Organoid Maturation by Circadian Entrainment
Abstract
Stem cell-derived tissues that recap endogenous physiology are key for regenerative medicine. Yet, most methods yield products that function like fetal, not adult tissues. Organoids are typically grown in constant environments, while our tissues mature along with behavioral cycles. Here, we show that inducing circadian rhythms in pancreatic islet organoids, by entraining them to daily feeding-fasting cycles, elicits their metabolic maturation. Our results show that rhythms can be harnessed to further functional maturation of organoids destined for human therapeutics.
INTRODUCTION
Islet transplantation can cure insulin-dependent diabetes, but is curbed by scarcity of acceptable islets [1]. Stem cell-derived islet organoids offer an unlimited islet supply, yet an immature physiology limits their therapeutic use [2]. To find maturation-driving mechanisms, we studied regulatory dynamics during their stepwise formation from human pluripotent stem cells [3].
MATERIALS AND METHODS
Human cell lines, primary samples, and rodent strains are detailed in [3]. We devised flow sorting strategies to purify cells at successive stages of differentiation into the islet lineage [4]. These included stem cell-derived β (SC-β), insulin+ glucagon+ polyhormonal (PH) cells, and their progenitors. Cells were subject to whole-genome bisulfite sequencing (WGBS), assay for transposase-accessible chromatin by sequencing (ATAC-seq), chromatin immunoprecipitation sequencing for two histone marks (H3K27ac and H3K4me1), and directional total RNA sequencing (RNA-seq), as detailed in [3]. Computational methods are described elsewhere [3, 5–7]. Animal studies were conducted as described in [8]. Cell culture methods and assays are detailed in [3].
RESULTS
Our studies reveal mechanisms controlling cell fate during islet organoid development and show that organoids are amenable to functional improvement by circadian modulation.
Fig.1
Regulatory dynamics during organoid formation. (A) Islet organoid formation recapitulates human islet development [3]. Pluripotent stem cells are sequentially differentiated into endoderm, pancreatic and endocrine progenitors, which generate islet cells (insulin+ β or insulin+ glucagon+ polyhormonal counterparts). (B) We devised tools for purification of live islet lineage intermediates [4]. Sorting SC-β from PH cells by flow cytometry, based on staining for the indicated markers. Typical population purities, assessed by fixing sorted cells followed by staining for C-peptide and GLP2 (byproducts of proinsulin and proglucagon processing), are shown to the right. This enabled comprehensive molecular profiling during stepwise organoid differentiation. (C) We used H3K27ac enrichment to define enhancer domains, and examined their chromatin accessibility, modification, methylation and transcription dynamics during organoid formation. Tracks show normalized ATAC, H3K4me1, H3K27ac, RNA, and WGBS signal over a region 1.5x greater than the enhancer domain shown below. Heatmaps display relative signal over the enhancer domain. The data link enhancers to the control of stage-specific gene expression.
![Regulatory dynamics during organoid formation. (A) Islet organoid formation recapitulates human islet development [3]. Pluripotent stem cells are sequentially differentiated into endoderm, pancreatic and endocrine progenitors, which generate islet cells (insulin+ β or insulin+ glucagon+ polyhormonal counterparts). (B) We devised tools for purification of live islet lineage intermediates [4]. Sorting SC-β from PH cells by flow cytometry, based on staining for the indicated markers. Typical population purities, assessed by fixing sorted cells followed by staining for C-peptide and GLP2 (byproducts of proinsulin and proglucagon processing), are shown to the right. This enabled comprehensive molecular profiling during stepwise organoid differentiation. (C) We used H3K27ac enrichment to define enhancer domains, and examined their chromatin accessibility, modification, methylation and transcription dynamics during organoid formation. Tracks show normalized ATAC, H3K4me1, H3K27ac, RNA, and WGBS signal over a region 1.5x greater than the enhancer domain shown below. Heatmaps display relative signal over the enhancer domain. The data link enhancers to the control of stage-specific gene expression.](https://content.iospress.com:443/media/stj/2020/2-1/stj-2-1-stj209001/stj-2-stj209001-g001.jpg)
Fig.2
Epigenetic priming predicts lineage potential. (A-B) The competence to execute specific cell fates can be linked to a gain of H3K4me1 before H3K27ac deposition at lineage-specific enhancers (9). We find that α-specific enhancers are selectively H3K4me1-marked in PH cells (A). Following transplantation under the kidney capsule of immunocompromised mice, PH indeed resolve toward α cells, as evidenced by graft staining for insulin (green)/glucagon (red). SC-β cells (B) show the opposite trend, as expected. Thus, epigenetic priming steers PH toward an α cell fate.
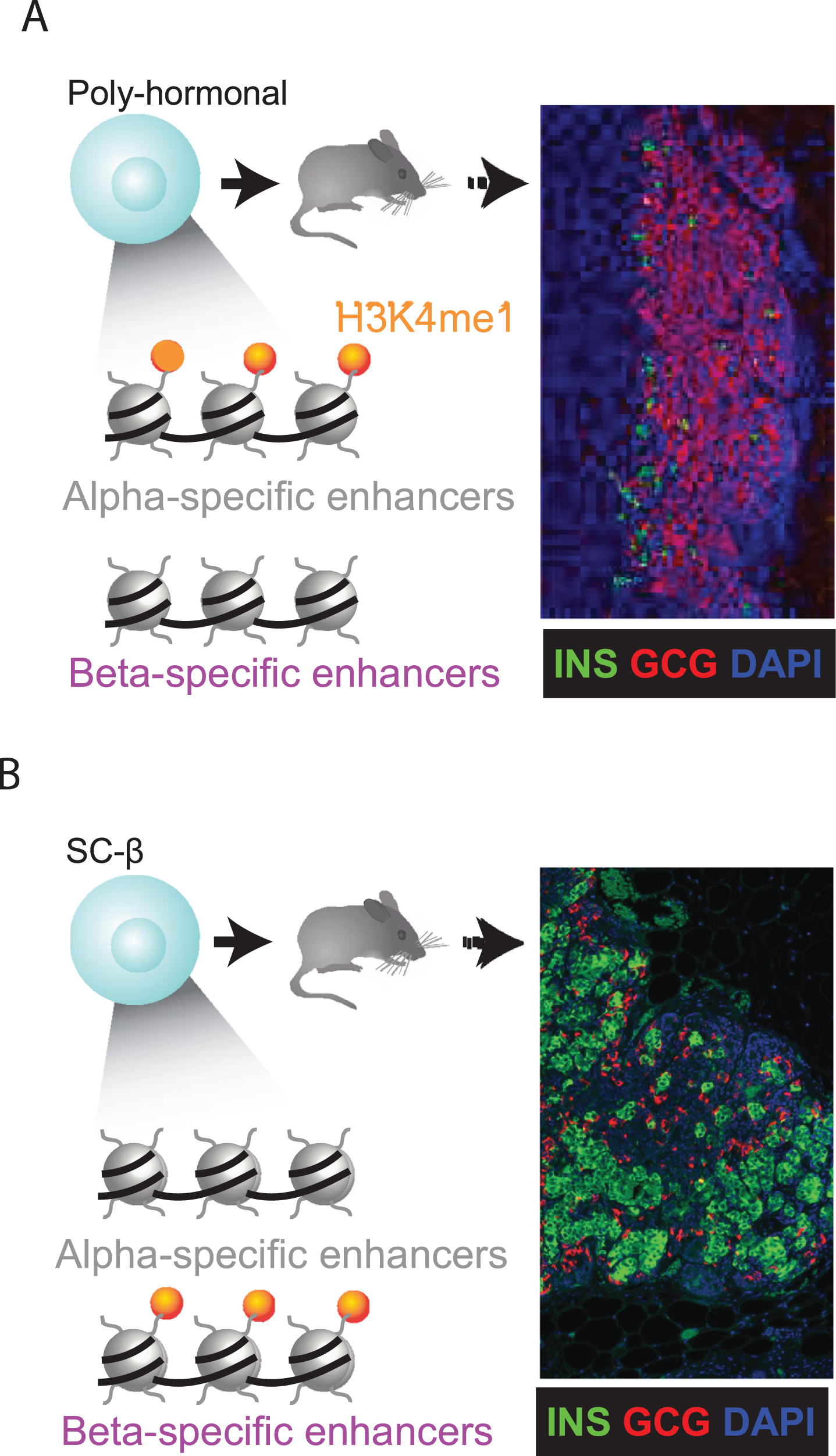
Fig.3
Organoid maturation pioneers & regulatory circuits. (A) Developmental competence is endowed by chromatin-opening pioneer factors (10). To find maturation pioneers, we examined chromatin openness over a 4-week time course when organoids develop glucose-coupled insulin secretion (center). Recognition motifs within newly opened chromatin sites (left) identify CLOCK and ARNTL/BMAL1 among the top-enriched TFs (right), casting them as pioneers during the onset and refinement of β cell function. (B) Stable cell states are set by TFs via autoregulatory loops involving joint formation of extended or super enhancers (SEs) (7). A cell’s core regulatory circuit (CRC) can thus be modeled by finding SE-driven TFs in interconnected autoregulatory loops (left). Using this logic, we generated CRC models that effectively capture known master β cell regulators (center). Nodes represent stage-specific TFs that partake (pink) or not (gray) in the β cell CRC. Edges represent predicted transcriptional regulatory relationships between TFs within the same CRC. TFs unique to β cells are highlighted in red. Ranking CRC TFs by their connectivity (right) highlights the circadian regulator DEC1/BHLHE40 as the mostinterconnected.
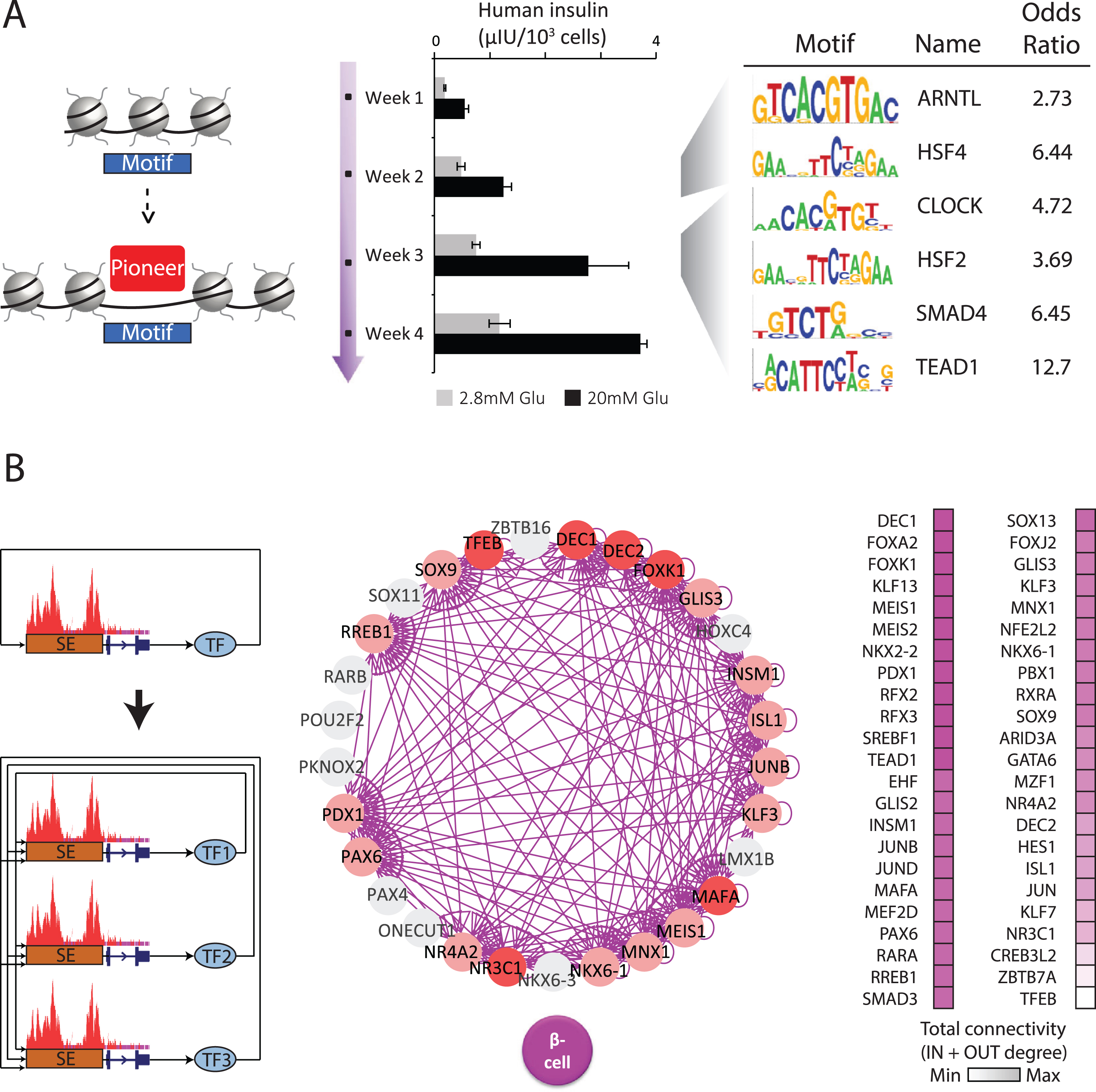
Fig.4
Circadian entrainment triggers organoid maturation. (A) We asked whether clock entrainment can foster in vitro organoid maturation. Entrainment to daily feeding/fasting rhythms using various stimuli (glucose, arginine, forskolin, or insulin) was followed by glucose-stimulated insulin secretion (GSIS) assays over 72 h (top). Following entrainment (middle), organoids gain rhythmic GSIS (red) with significantly expanded stimulation capacity relative to parallel mock-treated counterparts (gray). Oxygen levels in the medium of unstimulated cultures following entrainment also cycle (bottom), as measured by partial O2 pressure (blue). This links GSIS oscillations to metabolic rhythms. (B) Circadian entrainment enhances single-cell glucose responsiveness. Calcium staining using Fluo4-AM reveals that entrained cells (red) flux significantly more calcium in response to stimulation than most ( 75%) mock-treated counterparts (gray). Thus, enhanced responsiveness reflects new cellular states.
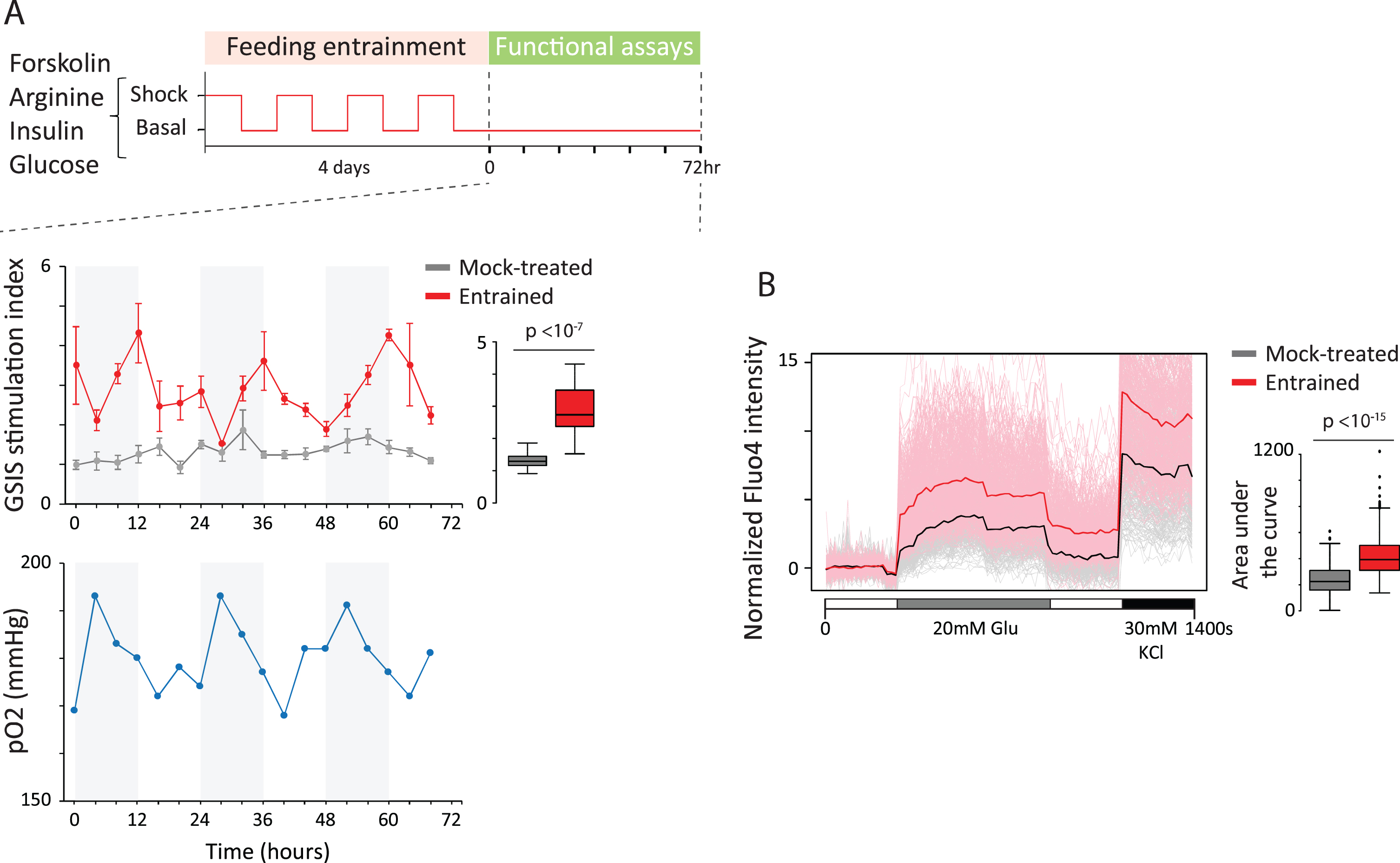
Fig.5
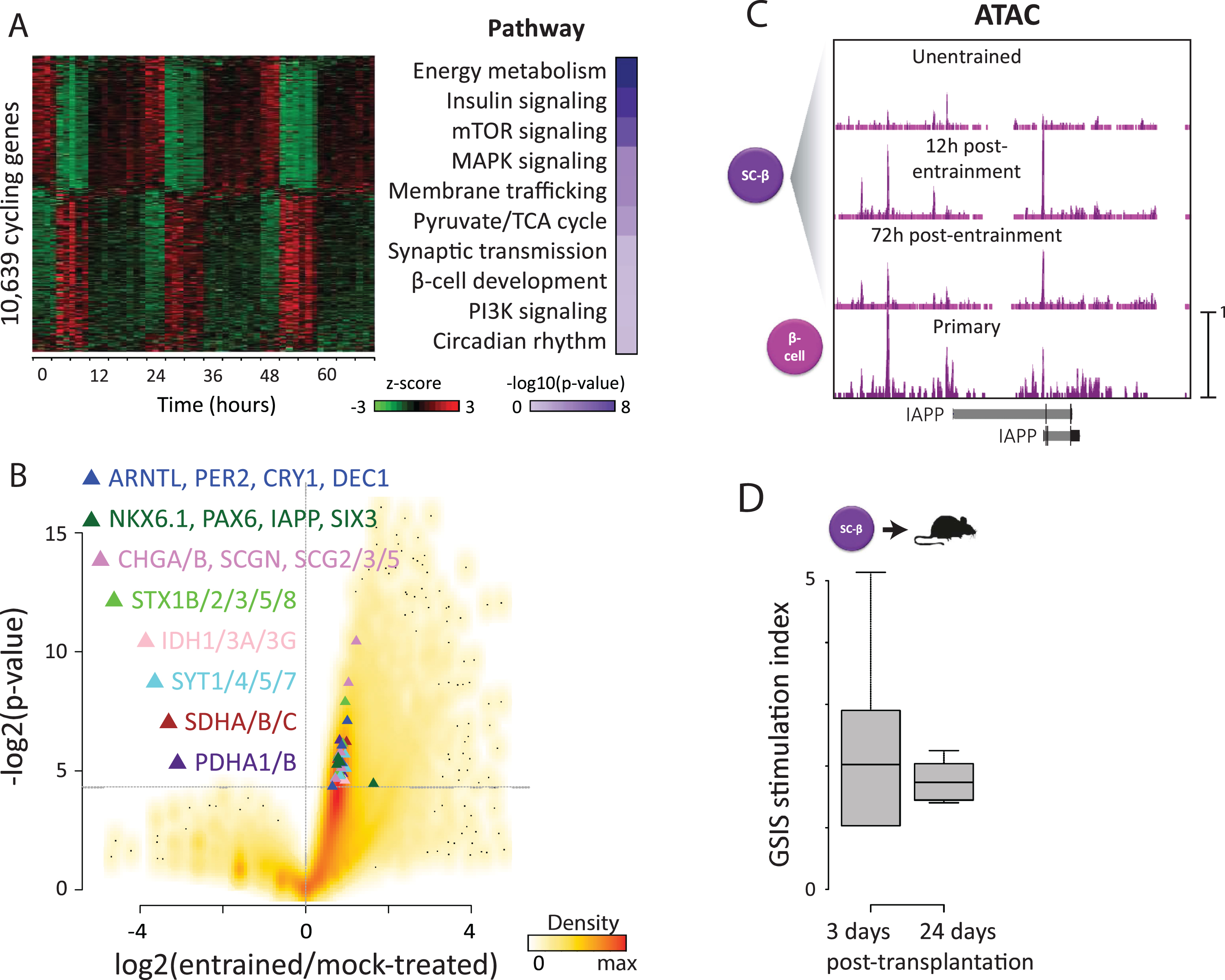
Maturity-driven genomic rhythms & in vivo function. (A) We investigated how circadian entrainment promotes organoid maturation. RNA-seq reveals >10,000 genes (31% of all detected) that oscillate (p < 0.05, harmonic regression test for rhythmicity) specifically upon entrainment (left). These enrich for functions in energy metabolism (right), consistent with metabolic rhythms. We also detect antiphasic insulin/ glucagon expression (top), recalling in vivo priming of their secretion in anticipation of diurnal/nocturnal demand. This provides a molecular basis for circadian variation in insulin responses, via rhythmic control of its synthesis, transport, and secretion. (B) Differential gene expression analysis reveals upregulation of core clock TFs (blue triangles) in entrained vs. parallel mock-treated counterparts, indicating that entrainment not only synchronizes but also activates islet organoid clocks. Entrainment also induces maturity-linked factors (green triangles) and machinery involved in energy metabolism and insulin secretion (rest of triangles), consistent with enhanced GSIS capacity. (C) To study how genomic/function rhythms persist following entrainment, we used ATAC-seq to detect thousands of newly opened chromatin sites 12 h post-entrainment, most of which remain after 72 h. These are overrepresented at maturity-linked genes such as IAPP (shown) and direct GSIS effectors, suggesting that circadian control of genes enabling mature GSIS persists as a result of stable chromatin changes. (D) We tested whether maturation in vitro leads to better function in vivo. Entrained organoids were transplanted under the kidney capsule of immunocompromised mice, and serum human insulin was assayed before/30 min after a glucose injection. Robust GSIS (>2 mIU/ml,>1.5-fold stimulation) was evident as early as 3 days post-transplant, and remained 24 days after.
DISCUSSION
This work provides a proof-of-principle that circadian control can drive functional maturation of stem cell-derived organoids destined for human therapeutics. Islet maturity develops between birth and weaning, along with onset of circadian behavioral (sleep, feeding) cycles. We show that recreating fasting/feeding cycles recapitulates metabolic maturation in islet organoids. Entrained organoids develop a capacity to anticipate diurnal changes in insulin demand, and are functional within days—rather than weeks—of transplantation. Circadian entrainment may be harnessed to further functional maturation of other stem cell-derived products, consistent with the ability of clock regulators to bind distinct targets in distinct tissues. Thus, our general approach may inform attempts to control the fate and function of any human cell type.
CONCLUSIONS
• Epigenome dynamics show how epigenetic priming steers endocrine cell fates.
• Modeling maturation regulatory circuits uncovers roles for circadian controllers.
• Circadian entrainment triggers organoid maturation via clock-controlled metabolic cycles.
• Entrained organoids gain stable genomic changes and function within days of transplant.
ACKNOWLEDGEMENTS
The authors thank Deanne Watson, Dani Swain, Jeff Davis, Ramona Pop, and Samantha Collins for reagents and assistance with experiments; the HSCRB histology and flow cytometry, BPF next-gen sequencing, and Bauer flow cytometry and sequencing core facilities at Harvard University for technical support and critical discussions. J.R.A-D. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. D.A.M. is an investigator of the Howard Hughes Medical Institute. This work was supported by the Max Planck Society and NIH grants DP3K111898 and P01GM099117 (A.M.) and by grants from the Juvenile Diabetes Research Foundation, Helmsley Charitable Trust, and the JPB Foundation.
AUTHORSHIP
J.R.A.-D., J.D., N.R., J.H.R.K., and A.H. performed experiments. J.R.A.-D., J.D., J.C., and J.R.S. conducted bioinformatics analyses. J.R.A.-D., J.D., J.H.R.K., A.M., and D.A.M. designed the research, interpreted results, and wrote the manuscript.
CONFLICT OF INTEREST/DISCLOSURE STATEMENT
D.A.M. is a founder and scientific advisory board member for Semma Therapeutics. All other authors have no conflict of interest to report.
CONFERENCE INFORMATION
15th Annual Harvard Stem Cell Institute Retreat: Translating Science to the Clinic, Virtual (Wednesday May 20, 2020).
REFERENCES
[1] | Shapiro AM , Pokrywczynska M , Ricordi C . Clinical pancreatic islet transplantation. Nat Rev Endocrinol. (2017) ;13: (5):268–77. |
[2] | Sneddon JB , Tang Q , Stock P , Bluestone JA , Roy S , Desai T , et al. Stem Cell Therapies for Treating Diabetes: Progress and Remaining Challenges. Cell Stem Cell. (2018) ;22: (6):810–23. |
[3] | Alvarez-Dominguez JR , Donaghey J , Rasouli N , Kenty JHR , Helman A , Charlton J , et al. Circadian Entrainment Triggers Maturation of Human In Vitro Islets. Cell Stem Cell. (2020) ;26: (1):108-22 e10. |
[4] | Rasouli N , Melton DA , Alvarez-Dominguez JR . Purification of Live Stem-Cell-Derived Islet Lineage Intermediates. Curr Protoc Stem Cell Biol. (2020) ;53: (1):e111. |
[5] | Alvarez-Dominguez JR , Zhang X , Hu W . Widespread and dynamic translational control of red blood cell development. Blood. (2017) ;129: (5):619–29. |
[6] | Alvarez-Dominguez JR , Knoll M , Gromatzky AA , Lodish HF . The Super-Enhancer-Derived alncRNA-EC7/Bloodlinc Potentiates Red Blood Cell Development in trans. Cell Rep. (2017) ;19: (12):2503–14. |
[7] | Saint-Andre V , Federation AJ , Lin CY , Abraham BJ , Reddy J , Lee TI , et al. Models of human core transcriptional regulatory circuitries. Genome research. (2016) ;26: (3):385–96. |
[8] | Pagliuca FW , Millman JR , Gurtler M , Segel M , Van Dervort A , Ryu JH , et al. Generation of functional human pancreatic beta cells in vitro . Cell. (2014) ;159: (2):428–39. |
[9] | Creyghton MP , Cheng AW , Welstead GG , Kooistra T , Carey BW , Steine EJ , et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. (2010) ;107: (50):21931–6. |
[10] | Iwafuchi-DoiM, ZaretKS. Pioneer transcription factors in cell reprogramming. Genes Dev. (2014) ;28: (24):2679–92. |




