Human Pluripotent Stem Cell-Derived Organoids as a Model of Intestinal Xenobiotic Metabolism
Abstract
Background:
The human intestine is the site of absorption and first-pass metabolism for oral intake. Assessment of absorption, distribution, metabolism, excretion, and toxicity (ADMET) of xenobiotics has transformed the understanding of in vivo pharmacology. However, these processes are difficult torecapitulate in vitro.
Objective:
We have developed a simple protocol for the generation of mature functional intestinal organoids from human pluripotent stem cells (hPSCs)under xenogeneic-free conditions. We sought to characterize transcription level in drug transporters and metabolism and evaluate CYP3A4 catalytic function of the organoids.
Methods:
Human pluripotent stem cell-derived intestinal organoids were generated and evaluated the expression of drug transporters and metabolizing enzymes. We examined the induction of CYP3A4 and ABCB1 gene expression in the organoids. Furthermore, we analyzed the CYP3A4 enzyme activity of the organoids by the p450-Glo CYP3A4 assay kit with luciferin isopropyl acetal.
Results:
Stem cell-derived intestinal organoids had an outward polarized intestinal epithelial layer and showed similar expression levels of drug transporters and metabolism genes as the adult healthy intestine. They also exhibited CYP3A4 enzymatic function in vitro.
Conclusion:
This model provides a novel platform for pharmacological testing and can enhance human ADMET studies in drug development.
INTRODUCTION
The processes of absorption, distribution, metabo-lism, excretion, and toxicity (ADMET) related to oral drugs are of great importance. The intestine is the first site of drug disposition through absorption and metabolism upon oral administration. It is also involved in the toxicity of compounds. Various models have been developed for the prediction of ADMET in the intestine. Among these, animal models are often used to estimate the bioavailability of a compound. However, species differences result in differences in the expression of metabolic enzymes and transporters, thereby leading to poor clinical predictions [1, 2]. In vitro cell culture models are also frequently used to study intestinal bioavailability and toxicity.The colorectal tumor-derived Caco-2 monolayer culture is the most established model to study permeability and to predict the intestinal absorption of drugs. However, the expression patterns of xenobiotic metabolizing enzymes and transporters in Caco-2 cells are different from those of human intestinal cells, potentially leading to misrepresentation of the native response [3–5]. It is thus difficult to accurately predict the absorption rate in humans, and model systems have limitations.
The development of the 3D intestinal stem cell cultures has great applicability as a human intestinal epithelial model in vitro. Human intestinal organoids can be propagated long term and differentiate into diverse epithelial cell types, which have crypt-villous tissue structures [6]. A defining feature of enteroids is their cystic tissue structure with the apical side of the epithelium oriented inward. Recently, the use of pluripotent stem cells (PSCs) differentiated into intestinal epithelium in vitro has been added to this spectrum of organoid technologies [7]. Human intestinal organoids (HIOs) can be differentiated from human PSCs and have more complex tissue structures. HIOs also have cyst-like structures oriented inward towards the apical side of the epithelium [8]. Extensive application for drug screening might require extra-technical adaptations to address the luminal epithelium of these reported organoids, which might limit such application to either the apical or basal surfaces [8]. Furthermore, it is unclear whether such reported intestinal organoids have transporters/metabolic property. The generation ofmore functional and unique-featured intestinal organoids from human PSCs under xenogeneic-free culture conditions in vitro has been reported previously [9]. Xenogeneic-free human intestinal organoids(XF-HIOs) are morphologically unique because of extroversion of the mucosal epithelium, thereby facilitating xenobiotic screening and with potential use in the testing of intestinal infectious agents. Moreover, XF-HIOs contain mesodermal smooth muscles and ectodermal intestinal nerves, in addition to endodermal epithelial layers, thus enabling their self-moving peristalsis-like movements.
Here, to investigate whether PSCs-derived organoids exert catalytic activity on xenobiotics, we examined the expression levels of the xenobiotic transporters in XF-HIOs and their metabolic properties. Moreover, we performed a chemical-mediated CYP3A4 induction test and explored the response dynamics of drug-mediated CYP3A4 induction in XF-HIOs with Caco-2 cells as the control.
MATERIALS AND METHODS
Cell culture and generation of intestinal organoids
Human embryonic stem cells (hESCs) SEES2 [10] were maintained in StemFlex™ medium (Thermo Fisher Scientific, Waltham, MA, USA) or StemFitR medium (Ajinomoto, Tokyo, Japan) on vitronectin (VTN-N; Thermo Fisher Scientific). The medium was changed every other day, and cells were sub-cultured approximately once a week using TrypLE™ Select enzyme (Thermo Fisher Scientific). In our previous studies [9], we generated highly functional intestinal organoids using a defined xenogeneic-free (XF) differentiation medium containing 85% Knockout DMEM, 15% Knockout Serum Replacement XF CTS (XF-KSR; Thermo Fisher Scientific), 2 mM GlutaMAX-I, 0.1 mM NEAA, Pen-Strep, 50 μg/mL L-ascorbic acid 2-phosphate (Sigma-Aldrich, St. Louis, MO, USA), 10 ng/mL heregulin-1β (recombinant human NRG-beta 1/HRG-beta 1 EGF domain; R&D Systems, Minneapolis, MN, USA), 200 ng/mL recombinant human IGF-1 (LONG R3-IGF-1; Sigma-Aldrich), and 20 ng/mL human bFGF (Thermo Fisher Scientific). Briefly, undifferentiated hESCs were dissociated and plated on a cell-patterning glass substrate CytoGraph (Dai Nippon Printing Co., Ltd., Tokyo, Japan) coated with VTN-N. The cells (4×106 in 3 mL of culture medium) were plated onto the patterned substrate areas, and 10 mL of fresh medium was added gently. The XF differentiation medium was changed every 3–4 days. After 40 days, gut-like peristaltic organoids were collected and cultured together in 100 mm-tissue culture dish (Corning, NY, USA) in the XF differentiation medium.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis
RNA was isolated from the organoids using the RNeasy Mini Kit (Qiagen, Hilden, Germany), and DNA was removed using DNase (Thermo Fisher Scientific). First-strand complementary (cDNA) was synthesized using SuperScript III reverse transcriptase and an oligo-dT primer (Life Technologies) according to the manufacturer’s instructions. qRT-PCR was performed in triplicate using the SYBR Green PCR Master mix and a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific); the primer sequences are listed in the Supplementary Table. After amplification, dissociation curves were obtained to ensure that every PCR product was amplified. QuantStudio 12K Flex software v1.0 was used to quantify the relative mRNA levels of the target genes after normalization against the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Three different healthy human adult small intestinal tissues (R1234226-50 and C1234226, BioChain Institute, CA, USA; #637235, Takara Bio, Shiga, Japan) were used as a positive control.
Immunocyto chemistry
The organoids were fixed with 4% paraformaldehyde in Dulbecco’s phosphate-buffered saline (PBS; Thermo Fisher Scientific) for 5 min at 4°C, permeabilized with 0.2% Triton X-100 for 2 min at room temperature, and blocked with PBS containing 5% normal serum as appropriate for each antibody. The cells were then incubated overnight at 4°C with primary antibodies against E-cadherin (610181, 1:50, BD Pharmingen, CA, USA), CDX2 (ab76541, 1:100, Abcam, Cambridge, UK), chromogranin A (ab16007, 1:100, Abcam), Mucin 2 (sc-7314, 1:50, SANTA CRUZ, TX, USA)and Na+/K+-ATPase (NB300-146, 1:100, Novus Biologicals, CO, USA). Alexa 488- or Alexa 546-conjugated anti-mouse, anti-rabbit, or anti-goat secondary antibodies (BD Biosciences) were used; the nuclei were counterstained with 4, 6-diamidino-2-phenylindole (DAPI). Cell fluorescence was analyzed using the LSM 510 Meta Laser Scanning Confocal Microscope (Carl Zeiss Microscopy, Jena, Germany).
Histochemical analysis
The organoids were fixed in 4% paraformaldehyde, embeddedin paraffin,and seriallycut into 4-μm sections. Alternate sections were mounted on slides, stained with hematoxylin and eosin, and analyzed. Immunohistochemical analysis of the developed organoids was carried out using sections stained with antibodies against SLC10A2 (HPA004795, 1:100, Sigma-Aldrich), SLC15A1 (SC20653, 1:200, SANTA CRUZ), ABCB1 (ab170904, 1:100, Abcam), CYP3A4 (SAB1400064, 1:100, Sigma-Aldrich), Ezrin (26056-1-AP, 1:2000, proteintech, IL, USA), CES1 (ab45957, 1:20, Abcam), and CES2 (ab126970, 1:100, Abcam). The secondary antibody used was polyclonal rabbit anti-mouse immunoglobulin HRP (1:100; Dako, CA, USA).
Drug transporter and metabolizing enzyme PCR arrays
RNA was isolated from the organoids and hESCs using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA templates for quantitative RT–PCR analysis were prepared using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific), following the manufacturer’s protocol. Single-stranded cDNA was synthesized using the Superscript III First Strand cDNA Synthesis System (Thermo Fisher Scientific).
We examined the expression of multiple genes encoding drug transporters and metabolizing enzymes using RT2 Profiler PCR Arrays (array numbers: PAHS-070Z and PAHS-002Z; Qiagen). ACTB(Actin Beta)and GAPDH were used as endogenous controls. The data were analyzed using the RT2 Profiler PCR Array Data Analysis online tool provided by the manufacturer. Three different healthy human adult small intestine cDNA samples (R1234226-50 and C1234226, BioChain Institute; #637235, Takara Bio) were averaged to 1.0 as the positive control.Five organoids were independently performed qRT-PCR as five biological replicates. Gene expression was calculated by the ΔΔCt method and expressed as 2-ΔΔCt.
Induction assay of CYP3A4 and ABCB1 (P-glycoprotein) in XF-HIOs and Caco2-cells
The organoids and the colonic cancer cell line Caco-2 cells were treated with 100 nM of 1,25-dihydroxyvitamin D3 (1,25-VD3; Sigma-Aldrich) for 24 h. The control organoids and Caco-2 cells were treated with 0.1% dimethyl sulfoxide (DMSO; Sigma-Aldrich). After incubation, the samples were collected and washed two times with PBS. Total RNA was extracted using the RNeasy Mini kit (Qiagen), and first-strand cDNA was synthesized using SuperScript IV VILO Master Mix according to the manufacturer’s instructions (Thermo Fisher Scientific). Quantitative RT-PCR was performed in triplicate using the SYBR Green PCR Master mix. Transcript levels were determined using the QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific). The CYP3A4 and ABCB1 primer sequences are listed in the Supplementary Table. QuantStudio 12K Flex software v1.0 was used to quantify the relative mRNA levels of the target genes after normalization against the housekeeping gene GAPDH. Normal human adult small intestinal tissue cDNA samples (R1234226-50 and 1234226, BioChain; #637235, Takara Bio) were used as control.
CYP3A4 catalytic function Assay
We prepared D60-70 organoids derived from hESCs, and CYP3A4 catalytic function was analyzed using the p450-Glo CYP3A4 assay kit with luciferin isopropyl acetal (luciferin-IPA; Promega,WI, USA) according to the manufacturer’s protocol [11]. The organoids were placed onto a 24-well plate (Nunc, MA, USA) with a single organoid per well and then incubated with 500 μL of the medium supplemented with luciferin-IPA and/or 1.0 μM Ketoconazole (Tokyo Chemical Industry, Tokyo, Japan) as a CYP3A4 inhibitor. After 2 h of incubation, the supernatants and the organoid-fluidic contents were collected with a 34 G microneedle (Unisis, Tokyo, Japan) in a 1 mL syringe (Terumo, Tokyo, Japan) and analyzed according to the manufacturer’s instruction. Luminescence was measured as relative light units (RLU) using the Synergy H1 hybrid multi-mode plate reader and Gen 5 data software (BioTek, VT, USA). The culture medium was used as control.
Statistical analysis
Quantitative data are reported as means±standard error of the mean from at least three independent experiments. Statistical analysis was performed using either an unpaired ortwo-tailed t-test. P < 0.05 indicates statisticalsignificance.
Ethics approval
Human ESC experiments were performed following the hESC guidelines of the Japanese government and were approved by the Institutional Review Board on hESC research at the National Centre for Child Health and Development (NCCHD; Notification No. 232). Surgical specimens for analysis were obtained from patients after informed consent; and parental written informed consent was obtained from all families. The use of intestinal tissues was approved by the Institutional Review Board (IRB) of the NCCHD (IRB permission #927), in adherence to the Declaration of Helsinki.
RESULTS
Morphological structure of XF-HIOs
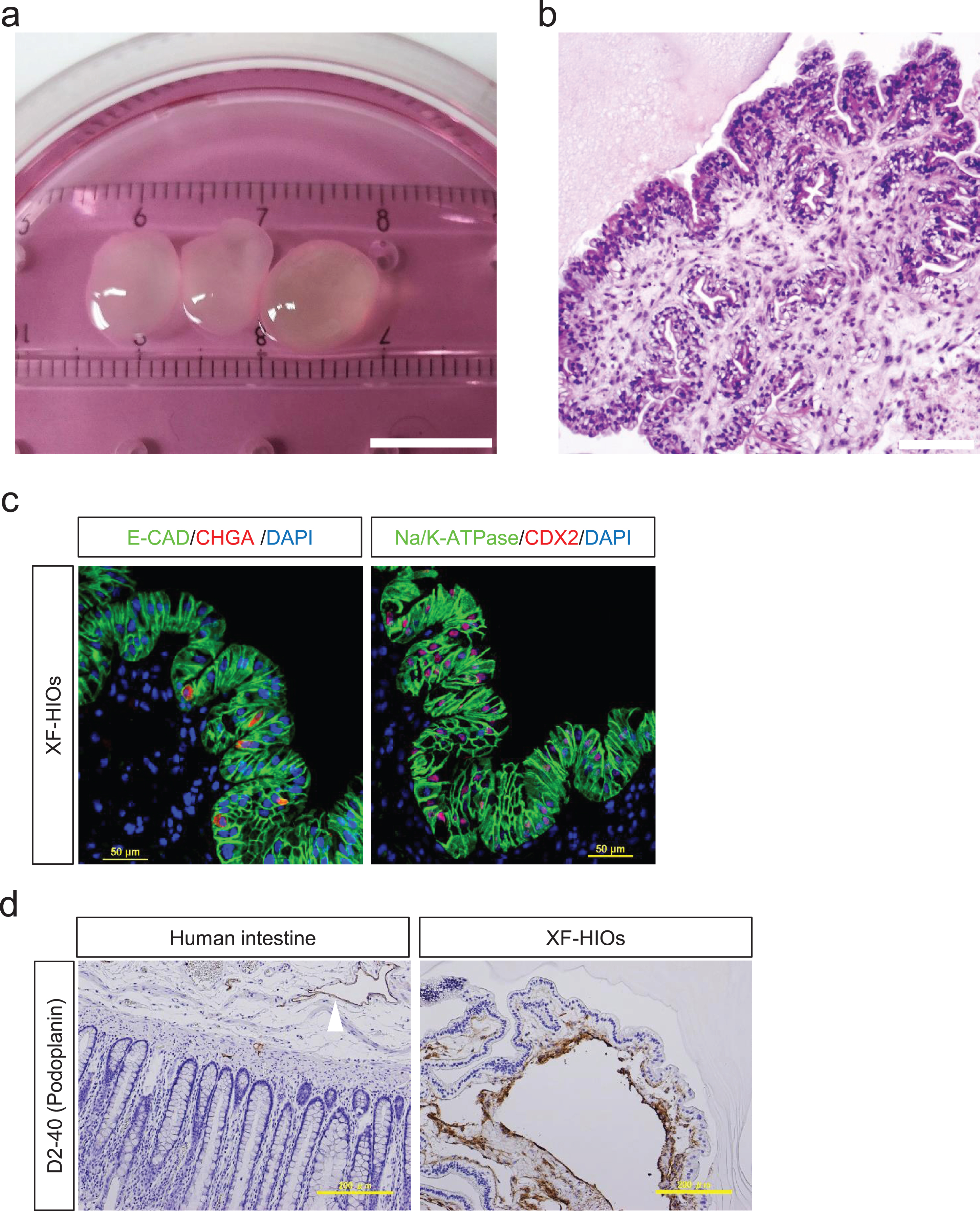
We have previously reported the successful development of an intestinal organoid system in vitro under xenogeneic-free conditions using hESCs or induced pluripotent stem cells (hiPSCs) [9]. In this study, we applied this procedure to generate organoids and examined their xenobiotic metabolic functions. The culture yielded approximately 10-mm-sized XF-HIOs with cyst-like structures (Fig. 1a), which were relatively larger compared to previously reported intestinal organoids [6, 7]. Their histological features were uniquely structured outwards and oriented towards the epithelial layers (Fig. 1b). Immunofluorescence staining for an epithelial cell-specific E-cadherin (E-CAD), an intestinal epithelial enteroendocrine cell marker chromogranin A (CHGA), a Na+/K+-ATPase, and an intestinal transcription marker CDX2 showed that the outward-oriented layers exhibited human intestinal epithelium structure with the correct expressions of intestinal epithelium (E-CAD and Na+/K+-ATPase) and epithelial specific cell markers (CHGA and CDX2; Fig. 1c). Cell counting revealed that the epithelium contained approximately 95% CDX2 positive cells, and 10% chromogranin-A expressing enteroendocrine cells (Fig. 1c and Supplementary Fig. 1a, b). The epithelial layers of the XF-HIOs, cyst-like structures were lined underneath with lymphatic endothelial marker, D2-40 positive cells (Fig. 1d). Ezrin, an apical surface marker, was positively stained in the epithelial layers of the XF-HIOs (Supplementary Figure 1c).
Fig. 1
Characterization ofintestinal organoids from human embryonic stem cells. (a) In vitro culture of Xenogeneic-free human intestinal organoids (XF-HIOs) generated from hESCs. The XF-HIOs are stably maintained in a xenogeneic-free medium. Scale bar corresponds to 10 mm. (b) Hematoxylin and eosin (HE) staining of organoids on day 100. Scale bar corresponds to 200 μm. The XF-HIOs are structured outwards andoriented towards the epithelial layers. (c) Immunostaining for Chromogranin A (CHGA), caudal type homeobox 2 (CDX2), and Na/K-ATPase. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI). Scale bars correspond to 50 μm. (d) Under the epithelial layers of the XF-HIOs, cyst-like structures were lined with the lymphatic endothelial marker, D2-40 positive cells (Fig. 1d). Immunohistochemical staining for lymphatic endothelium marker Podoplanin (D2-40) in XF-HIOs and in healthy adult human intestine (white arrowhead) as control. Cystic structures of the XF-HIOs lined by D2-40 positive cells. Scale bars correspond to 200 μm.
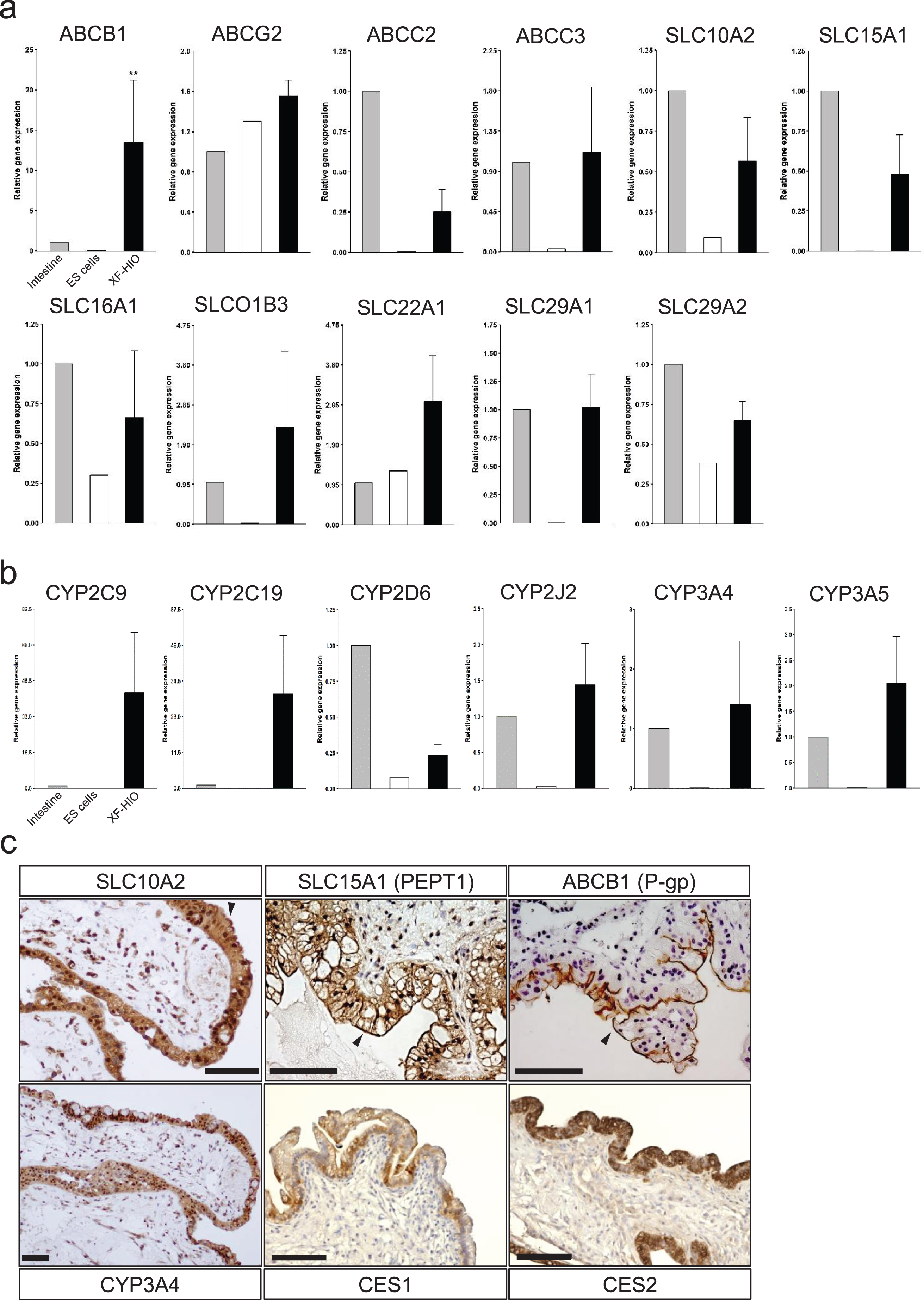
Gene expressions of drug transporters and metabolizing enzymes
The gene expressions of 86 human drug transporters (Supplementary Figure 2) and 84 human drug metabolizing enzymes (Supplementary Figure 3) were examined by qRT-PCR of a single XF-HIO as a control for the adult intestine. Among the examined drug transporters, 4 ATP-binding cassette (ABC; ABCB1, ABCG2, ABCC2, and ABCC3) and 7 solute carrier (SLC; SLC10A2,SLC15A1, SLC16A1, SLCO1B3, SLC22A1, SLC29A1, and SLC29A2) transporters are shown in Fig. 2a. These transporters are proposed by the International Transporter Consortium (ITC) as important for evaluation during drug development [12] and are similar to those expressed in adult intestines (Fig. 2a). Cytochromes P450 monooxygenases (CYPs) constitute a superfamily of heme-thiolate proteins and are the dominant enzyme system responsible for xenobiotic detoxification and drug metabolism. Among the metabolizing enzymes in the human intestine, CYPs are of particular importance in the intestinal first-pass metabolism of orally administered drugs [11], of which the CYP3A subfamily (CYP3A4 and CYP3A5) was reported as the most abundantly expressed in the small intestine [13]. CYP3A, CYP2C9, CYP2C19 and CYP2J2 were highly expressed in XF-HIOs compared to the adult intestine (Fig. 2b). To analyze the transporters and drug metabolic enzymes at the protein level, immunohistochemical staining was performed on the XF-HIOs. Intestinal transporters, i.e.,SLC10A2, SLC15A1, and ABCB1 (P-glycoprotein; P-gp), and catabolic enzymes, i.e., CYP3A4 and carboxylesterase (CES) 2, were located in the mucosal layers (Fig. 2c). SLC10A2, SLC15A1, ABCB1 and Ezrin were strongly stained at the apical part of the epithelial cells (Fig. 2c and Supplementary Figure 1c). The XF-HIOs exhibited strong staining for an intestinal-type enzyme, CES2,but not for a colorectal-type enzyme CES1 (Fig. 2c and Supplementary Figure 4). Isotype control antibodies in the immunohistochemical analysis were presented (Supplementary Figure 5).
Fig. 2
Gene expression levels of drug transporters and metabolizing enzymes in XF-HIOs. (a-b) Relative gene expressions of drug transporters (a) and metabolizing enzyme (b) of per the organoid (black bars) and inthe control, i.e., adult intestine (gray bars) and human ES cells (white bars). Values were normalized against GAPDH. The data are reported as means±SE. Statistical significance was identified using Student’s t-test (*P < 0.05, **P < 0.01; n = 5) (c) Immunohistochemical staining for drug transporters, i.e., SLC10A2, SLC15A1, and ABCB1, and metabolic enzymes, i.e., CYP3A4, CES1, and CES2, in D60-70 XF-HIOs. The epithelial layer of the XF-HIOs exhibited strong staining for CES2, an intestinal-type enzyme. SLC10A2, SLC15A1 and ABCB1 were positively stained apical membranous in epithelial layers of the XF-HIOs (black arrowheads). Scale bars correspond to 100 μm.
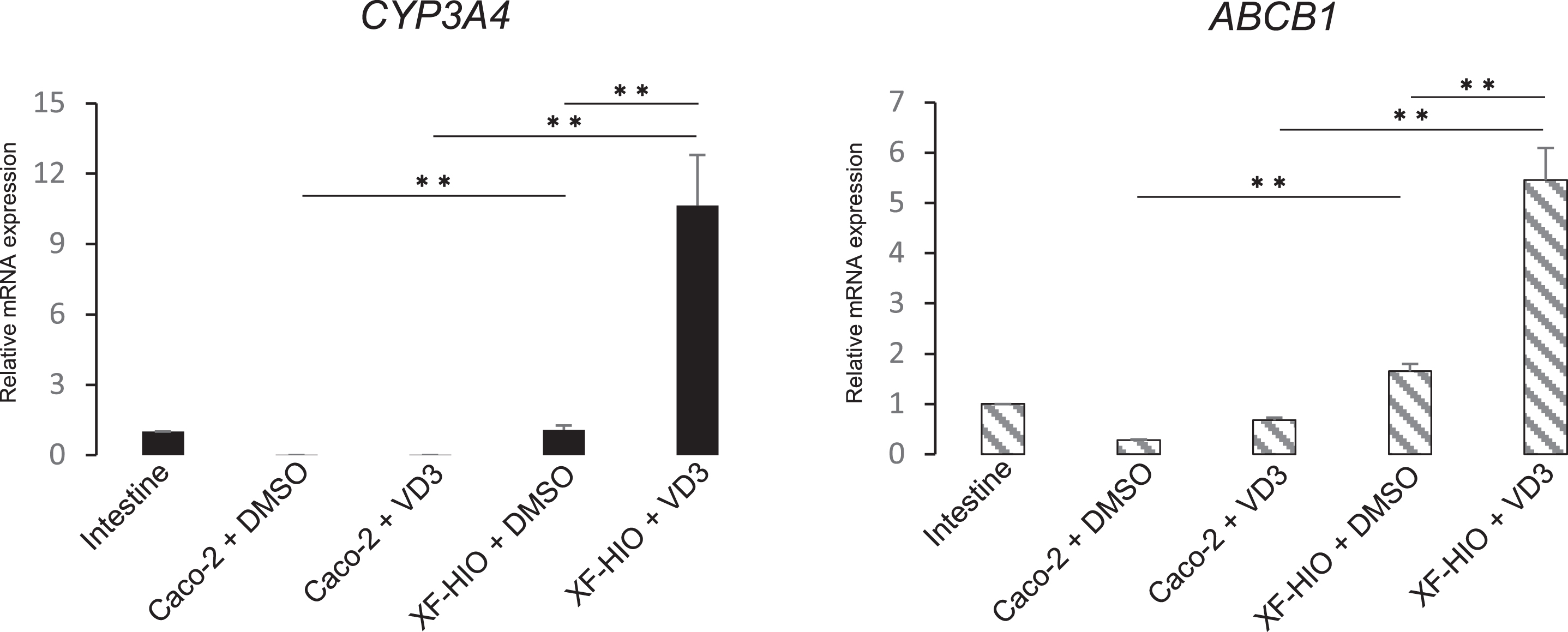
CYP3A4 and ABCB1 activities and inducibilities
To examine CYP3A4 and ABCB1 induction, we used 1,25-dihydroxyvitamin D3 (1,25-VD3) because it can induce the expression of CYP3A [14]. The transcript expression levels of CYP3A4 and ABCB1 were significantly higher in the XF-HIO streated with 1,25-VD3 than in the control organoids (Fig. 3). 1,25-VD3 induced the expression of CYP3A4 significantly.
Fig. 3
Induction of CYP3A4 and ABCB1 in XF-HIOs and Caco2-cells. The transcript expression levels of CYP3A4 (black bars) and ABCB1 (hatched bars) in XF-HIOs were significantly higher than those in the adult intestinesafter treatment with1,25-Dihydroxyvitamin D3 (VD3). Three different adult intestines were used for RT-PCR and set to 1.0 as the reference sample. The control organoids or Caco-2 cells were treated with 0.1% Dimethyl sulfoxide (DMSO). Data are reported as means±SE, and statistical significance was identified using Student’s t-test (**P < 0.01; n = 3).
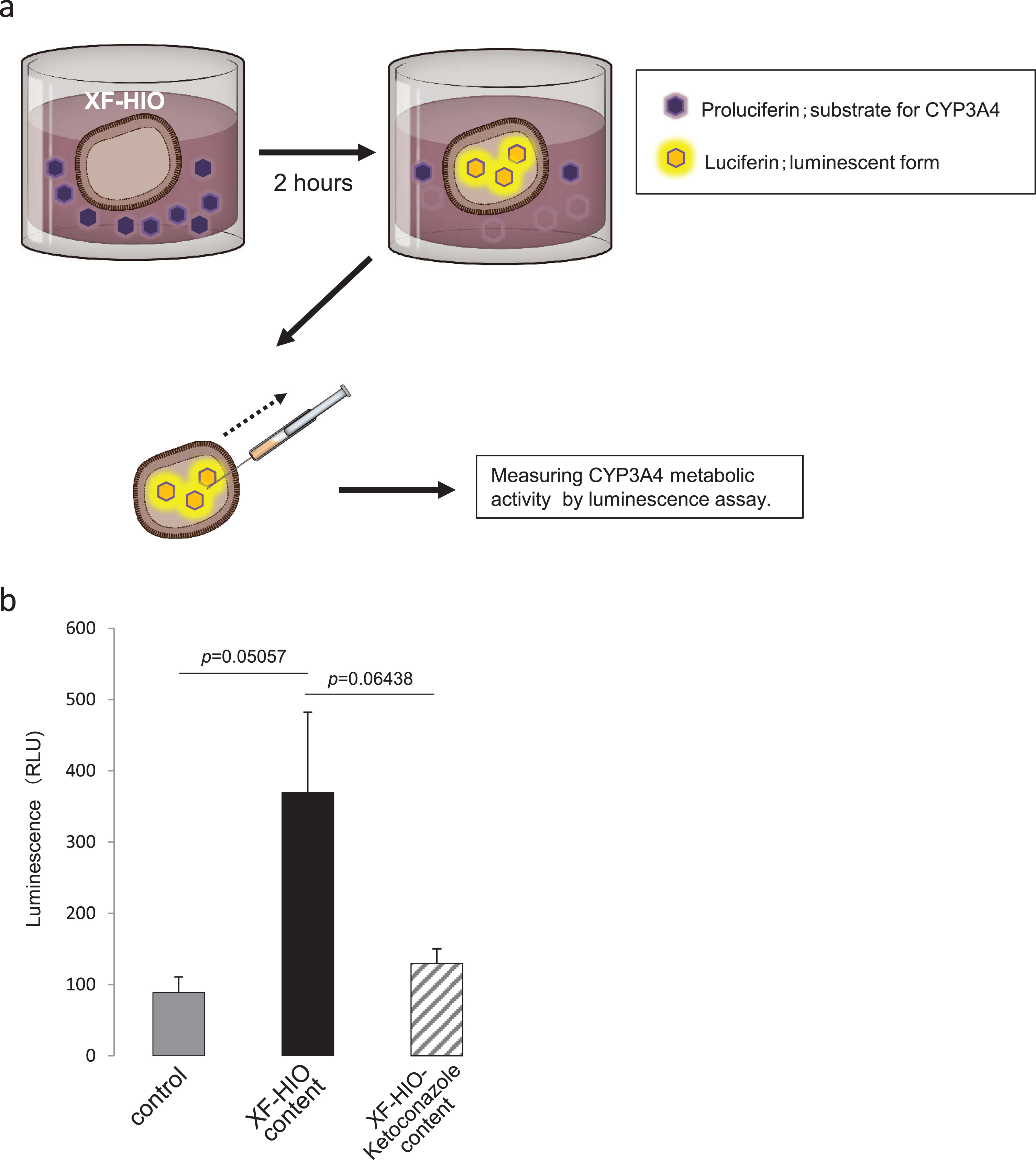
CYP3A4 metabolic activity
To analyze the CYP3A4 enzyme activity of the XF-HIOs, single organoids were treated with proluciferin-IPA and/or Ketoconazole for 2 h. Because XF-HIOs have cyst-like structures and have outward oriented epithelium layers, we collected their contentsusing a 34 G microneedle to analyze luminescence (Fig. 4a). Luminescence was considerably higher in proluciferin-IPA treated XF-HIOs than both in the control without organoids and in the XF-HIOs treated with Ketoconazole, though the difference was not statistically significant (Fig. 4b; n = 5).
Fig. 4
Metabolic activity of CYP3A4 in XF-HIOs. (a) Schematic diagram of measuring CYP3A4 catalytic activity in XF-HIOs. Single organoids were treated with proluciferin-IPA, and their content was collected using a 34 G microneedle for luminescence assay. (b) CYP3A4 catalytic activity was scored as relative light units (RLU). Luminescence was significantly higher in proluciferin–IPA-treated XF-HIOs than in control without XF-HIOs. Data are reported as means±SE. Statistical significance was identified using Student’s t-test (n = 5).
DISCUSSION
In this study, the xenogeneic-free human intestinal organoids derived from hPSCs showed expression of xenobiotic transporters and their metabolic properties, as well as the activity of CYP3A4 enzyme. It has been reported that gastrointestinal organoids were derived either from intestinal tissues or hPSCs and that these organoids showed similarity with the human intestine based on tissue-specific gene expression and their epithelial tissue properties [9]. However, it is still unclear whether drug metabolism functions at a similar magnitude as in human intestinal tissues. Therefore, the variability of stem cell-derived organoids in bioassays on xenobiotics has to be considered and resolved in future applications.
The use of cellular organoids is a developing research technology and provides another in vitro model for the intestine [16, 17]. Therefore, organoids generated from gastrointestinal tissue progenitors or PSCs can be applied to intestinal research. However, their xenobiotic metabolism is not yet analyzed. Furthermore, their closed lumens with inward epithelial orientation are relatively inaccessible for absorption, distribution, metabolism, and excretion assays [18]. As previously reported, XF-HIOs have a well-organized structure and similar functional characteristics to mature intestines [9]. Here, the XF-HIOs obtained have a morphologically unique organoid structure with outward epithelial orientation unlike other intestinal organoids, therebyfacilitating its applicability in bioassays of chemical substances or drugs for ADMET analysis.They exhibited substantial mRNA expression of ABCB1 and ABCG2, which are recommended for drug evaluation in the 2012 US Food and Drug Administration (FDA) Draft Guidance on Drug Interactions [11]. In addition, intestinal drug metabolic enzymes, such as CYP3A,are expressed in XF-HIOs, similar to human intestine.
Our immunostaining analysis indicated that SLC10A2, SLC15A1, ABCB1, CYP3A4, and CES2 were localized along the lumen of the polarized epithelium of XF-HIOs, suggesting that the simple addition of CYP3A4 substrates in XF-HIOs cultures could enable evaluation of their enzymatic activities. In a functional assay, XF-HIOs can metabolize a CYP3A4 substrate, and the metabolites accumulated in their cystic structure.The presence of functional key CYP3A4 metabolic enzymes was also confirmed. To our knowledge, this is the first study to show that human gastrointestinal organoids exert catalytic activity on xenobiotics.The human intestine has diverse physiological functions related to its complex tissue formation including the immune system. To understand the intestinal physiological conditionsproperly, the organoids bio-modelshould be integrated with immune cells in future studies.
In conclusion, we have generated intestinal organoids that can metabolize xenobiotics in a xenogeneic-free system. To overcome the existing limitations of the current in vitro systems in human gastrointestinal organoids, the XF-HIOs generated in this study could be used to develop a novel system with ADMET applications for drug development.Therefore, this study enables us to use the intestinal organoids system as a chemical screen for drug development and as a toxicological prediction model to fill out the gap between in vivo and in vitro systems.
CONFLICT OF INTEREST STATEMENT
H.A. receives a research grant from Dai Nippon Printing Co., Ltd. (DNP). H.A. and DNP are inventors on several patents related to intestinal organoid technology. M.I. is an employee of DNP. Other authors have no conflict of interest to report.
ACKNOWLEDGMENTS
We are grateful to the members of the Life Science Laboratory, Dai Nippon Printing Co. (DNP), and to the members of our laboratory for their helpful discussions. We thank Minoru Ichinose for preparing histological samples. This work was funded by the Ministry of Health, Labour, and Welfare Sciences (MHLW) to HA and AU (25670710,21390456 and 26293364); the National Center for Child Health and Development (26-1 and 24-6) to HA. The funders had no role in the study design, data collection and analysis, decision to publish, nor the preparation of the manuscript.
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/STJ-200001.
STEMJOURNAL OPEN REVIEW
The evaluations from peer reviewers for this article are freely available and can be found as supplementary material here: https://dx.doi.org/10.3233/STJ-200001.
REFERENCES
[1] | Peters SA , Christopher RJ , Anna-Lena U , Oliver JDH . Predicting Drug Extraction in the Human Gut Wall: Assessing Contributions from Drug Metabolizing Enzymes and Transporter Proteins using Preclinical Models. Clin Pharmacokinet. (2016) ;55: (6):673–96. doi: 10.1007/s40262-015-0351-6 |
[2] | Komura H , Iwaki M . In vitro and in vivo small intestinal metabolism of CYP3A and UGT substrates in preclinical animal species and humans: Species differences. Drug Metab Rev. (2011) ;43: (4):476–98. doi: 10.3109/03602532.2011.597401 |
[3] | Le Ferrec E , Chesne C , Artusson P , Brayden D , Fabre G , Gires P , et al. In vitro models of the intestinal barrier. The report and recommendations of ECVAM Workshop 46. European Centre for the Validation of Alternative methods. Altern Lab Anim. (2001) ;29: (6):649–68. doi: 10.1177/026119290102900604 |
[4] | Bentz J , O’Connor MP , Bednarczyk D , Coleman J , Lee C , Palm J , et al. Variability in P-glycoprotein inhibitory potency (IC50) using various in vitro experimental systems: Implications for universal digoxin drug-drug interaction risk assessment decision criteria. Drug MetabDispos. (2013) ;41: (7):1347–66. doi: 10.1124/dmd.112.050500 |
[5] | Harwood MD , Achour B , Neuhoff S , Russell MR , Carlson G , Warhurst G , Rostami-Hodjegan A . In Vitro-In Vivo Extrapolation Scaling Factors for Intestinal P-Glycoprotein and Breast Cancer Resistance Protein: Part I: A Cross-Laboratory Comparison of Transporter-Protein Abundances and Relative Expression Factors in Human Intestine and Caco-2 Cells. Drug MetabDispos. (2016) ;44: (3):297–307. doi: 10.1124/dmd.115.067371 |
[6] | Sato T , Vries RG , Snippert HJ , van de Wetering M , Barker N , Stange DE , et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. (2009) ;459: (7244):262–5. doi: 10.1038/nature07935 |
[7] | Spence JR , Mayhew CN , Rankin SA , Kuhar MF , Vallance JE , Tolle K , et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. (2011) ;470: (7332):105–9. doi: 10.1038/nature09691 |
[8] | Tsuruta S , Uchida H , and Akutsu H . Intestinal organoids generated from human pluripotent stem cells. Japan Med Assoc J. (2020) ;3: (1):9–19. doi: 10.31662/jmaj.2019-0027 |
[9] | Uchida H , Machida M , Miura T , Kawasaki T , Okazaki T , Sasaki K , et al. A xenogeneic-free system generating functional human gut organoids from pluripotent stem cells. JCI Insight. (2017) ;2: (1):e86492. doi: 10.31662/jmaj.2019-0027 |
[10] | Akutsu H , Machida M , Kanzaki S , Sugawara T , Ohkura T , Nakamura N , et al. Xenogeneic-free defined conditions for derivation and expansion of human embryonic stem cells with mesenchymal stem cells. Regen Ther. (2015) ;1: :18–29. doi: 10.1016/j.reth.2014.12.004 |
[11] | Cali JJ , Ma D , Sobol M , Simpson DJ , Frackman S , Good TD , et al. Luminogenic cytochrome P450 assays. Expert Opin Drug MetabToxicol. (2006) ;2: (4):629–45. doi: 10.1517/17425255.2.4.629 |
[12] | Hillgren KM , Keppler D , Zur AA , Giacomini KM , Stieger B , Cass CE , et al. Emerging transporters of clinical importance: An update from the International Transporter Consortium. Clin PharmacolTher. (2013) ;94: (1):52–63. doi: 10.1038/clpt.2013.74 |
[13] | Thelen K , Dressman JB . Cytochrome P450-mediated metabolism in the human gut wall. J Pharm Pharmacol. (2009) ;61: (5):541–58. doi:10.1211/jpp/61.05.0002 |
[14] | Paine MF , Hart HL , Ludington SS , Haining RL , Rettie AE , Zeldin DC . The human intestinal cytochrome P450 “pie”. Drug MetabDispos. (2006) ;35: (5):880–6. doi: 10.1124/dmd.105.008672 |
[15] | Makishima M , Lu TT , Xie W , Whitfield GK , Domoto H , Evans RE , et al. Vitamin D receptor as an intestinal bile acid sensor. Science. (2002) ;296: (5571):1313–6. doi: 10.1126/science.1070477 |
[16] | Dedhia PH , Bertaux-Skeirik N , Zavros Y , Spence JR . Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology. (2016) ;150: (5):1098–112. doi: 10.1053/j.gastro.2015.12.042 |
[17] | Lancaster MA , Knoblich JA . Organogenesis in a dish: modeling development and disease using organoid technologies. Science. (2014) ;345: (6194):1247125. doi: 10.1126/science.1247125 |
[18] | Madden LR , Nguyen TV , Garcia-Mojica S , Shah V , Le AV , Peier A , et al. Bioprinted 3D Primary Human Intestinal Tissues Model Aspects of Native Physiology and ADME/Tox Functions. iScience. (2018) ;2: :156–67. doi: 10.1016/j.isci.2018.03.015 |




