appropriate controls which included iPSC line PB33 (Control iPSC without Ph1 chromosome), K562 cells, the UT7 cell line and its counterpart expressing BCR-ABL (UT7-11) as well as hESC line H1. K562 cells were acquired from ATCC whereas the UT7-11 cell line has been generated in our laboratory as previously described [11] by transduction of BCR-ABL oncogene into the GM-CSF-dependent UT7 line provided by Dr Komatsu’s laboratory [12]. Both K562 and UT7-11 cells were used as positive controls for BCR-ABL expression. As can be seen in Fig. 1A, BCR-ABL was not detected in control embryonic stem cell line H1, IPS cell line PB33 and the parental UT7 cell line (Fig. 1A). On the other hand, the CML iPSC PB32 cells expressed BCR-ABL protein but at much lower levels as compared to K562 and UT7-11 cell lines expressing BCR-ABL (Fig. 1A). The analysis performed after Imatinib Mesylate (IM) treatment did not reveal any difference in protein expression. A quantification of BCR-ABL gene expression levels with and without IM in CML-IPSC showed no significant difference (Fig. 1B).
Fig. 1
(A) Western blot analysis for the evaluation of BCR-ABL expression in CML iPSC line PB32 as compared to control cell lines. Lane 1: BCR-ABL expressing K562 cell line; Lane 2: Control UT7 cell line; lane 3.:UT7-11 cell line expressing BCR-ABL; Lane 4 and 5: CML IPSC line PB32 before and after exposure to Imatinib (PB32-IM); (lane 6 to 9): control hESC line H1 and control IPS cell line PB33 before and after exposure to imatinib. (B) Expression of BCR-ABL in CML IPS PB32 by RT-q-PCR before and after exposure to Imatinib.
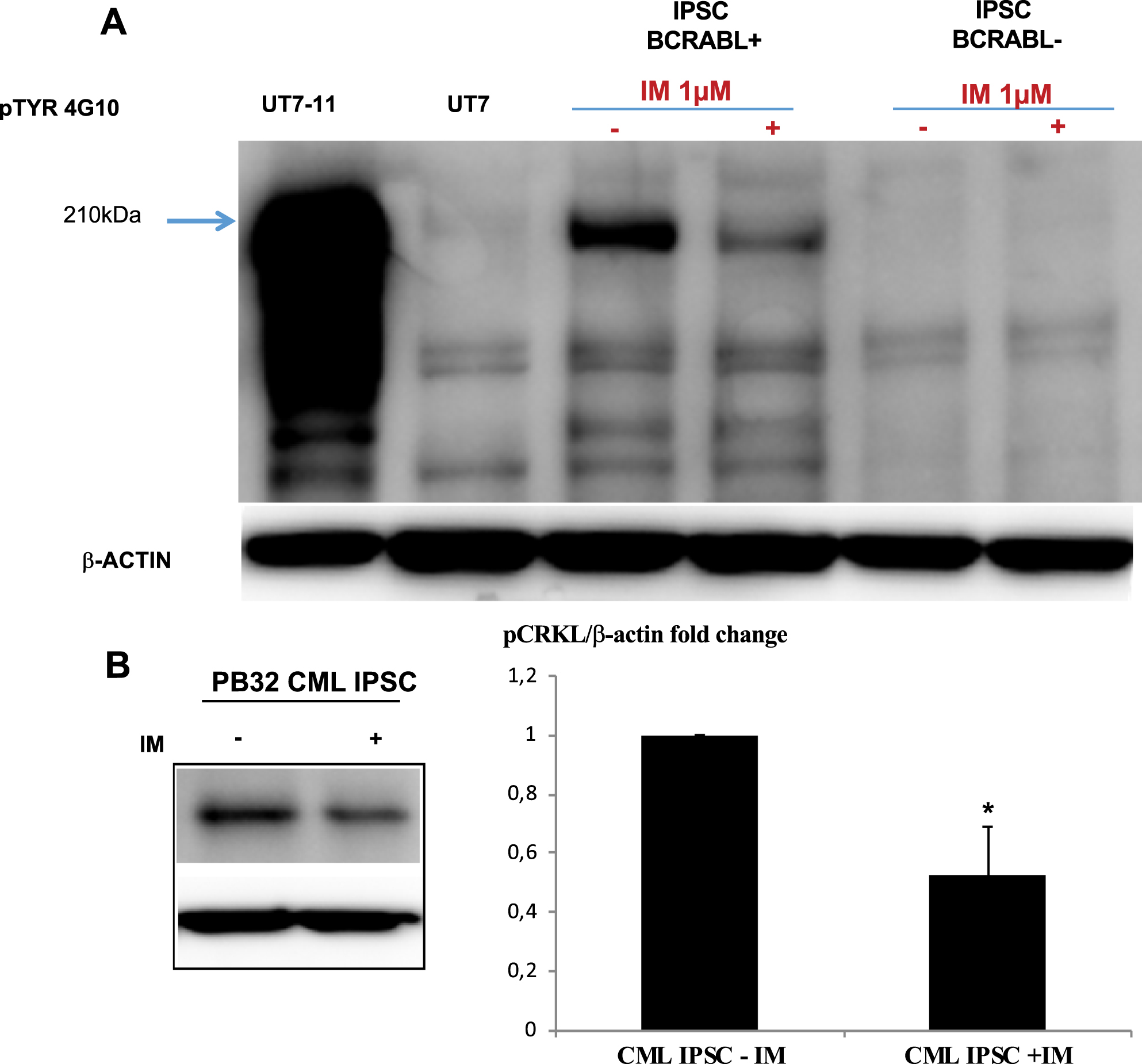
BCR-ABL signaling in CML iPSC
The next question we asked was to determine if at the pluripotent stage, BCR-ABL-expressing CML iPSC were able to generate a tyrosine kinase activity, by either autophosphorylation or by phosphorylation of the Crk-L protein. To evaluate this hypothesis, we used Western blots using the pTYR4G10 phosphotyrosine antibody. As can be seen in Fig. 2A, PB32 cells showed a highly phosphorylated BCR-ABL p210 protein as compared to control IPSC. Furthermore, this phosphorylation level corresponding to p210 BCR-ABL was decreased after imatinib treatment suggesting the detection of an autophosphorylated BCR-ABL in PB32 cells (Fig. 2A). We then looked for the phosphorylation of the surrogate BCR-ABL targets CrkL. We used an antibody against its phosphorylated form in CML-IPSC (4 independent experiments). As can be seen in Fig. 2B, Tyrosine phosphorylated CrkL proteins were present in CML-IPSC and the level of them was significantly decreased after imatinib treatment (Mann-Whitney test p-value = 0.03). We confirmed this observation with ImageJ quantification relative to the level of β-Actin (Fig. 2B). These results therefore suggested that BCR-ABL signaling is preserved in CML iPSC, at least as assessed by its phosphotyrosine generation potential.
Fig. 2
Evaluation of BCR-ABL phosphorylation in CML iPSC line PB32. (A) Evaluation of BCR-ABL phosphorylation levels by Western blot analysis using a phospho-tyrosine antibody (4G10 clone). Lane 1 and 2: BCR-ABL-positive cell line UT7-11 and its negative counterpart control UT7; Lane 3 and 4: PB32 CML iPSC with and without Imatinib; Lane 5 and 6: A control iPSC line without Ph1 chromosome, cultured with and without Imatinib. (B) ImageJ quantification of pCRKL protein in CML IPS cell line with and without Imatinib.
CML-derived iPSC generate high numbers of blast CFC in vitro
Bl-CFC, generated according to the technique previously published (Lu et al., 2007) were quantified in the methylcellulose from day 5 to day 7. At day 5, we observed different types of small, either tight or diffuse colonies comprised of uniformly round cells corresponding top Bl-CFC (Fig. 3B). As compared to H1 control, we observed a 4-fold increase of the Bl-CFC clonogenic potential in the IPSC derived from CML (unpaired t.test with Welch’s correction p = 0.0037) in > 3 independent experiments in duplicate) (Fig. 3A). The control human IPS cell line PB33 failed to generate Bl-CFC in most experiments.
Fig. 3
Impact of BCR-ABL expression in hematopoietic potential of CML IPS derived Bl-CFC. (A) Blast colony forming cells (Bl-CFC) derived from control embryonic stem cell line H1 and PB32 CML iPSC. (B) Representative photos of Bl-CFC. (C) Flow cytometry analysis of hematopoietic markers CD34 and CD45 in Bl-CFC derived from control ESC (H1) and PB32 CML iPSC.
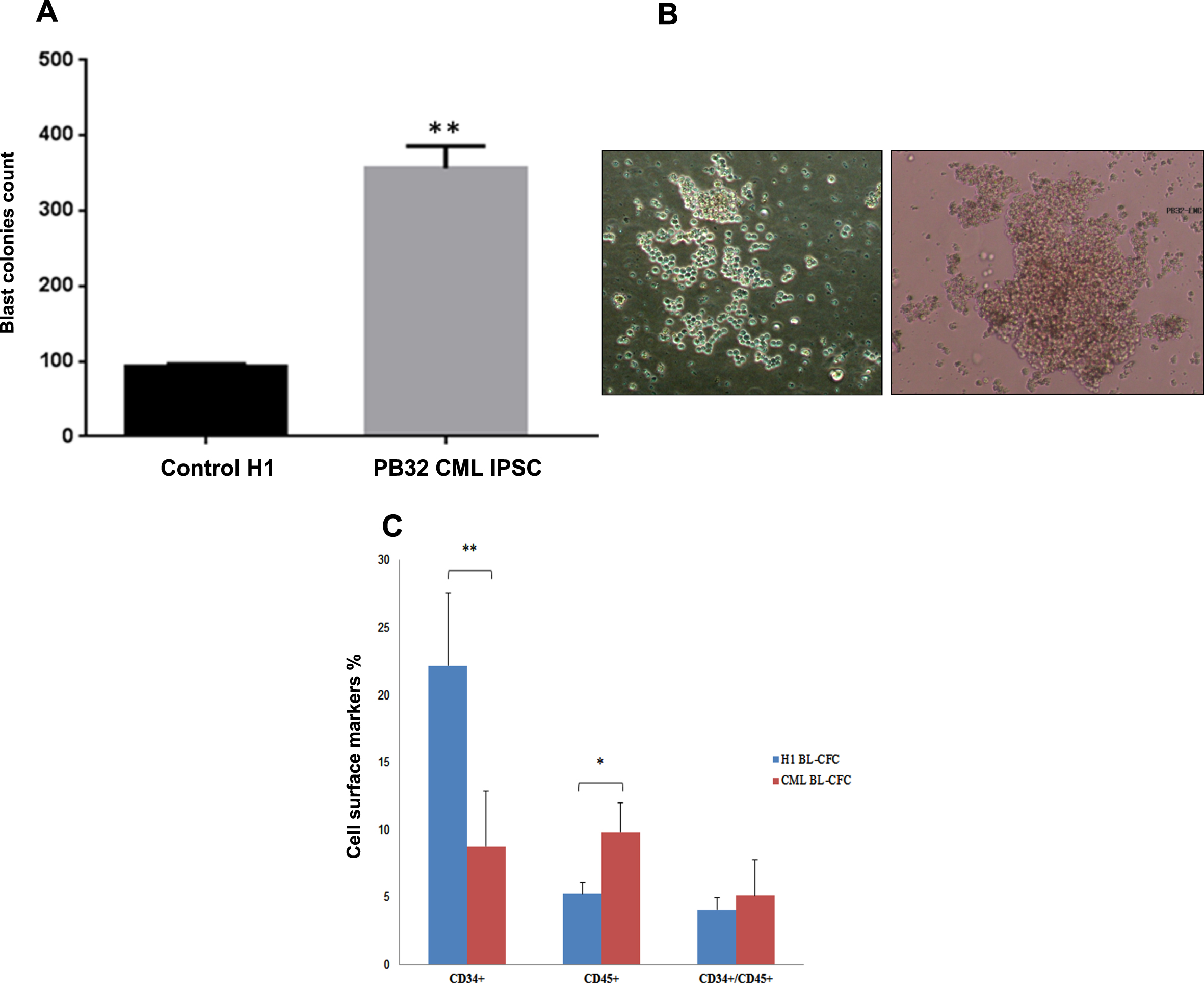
To characterize these blast cell colonies, we performed FACS analysis after collecting day 5 Bl-CFC from methylcellulose cultures (n = 4 experiments). As compared to Bl-CFC generated from H1-ESC, Bl-CFC derived from CML-IPSC expressed significantly less CD34 marker (mean % of 9% versus 22% in H1 p = 0.0069) and significantly more CD45 (10% vs 5%, respectively (p = 0.0156) (Fig. 3C). These results suggested that CML Bl-CFC had higher proliferative potential and were more committed to hematopoietic lineage.
In addition, FACS analyses of CML-Bl-CFC showed expression of both hematopoietic and endothelial markers (supp Figure 1). These cells co-expressed CD309, VEGF-R, CD34, as well as CD31. Cells co-expressing the VE-cadherin (CD144) with CD309, BB9 or CD34 were also present. In addition, a clear hematopoietic nature of these cells was identified by the generation of CD45+ cells expressing CD43, CD34, were detected.
We also showed that CML Bl-CFC expresses IL1RAP which has been shown to be expressed in the primitive CML stem cells [13]. IL1-RAP+ cells co-expressed CD45.
Bl-CFC and embryoid body-mediated hematopoietic clonogenic cell potential is increased in CML-iPSC
We performed hematopoietic differentiation using CFU assay from Bl-CFC or from EB (n = 3 experiments) (Fig. 4). These experiments showed a major and reproducible increase of CFC number EB and Bl-CFC generated from PB32 as compared to H1cell line, which is a reference for its hematopoietic potential [14]. As a matter of fact, seeding of 10x10∧3 CML Bl-CFC, generated approximately 5 times more hematopoietic colonies of all types (Fig. 4A-B) as compared to those produced from H1 Bl-CFC. Similarly, seeding of PB32 EB’s generated 5 to 8 times more hematopoietic colonies as compared to control EB, this increase being highly significant (ANOVA test with multiple comparison on PRISM software p < 0.0001)
Fig. 4
Hematopoietic differentiation from Bl-CFC and EB derived from PB32 CML iPSC. (A) Colony forming cell (CFC) count after seeding 104 control cells from CML Bl-CFC or EB. (B) Photos of hematopoietic cell colonies.
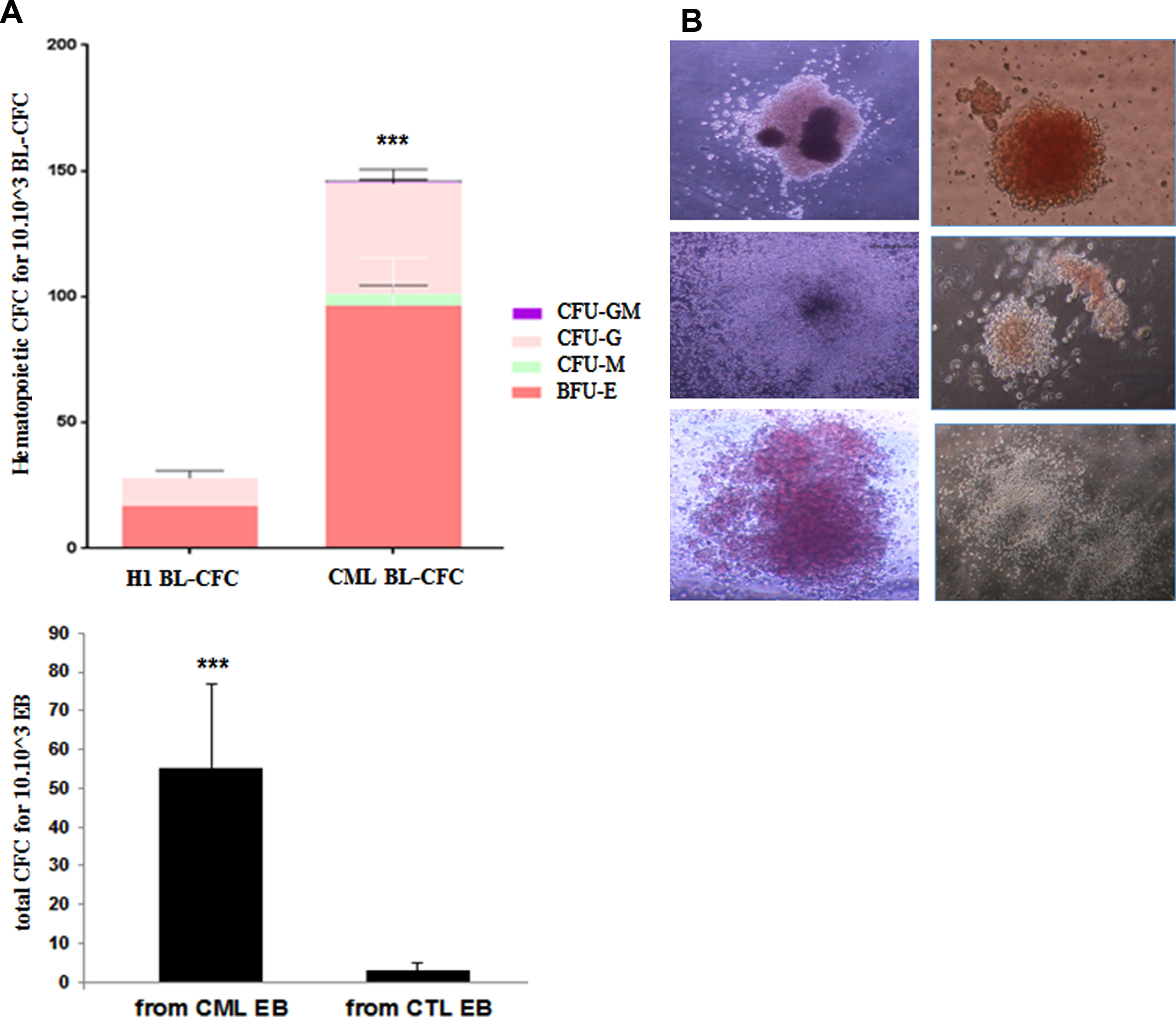
One interesting finding was the fact that we have observed constantly a higher hematopoietic potential when Bl-CFC were used as compared to EB-derived hematopoiesis (Fig. 4A)
Evaluation of BCR-ABL expression in progenitor cells
As we have generated Bl-CFC and EB-derived hematopoietic cells from the iPSC expressing BCR-ABL as described above, the next question was to determine if these primitive cells expressed also BCR-ABL after differentiation. These experiments showed the expression of BCR-ABL mRNA in both Bl-CFC and EB (Fig. 5A). This expression was also confirmed at the protein level using Western blots (Fig. 5B). CML-IPSC-derived Bl-CFC expressed significantly more BCR-ABL mRNA than the amount observed in EB (unpaired t.test with Welch’s correction p = 0.0006) (Fig. 5A).
Fig. 5
BCR-ABL expression in embryoid bodies (EBs) and Bl-CFC cells derived from CML iPSC. 5A (Left panel): PCR analysis to detect BCR-ABL in CML iPSC-derived blast colony forming cells (Bl-CFC) and embryoid bodies (EB). 5A (Right panel): Expression profile of BCR-ABL gene by RT-qPCR in CML iPSC-derived Bl-CFC and EB. 5B (Left panel): Western blot analysis to detect BCR-ABL phosphorylation level using a phospho-tyrosine antibody. 5B (Right panel): ImageJ quantification of pTYR in iPSC-derived EB and Bl-CFC.
The use of P-Tyr antibodies showed also a higher expression of phosphorylated BCR-ABL, in CML Bl-CFC (Fig. 5B).
Evaluation of the integrity of the BCR-ABL signaling at the level of embryoid bodies
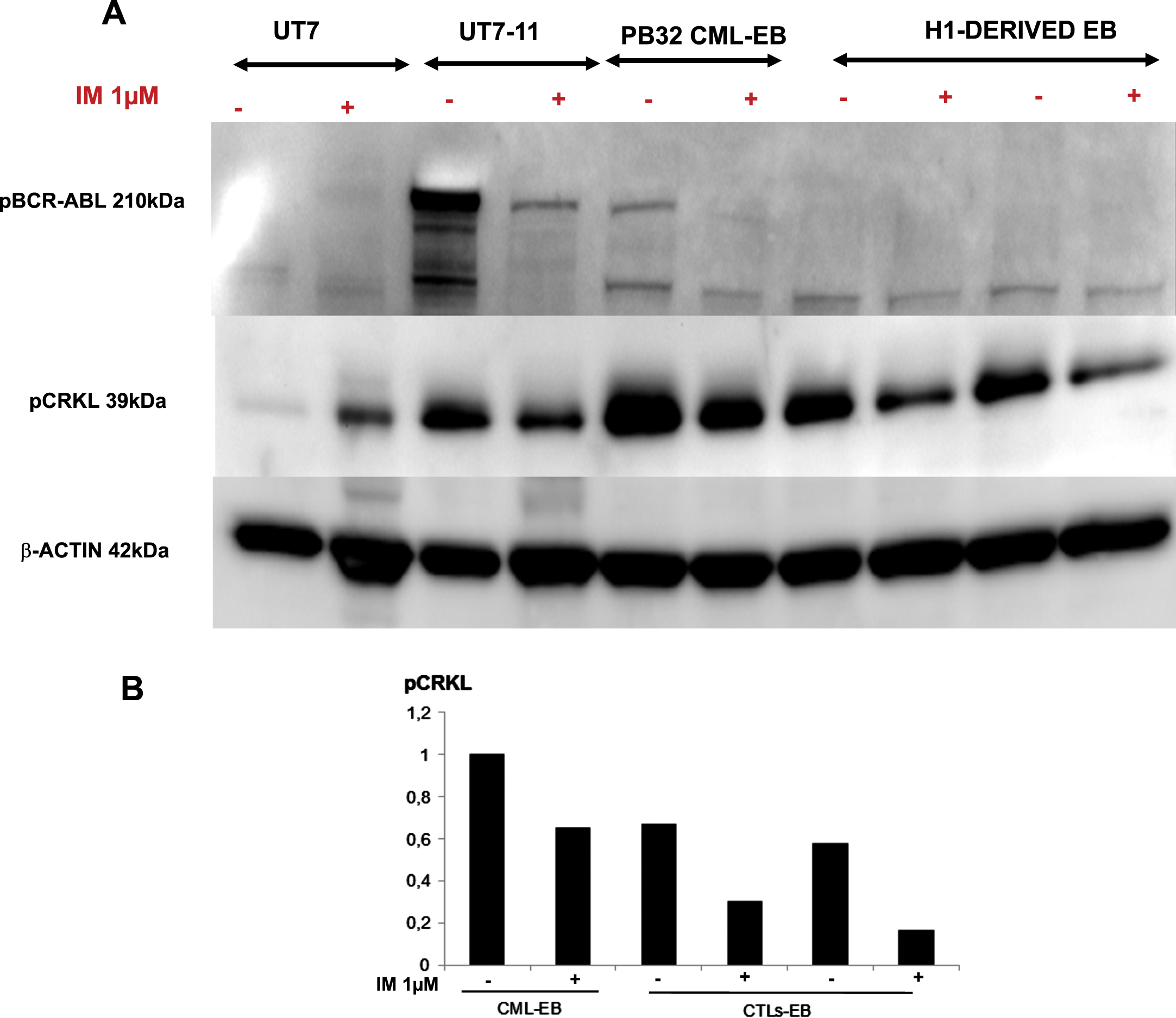
The next question we have addressed was to determine if at the stage of EB’s where hematopoietic commitment occurs [15], the tyrosine kinase activity of BCR-ABL was sufficiently active to phosphorylate the surrogate marker Crk-L. To determine this event, we first evaluated using Western blots the detection of phosphorylated forms of BCR-ABL (pBCR-ABL) using western blot analysis. As shown in Fig. 6A, the phosphorylated p210 band was detected only in the PB32-CML EBs as well as in the control BCR-ABL-expressing cell line UT7-11 (Fig. 6A). The use of Imatinib induced a major decrease of pBCR-ABL band in PB32-CML EBs (Fig. 6A, lane 6). Similarly, phospho-CrkL expression was highly increased in PB-32-CML EB protein extracts, (Fig. 6A, lane 5) and it was reduced upon IM treatment of the cells (Fig. 6A, Lane 6 and Fig. 6B). Interestingly, Imatinib also induced a decrease of CrkL phosphorylation in CTLs EB (Fig. 6A, Lanes 8 and 10 and Fig. 6B).
Fig. 6
Western blot analyses to evaluate BCR-ABL phosphorylation levels in PB32-CML-iPSC and control cells. (A) Western blot analysis of BCR-ABL phosphorylated protein and Crk-L phosphorylation in CML and control EBs before and after exposure to 1μM Imatinib compared to BCR-ABL negative and and BCR-ABL-positive control cell lines UT7 and UT7/11, respectively. (B) ImageJ quantification of pCrk-L signal compared to β-Actin.
Evaluation of the sensitivity of CML-iPSC-derived progenitors to tyrosine kinase inhibitors
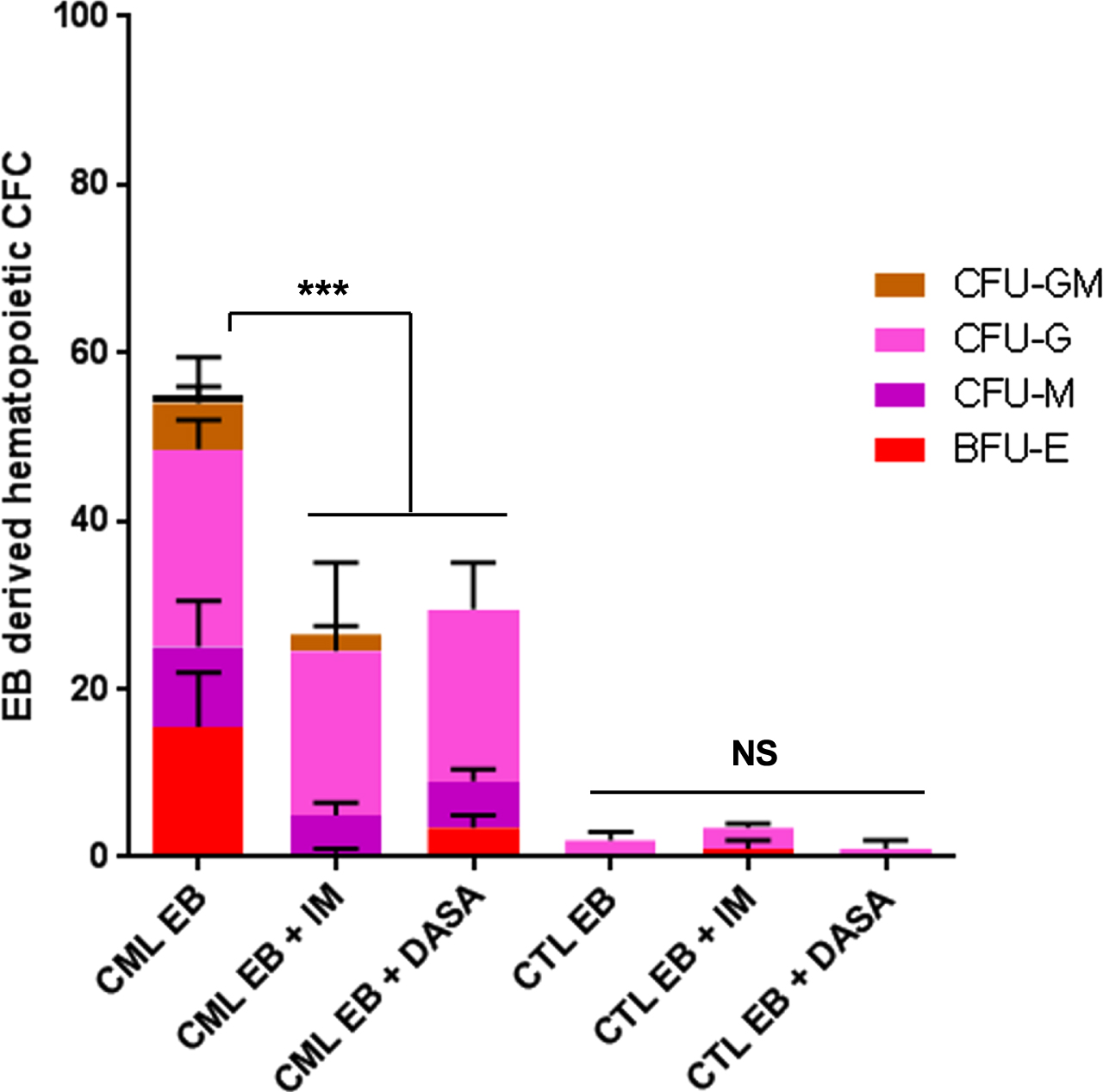
As BCR-ABL was functional in our CML-iPSC at different stages of differentiation, we next asked whether we could determine if TKI could influence the growth of hematopoietic progenitors. We have then performed CFC assays in the presence or in the absence of two tyrosine kinase inhibitors (TKI), Imatinib and Dasatinib. As can be seen in Fig. 7, the numbers of PB32-CML-EB derived CFC were highly increased as compared to CFC numbers derived from control EBs (Fig. 7). The use of IM and Dasatinib reduced the CFC growth by approximately 50% (n = 3) and this inhibition was highly significant (ANOVA p = 0.002) (Fig. 7).
Fig. 7
TKI sensitivity of PB32 CML iPSC and control hIPSC-derived hematopoietic cells. Hematopoietic cell colonies (CFC) counts from CML and EB derived from control hIPSC (PB33) in the presence or in the absence of 1μM Imatinib or 5nM Dasatinib. NS: Not significant.
Further characterization of CML-iPSC derived hematopoietic cells by analysis of the expression of genes involved in hematopoietic development
Hematopoietic commitment in adult is under the control of a well-defined set of genes. We analyzed the expression of some of these genes during differentiation from IPSC to Bl-CFC or EB derived hematopoietic progenitors (Fig. 8 gene expression and Table 1). As shown in Fig. 8, the expression of PU.1, Gfi.1 and C-myb was found to be increased in Bl-CFC compared to EB. Furthermore, Ikaros and GATA3 which are usually expressed in T lymphocyte lineage, were also found to be highly expressed in hESC (H1)-derived and PB32-CML derived Bl-CFC. LMO2 was expressed both in CML and control Bl-CFC albeit at lower levels in the latter (Fig. 8). CXCR4 expression as found to be downregulated in CML Bl-CFC or EB (Fig. 8).
Fig. 8
Hematopoietic gene expression profile. RT-PCR analysis of hematopoietic genes involved in hematopoietic development and differentiation. Lanes 1: Human ESC cell line (H1). Lane 2: PB32 CML iPSC; Lane 3: Normal control PBMC; Lane 4 and 5: BL-CFC derived from H1 cell line or PB32 CML IPSC; Lane 6 and 7: EB derived from H1 cell line and CML IPSC; Lane 8: Bl-CFC derived from a control iPSC (PB33) derived from a normal donor.
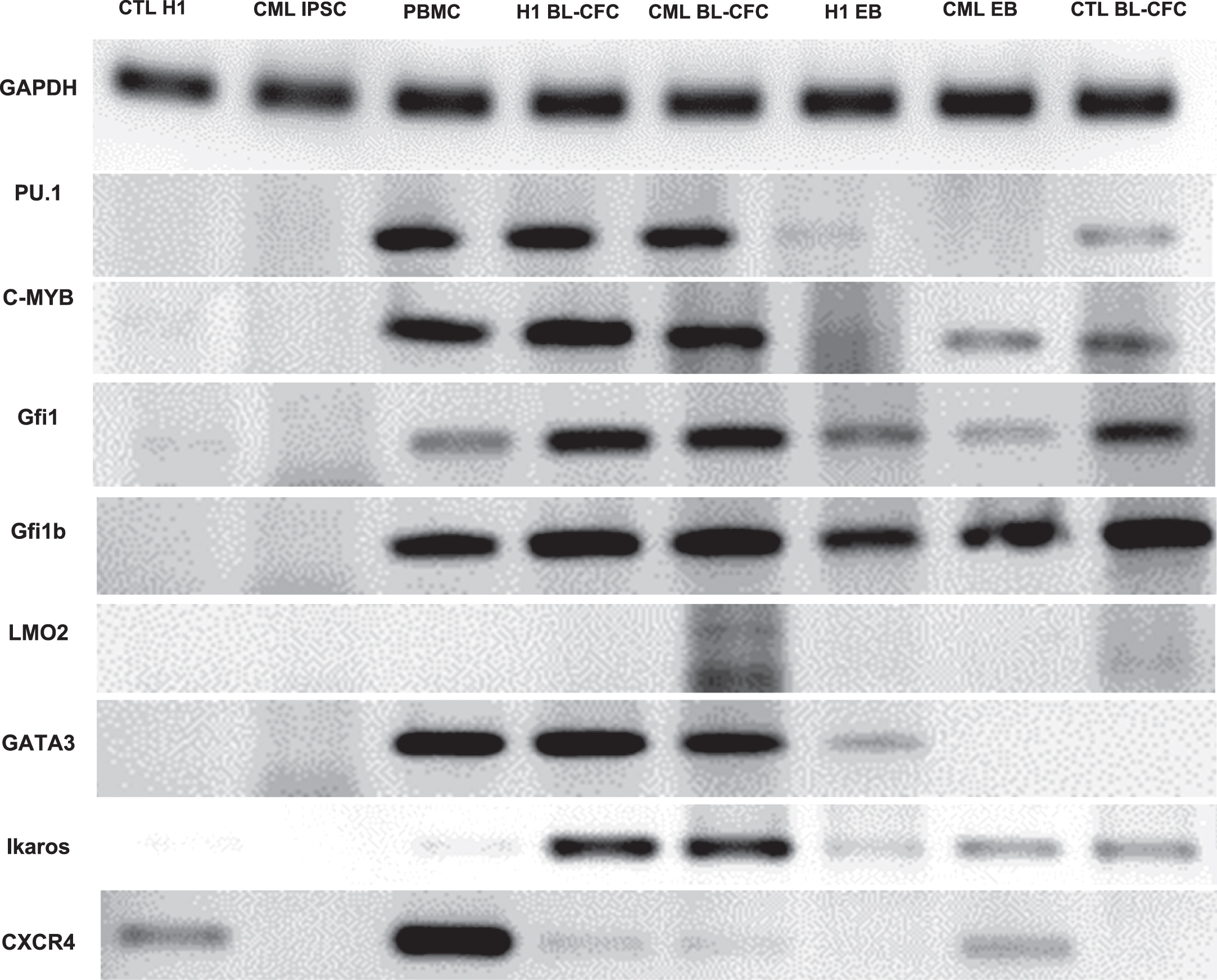
Table 1
Expression of selected genes involved in hematopoietic development and differentiation
| H1 IPS | PB32 | PBMC IPS | BL-CFC from H1 | BL-CFC from PB32 | EB from H1 | EB from PB32 | BL-CFC from PB33 | |
| LM02 | – | – | – | – | ++ | – | – | +/– |
| GFI1 | – | – | +/– | ++ | ++ | + | +/– | ++ |
| GFI1b | – | – | ++ | ++ | ++ | + | + | ++ |
| PU.1 | – | – | ++ | ++ | ++ | +/– | – | + |
| GATA3 | – | – | ++ | ++ | ++ | +/– | – | – |
| C-myb | – | – | ++ | ++ | ++ | +/– | + | + |
| Ikaros | – | – | +/– | ++ | ++ | +/– | + | + |
| CXCR4 | + | – | ++ | +/– | +/– | – | + | – |
| BCR-ABL | – | + | – | – | + | – | + | – |
H1: human ES cell line; PBMC: Peripheral blood mononuclear cells from normal blood. PB33: Control iPSC from a normal donor (PB33).
Evaluation of AHR expression in CML and control pluripotent cells
Aryl hydrocarbon receptor signaling has been shown to be involved in hematopoietic cell cycle and particularly HSC quiescence [16]. We have previously showed that AHR expression is downregulated in primary CML cells, potentially contributing to the myeloproliferative phenotype of CML [17]. We have therefore analyzed the expression of AHR in CML iPSC as compared to control iPSC. As can be seen in Fig. 9A, CML iPSC expressed significantly lower levels of AHR mRNA as compared to control iPSC but this level was equivalent to that observed in hESC line H1 (n = 3, p = 0.0095) (Fig. 9A).
Evaluation of the effects of AHR antagonism
Inhibition of AHR signaling by the use of AHR antagonist StemRegenin (SR1) has been shown to lead to an expansion of normal [18] as well as CML progenitors [17]. To determine the effects of AHR antagonism in the context of CML iPSC expressing high levels of AHR mRNA, we have performed experiments at two different stages of differentiation, as described in Fig. 9B. To this purpose, we generated Bl-CFC either in the presence or in the absence of SR1 added to the cultures during this culture period (Fig. 9B). From Bl-CFC generated in the absence or in the presence of SR1, we have then induced hematopoietic differentiation using CFC assays. During this clonogenic assays, we also tested the effects of adding or not adding of SR1 in methylcellulose cultures.
We first analyzed the effects of SR1 on the Bl-CFC potential of PB32 CML iPSC. As can be seen in Fig. 9B, there was an increase of Bl-CFC numbers when SR1 was added to the cultures during Bl-CFC generation (Fig. 9B). We next analyzed the potential of the Bl-CFC generated using SR1 towards hematopoietic differentiation. Quantitative and qualitative analyses of hematopoietic cells generated in these cultures in the absence or in the presence of SR1 is shown in Fig. 9C. For these experiments cells were collected from methylcellulose cultures at day +14, counted and washed before staining with a CD45 antibody.
As can be seen in Fig. 9C, there was a significant increase of CD45+ hematopoietic cell proliferation only when SR1 was added in CFC cultures without requirement of SR1 addition during Bl-CFC generation (n = 3 Experiments) (Fig. 9C). After phenotypic characterization, we also showed that SR1 addition yielded increased numbers of hematopoietic cells expressing CD45 + CD33+ (Suppl Figure 2). Cells with CD45 + CD31+ markers were also increased by AHR signaling inhibition (Suppl Figure 2).
DISCUSSION
The elusive nature of the most primitive CML stem cells which are very difficult to study in human samples stimulated a major research during the last 10 years to generate models of CML stem cells [19]. As matter of fact, primary CML stem cells which are very difficult to isolate and expand, are at the origin of relapses upon discontinuation of TKI therapies [20–24]. Several murine CML models have been developed to this purpose but they do not recapitulate faithfully the most primitive human CML cells. The use of embryonic stem cell models is of interest as the overexpression of BCR-ABL has been shown to be a major stimulus for generation of long-term leukemic hematopoiesis from murine ES cells [25–27]. However, one of the limitations of the ES models is the need to overexpress BCR-ABL under the control of different promoters potentially leading to results not relevant to the pathophysiology of human CML. With the advent of iPSC technology, it became of major interest to use human CML cells for reprogramming with the goal of reproducing the earliest stages of human CML and potentially to use them for drug screening purposes. The first work published in this field showed the feasibility of reprogramming the human CML cell line KBM to pluripotency [28] with the possibility of generating BCR-ABL-expressing CD45+ cells. Other groups used primary CD34+ leukemic cells which have been shown to be induced to pluripotency using different methodologies [5, 7]. The usefulness of this technology lies on the fact it becomes possible to obtain an unlimited number of primitive cells using the background of the primary leukemic cells in any given patient. However, experiments aiming to generate increased hematopoietic potential have been disappointing. In fact some groups showed that, CML iPSC expressed very low levels of BCR-ABL and did not generate a myeloproliferative phenotype [5, 7]. One group showed the possibility of using CML iPSC to discover a novel signaling pathway operational in human CML [8].
The iPS cell line used in the experiments reported here was generated from the primary leukemic cells of a 14-year old patient with TKI-resistant leukemia [9]. This patient had no evidence of ABL-kinase domain mutation and the reason of Imatinib resistance was not determined. The first goal of the reprogramming of the leukemic cells of the patient was to determine if BCR-ABL expression and induced signaling could be intact at different stages. We show here that BCR-ABL expression is present at the most primitive iPSC stage and BCR-ABL autophosphorylation is present in this context as it would have been expected in an adult leukemic cell context. Similarly, we show here that during the differentiation process through the stages of hemangioblasts and EB’s, BCR-ABL expression remains intact with evidence of tyrosine-phosphorylation of a target protein in both CML Bl-CFC and EB’s (Fig. 5B). Indeed, in PB-32 CML EB’s, the downstream target Crk-L was phosphorylated (Fig. 6A, 6B) and BCR-ABL autophosphorylation as well as Crk-L phosphorylation were reduced by exposure of cells to IM (Fig. 6A). This allowed us to perform TKI sensitivity experiments showing that these cells indeed respond to TKI inhibition at the level of CFC and this inhibition was seen with both IM and Dasatinib (Fig. 7). The patient sample at the origin of the iPSC was shown to be clinically resistant to IM but the sensitivity of his hematopoietic progenitors was not evaluated at the time of diagnosis.
To further characterize CML iPSC-derived hematopoiesis, we have analyzed the expression of hematopoietic genes involved in hematopoietic development at different stages of differentiation as compared to H1 ESC and adult hematopoietic cells. As can be seen in Fig. 8, CML iPSC at the pluripotent stage did not express any of the hematopoietic genes involved in hematopoietic development (PU1, c-MYB, Gfi1, Gfi1b, LMO2, GATA3, Ikaros) nor CXCR4 (Fig. 8). The expression of these genes was detected during differentiation and as compared to H1-derived EB’s, CML EB’s showed a slight increase of Gfi1b, CXCR4 and c-MYB expression (Fig. 8). At Bl-CFC stage, the expression of GATA3 was found to be higher as compared to EB stage and LMO2 was expressed both in CML and control Bl-CFC albeit at lower levels in the latter (Fig. 8). Interestingly, c-MYB has been shown to play a role in BCR-ABL-induced leukemogenesis [29].
We show here for the first time that the AHR expression, which we have reported to be downregulated in adult CML cells [17] is also downregulated in CML iPSC. It is not known at this time if this downregulation is specific to CML-iPSC, as ES cells have been shown to exhibit a downregulation of AHR under the influence of OCT4 [30]. We have used the antagonism of this pathway using SR1, to show that it is possible to further expand hematopoietic cells of the CML iPSC by this strategy. As we have shown in a previous work, data reported here suggest also that AHR pathways could represent a druggable target as the AHR agonist FICZ inhibited the Bl-CFC growth (Fig. 9B). This inhibitory effect was previously reported using primary CML stem cells [17].
In summary, we have generated and characterized a unique CML iPSC from of TKI-resistant patient’s leukemic cells. These cells partially recapitulate some features of the disease with essentially a myeloproliferative phenotype. This phenotype could be modulated by the antagonism of the AHR pathway which we have shown to be efficient in adult primary CML cells. CML-iPSC-derived hematopoietic cells express BCR-ABL with evidence of tyrosine- phosphorylation of downstream targets such as Crk-L and this phosphorylation is responsive to TKI inhibition. Finally further experiments will test the possibility of generating long-term hematopoiesis from these patient-specific CML iPSC after transplantation of iPSC-derived EB’s in immunodeficient mice.
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/STJ-210001.
STEMJOURNAL OPEN REVIEW
The evaluations from peer reviewers for this article are freely available and can be found as supplementary material here: https://dx.doi.org/10.3233/STJ-210001.
REFERENCES
[1] | Martin PJ , Najfeld V , Fialkow PJ . B-lymphoid cell involvement in chronic myelogenous leukemia: implications for the pathogenesis of the disease. Cancer Genet Cytogenet. (1982) ;6: :359–68. |
[2] | Fang B . Identification of human chronic myelogenous leukemia progenitor cells with hemangioblastic characteristics. Blood. (2005) ;105: :2733–40. |
[3] | Otten J , Schultze A , Schafhausen P , Otterstetter S , Dierlamm J , Bokemeyer C , Brummendorf TH , Fiedler W , Loges S . Blood outgrowth endothelial cells from chronic myeloid leukaemia patients are BCR/ABL1 negative. Br J Haematol. (2008) ;142: :115–8. |
[4] | Zeltner N , Studer L . Pluripotent stem cell-based disease modeling: current hurdles and future promise. Curr Opin Cell Biol. (2015) ;37: :102–10. |
[5] | Bedel A , Pasquet J-M , Lippert E , Taillepierre M , Lagarde V , Dabernat S , Dubus P , Charaf L , Beliveau F , de Verneuil H , et al. Variable behavior of iPSCs derived from CML patients for response to TKI and hematopoietic differentiation. PloS One. (2013) ;8: :e71596. |
[6] | Hu K , Yu J , Suknuntha K , Tian S , Montgomery K , Choi K-D , Stewart R , Thomson JA , Slukvin II . Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. (2011) ;117: :e109–119. |
[7] | Kumano K , Arai S , Hosoi M , Taoka K , Takayama N , Otsu M , Nagae G , Ueda K , Nakazaki K , Kamikubo Y , et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. (2012) ;119: :6234–42. |
[8] | Suknuntha K , Ishii Y , Tao L , Hu K , McIntosh BE , Yang D , Swanson S , Stewart R , Wang JYJ , Thomson J , et al. Discovery of survival factor for primitive chronic myeloid leukemia cells using induced pluripotent stem cells. Stem Cell Res. (2015) ;15: :678–93. |
[9] | Telliam G , Féraud O , Griscelli F , Opolon P , Divers D , Bennaceur-Griscelli A , Turhan AG . Generation of an induced pluripotent stem cell line from a patient with chronic myeloid leukemia (CML) resistant to targeted therapies. Stem Cell Res. (2016) ;17: :235–7. |
[10] | Bennaceur-Griscelli A , Turhan AG . Agence de Biomedecine Authorisation February 1st, 2017 (Official Journal of French Republic 20/06). |
[11] | Issaad C , Ahmed M , Novault S , Bonnet ML , Bennardo T , Varet B , Vainchenker W , Turhan AG . Biological effects induced by variable levels of BCR-ABL protein in the pluripotent hematopoietic cell line UT7. Leukemia. (2000) ;14: :662–70. doi:10.1038/sj.leu.2401730. PMID: 10764152. |
[12] | Komatsu N , Nakauchi H , Miwa A , Ishihara T , Eguchi M , Moroi M , Okada M , Sato Y , Wada H , Yawata Y , et al. Establishment and characterization of a human leukemic cell line with megakaryocytic features: dependency on granulocyte- macrophage colony-stimulating factor, interleukin 3, or erythropoietin for growth and survival. Cancer Res. (1991) ;51: :341–8. PMID: 1824823. |
[13] | Landberg N , Hansen N , Askmyr M , Ågerstam H , Lassen C , Rissler M , Hjorth-Hansen H , Mustjoki S , Järås M , Richter J , et al. IL1RAP expression as a measure of leukemic stem cell burdenat diagnosis of chronic myeloid leukemia predicts therapy outcome. Leukemia. (2016) ;30: :255–8. |
[14] | Kaufman DS , Hanson ET , Lewis RL , Auerbach R , Thomson JA . Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc Natl Acad Sci. (2001) ;98: :10716–21. |
[15] | Keller G , Kennedy M , Papayannopoulou T , Wiles MV . Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. (1993) ;13: :473–86. |
[16] | Singh KP , Casado FL , Opanashuk LA , Gasiewicz TA . The aryl hydrocarbon receptor has a normal function in the regulation of hematopoietic and other stem/progenitor cell populations. Biochem Pharmacol. (2009) ;77: :577–87. |
[17] | Gentil M , Hugues P , Desterke C , Telliam G , Sloma I , Souza LEB , Baykal S , Artus J , Griscelli F , Guerci A , Johnson-Ansah H , Foudi A , Bennaceur-Griscelli A , Turhan AG . Aryl hydrocarbon receptor (AHR) is a novel druggable pathway controlling malignant progenitor proliferation in chronic myeloid leukemia (CML). PLoS One. (2018) ;13: :e0200923. |
[18] | Boitano AE , Wang J , Romeo R , Bouchez LC , Parker AE , Sutton SE , Walker JR , Flaveny CA , Perdew GH , Denison MS , et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. (2010) ;329: :1345–8. |
[19] | Sloma I , Jiang X , Eaves AC , Eaves CJ . Insights into the stem cells of chronic myeloid leukemia. Leukemia. (2010) ;24: :1823–33. |
[20] | Lee S-E , Choi SY , Song H-Y , Kim S-H , Choi M-Y , Park JS , Kim H-J , Kim S-H , Zang DY , Oh S , et al. Imatinib withdrawal syndrome and longer duration of imatinib have a close association with a lower molecular relapse after treatment discontinuation: the KID study. Haematologica. (2016) ;101: :717–23. |
[21] | Mahon F-X , Réa D , Guilhot J , Guilhot F , Huguet F , Nicolini F , Legros L , Charbonnier A , Guerci A , Varet B , et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. (2010) ;11: :1029–35. |
[22] | Ross DM , Branford S , Seymour JF , Schwarer AP , Arthur C , Yeung DT , Dang P , Goyne JM , Slader C , Filshie RJ , et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. (2013) ;122: :515–22. |
[23] | Chomel JC , Bonnet ML , Sorel N , Bertrand A , Meunier MC , Fichelson S , Melkus M , Bennaceur-Griscelli A , Guilhot F , Turhan AG . Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood. (2011) ;118: :3657–60. |
[24] | Chomel JC , Bonnet ML , Sorel N , Sloma I , Bennaceur-Griscelli A , Rea D , Legros L , Marfaing-Koka A , Bourhis JH , Ame S , Guerci-Bresler A , Rousselot P , Turhan AG . Leukemic stem cell persistence in chronic myeloid leukemia patients in deep molecular response induced by tyrosine kinase inhibitors and the impact of therapy discontinuation. Oncotarget. (2016) ;7: :35293–301. |
[25] | Melkus M , Bennaceur-Griscelli A , Valogne Y , Flamant S , Chomel J-C , Sorel N , Bonnet M-L , Deininger MW , Mitjavila-Garcia M-T , Turhan AG . Biological effects of T315I-mutated BCR-ABL in an embryonic stem cell-derived hematopoiesis model. Exp Hematol. (2013) ;41: :335–345.e3. |
[26] | Perlingeiro RC , Kyba M , Daley GQ . Clonal analysis of differentiating embryonic stem cells reveals a hematopoietic progenitor with primitive erythroid and adult lymphoid-myeloid potential. Development. (2001) ;128: :4597–604. |
[27] | Peters DG , Klucher KM , Perlingeiro RC , Dessain SK , Koh EY , Daley GQ . Autocrine and paracrine effects of an ES-cell derived, BCR/ABL-transformed hematopoietic cell line that induces leukemia in mice. Oncogene. (2001) ;20: :2636–46. |
[28] | Carette JE , Pruszak J , Varadarajan M , Blomen VA , Gokhale S , Camargo FD , Wernig M , Jaenisch R , Brummelkamp TR . Generation of iPSCs from cultured human malignant cells. Blood. (2010) ;115: :4039–42. |
[29] | Lidonnici MR , Corradini F , Waldron T , Bender TP , Calabretta B . Requirement of c-Myb for p210BCR/ABL-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood. (2008) ;111: :4771–9. |
[30] | Kang B , Wang Y-J . Bidirectional talk between AHR and Oct4. Oncotarget. (2015) ;6: :15740–1. |




