Modeling COVID-19 positivity rates and hospitalizations in Texas
Abstract
The aim of this study was to jointly model COVID-19 test positivity rates and hospitalizations in Texas using Bayesian joinpoint regression. The data for both test positivity rates and hospitalizations were obtained from the Texas Department of State Health Services between April 5 and October 19, 2020. The stage 1 model identifies four significant shifts in test positivity rates, three of which occur roughly 9 days after documented policy or behavioral changes statewide. Estimated positivity rates from the first model were then used to predict hospitalization rates and to estimate lag time between changes in positivity and hospitalization. The resulting lag time is 9.056 days (
1.Introduction
The news is filled with speculation about the impact of policies such as shut-downs and mask orders in “flattening the curve” for COVID-19, while many ponder the reasons why increased (or decreased) incidence may not coincide with hospitalization or deaths. Several simulation models exist to estimate the impact of social distancing on COVID-19 spread based on hypothetical scenarios. See for example, (Keskinocak et al., 2020). In addition, predicting COVID-19 hospitalization rates is vital for medical centers to maintain proper staffing, medical, and device resources. A few studies exist that estimate the average time between onset of symptoms and hospitalization but they are limited in scope and sample size. For example, Yang et al. (2020) found that the median time between onset of symptoms and pneumonia was 5 days and the median time between onset of symptoms and ICU admission was 9.5 days for 52 patients admitted to Wuhan Jim Yin-Tan hospital with confirmed COVID-19. Linton et al. (2020) found a mean time from symptom onset to hospitalization between 3–9 days in Wuhan patients. To date no one has directly modeled the relationship between test positivity rates and hospitalization. We fit a two-stage Bayesian joinpoint model to study the relationship between the rate of COVID-19 confirmed cases and the rate of hospitalizations in Texas and estimate the associated lag time. The first stage model estimates and predicts the rate of confirmed cases, while the second stage model uses the estimated rates to estimate hospitalizations and lag time.
2.Methods
2.1Data
We used records on the 7-day COVID-19 positivity rate by specimen date and the number of persons hospitalized with COVID-19 provided by the Texas Department of State Health Services (2020a). This dataset comes with some caveats and limitations, in particular the disclaimer that “all data are provisional and subject to change.” Examples of limitations include:
• The dataset contains numerous case backlogs. Known backlogs are detailed in the dataset. Here we used the COVID-19 positivity rate by specimen collection date, so only unknown backlogs should affect the reliability of the dataset, and then only if the positivity rates of the backlogs differ markedly from the positivity rates already in the dataset. Additionally, we chose to exclude the most recent five days from our analysis, with the idea that most of the specimens collected on a given day are tested and reported in the dataset within 5–7 days.
• This dataset contains information only about the daily positivity rate of molecular COVID-19 tests. Texas does not publish the daily positivity rate of antigen or antibody tests, even though antigen tests look for active infections. Fortunately, to-date, the number of antigen tests has been small compared to the number of molecular tests, and we have not noticed any major differences between the 7-day positivity rate of antigen tests and the 7-day positivity rate of molecular tests after examining several earlier versions of the dataset that we have archived.
• Hospitalization data include only COVID-19 patients who have had a positive molecular test. We do not know if there are any people in Texas who have been hospitalized with a positive antigen test who have not undergone a molecular test. In addition, there is one hospitalization data point in the dataset (on July 23, 2020) that appears to be an outlier. Before running our analysis, we replaced this data point with the mean of its two neighbors. Otherwise, the hospitalization data is believed to be accurate.
2.2Models
Joinpoint Regression is a piecewise regression model that is widely used to capture trends in the data. The model identifies the significant points in the data where the trend changes. The model is popular in estimating the mortality and incidence of cancer trends. The two stage Bayesian joinpoint model applied in this study is an advancement of the previous models used to estimate cancer rates (Ghosh et al., 2009; Kafle et al., 2014). Let
(1)
where
The previous equation was modified to find the estimated rate of confirmed cases on a given day:
(2)
The assumption of joinpoints in our model is random so all parameters in the above models were fitted using the Bayesian methods. For
(3)
where
The parameters in the above models were fitted using the Bayesian methods as explained in Kafle et al. (2014). We estimated Daily Percentage Change (DPC) to detect the rate of change of confirmed cases in the model using the following formula:
(4)
We fitted the second stage model to study the rate of hospitalization in the population using the fitted COVID-19 rates from the first stage as one of the covariates and estimated the lag time in hospitalization. Let
(5)
Here
The above model is used to estimate the rate of hospitalization (
(6)
Here
(7)
3.Results
The data is analyzed using the freely available software packages, WinBUGS and R. For each model we ran 300,000 iterations giving 50,000 iterations as burn in periods for a wide range of initial values for different parameters. For each model, the posterior distribution of parameters is observed by monitoring the trace, iterations, Monte Carlo errors, standard deviations, and the density curves. The trace of each of the parameters satisfy the convergence criteria. Also, the Monte Carlo errors are within 3% of the posterior standard deviations.
3.1Test positivity rates
As shown in Fig. 1, the analysis identified four joinpoints that can be tied anecdotally to statewide initiatives and behavior changes: May 31, June 19, July 4 and September 21. Texas Department of State Health Services (2020b), reports a list of executive orders issued in response to the pandemic during this time period. According to the Centers for Disease Control (2020), the mean time from infection to symptom onset is 6 days, and the median number of days from symptom onset to COVID-19 test among positive patients is 3 days. Thus, changes in test positivity rates can be expected roughly 9 days after behavioral shifts.
Figure 1.
Daily rates of COVID-19 confirmed cases.
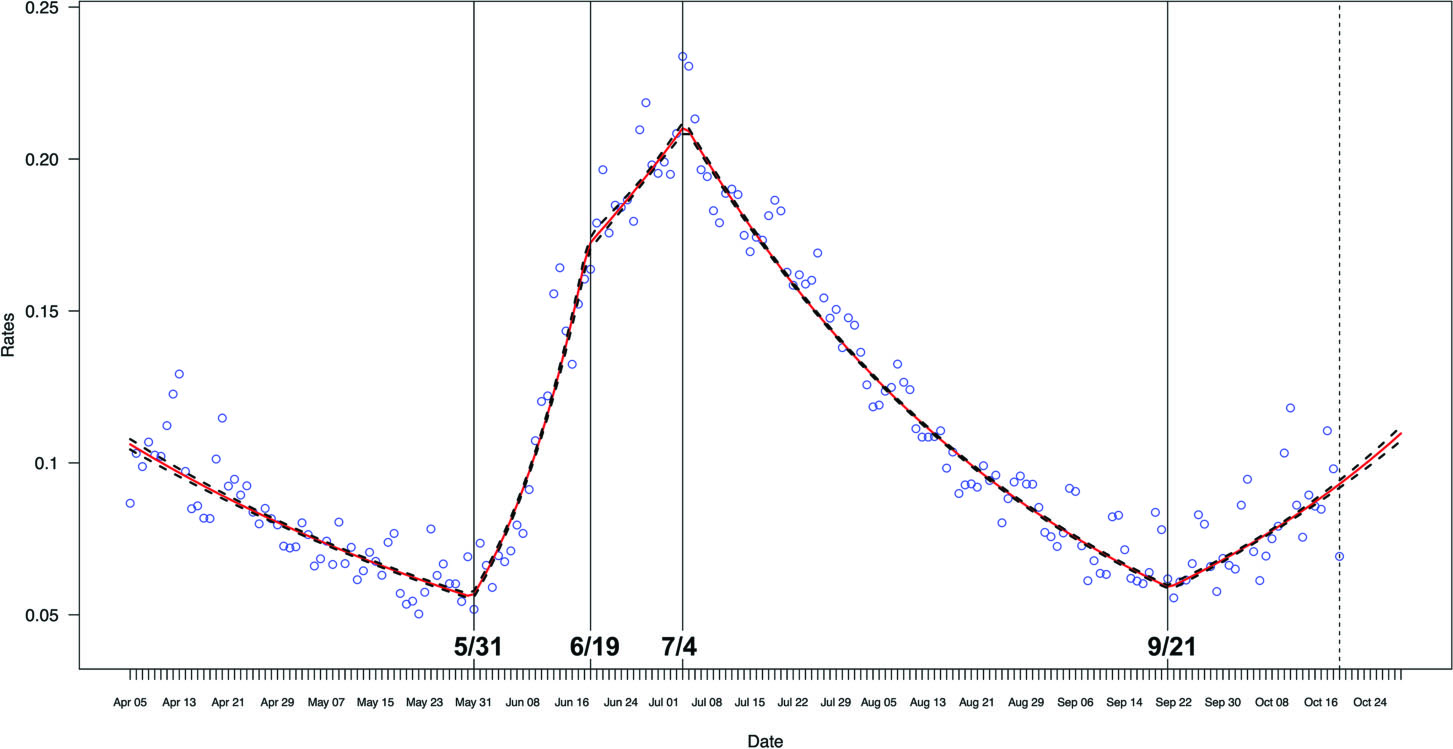
On April 2, a statewide stay at home order began. Bars, malls, retailers, and gyms shut down. Restaurants were allowed to stay open, but only for to-go orders. This order was highly effective in reducing the positivity rate of COVID-19 tests beginning April 5. On May 1, Texas began reopening. Texas reopened restaurants (to 25% capacity) on May 1, reopened offices and gyms (to 25% capacity) on May 18, and reopened bars (to 25% capacity) on May 22. Texas also expanded restaurants to 50% capacity on May 22. Nine days later, May 31, was identified as the first joinpoint in the model as the COVID-19 positivity rate in Texas began increasing steeply. Texas continued reopening in June as the positivity rate increased, although the slope shifted at a second joinpoint of June 19. On June 22, Governor Abbott implored Texans to distance and wear masks (Svitek, 2020). On June 26, bars in Texas were ordered to close except for delivery and take-out, and restaurants were reduced from 75% to 50% capacity. Eight days later, July 4, the COVID-19 positivity rate began to decline. Additionally, on July 2 the governor issued an order requiring masking in public spaces, “with few exceptions,” solidifying the downward trend. Universities across Texas reopened the weeks of August 17 and August 24 (ABC13 Eyewitness News, 2020, August 19), while K – 12 school districts opened for in person schooling on dates ranging from September 8 to October 13 (Swaby & Platoff, 2020, September 8). Not surprisingly, the model detected a reversal of the downward trend in incidence on September 21. Thus behavioral shifts do appear to pre-date breakpoints, although these observations are strictly anecdotal.
The mean, standard deviation, Monte Carlo errors, and associated 95% credible intervals for Test Positivity Rates model were presented in the table below.
3.2Hospitalization rates
In addition to the insights the first stage model provided for the impact of behavior changes, it informed the second stage model, using the estimated positivity rate to estimate the hospitalization rate, as well as the time to hospitalization, in the Texas population. The estimated positivity rate from the first stage model accurately modeled the hospitalization rates in the second stage (Fig. 2), with joinpoints at May 6, May 30, July 4, July 19 and September 22. As expected, both graphs have the same overall shape, with the exception of the first segments of each graph: The daily positivity rate of COVID-19 cases falls from April 5 to May 31, whereas the daily hospitalization numbers are roughly rising or flat over this time period. We believe that the primary reason for this deviation is the relatively small number of tests conducted during this time period: From April 5-30, Texas conducted an average of 11081 tests per day, whereas during May, Texas conducted an average of 24674 tests per day. In the months of June and July, the average number of tests per day were 44723 and 61671, respectively. Average daily tests have been above 35000 in each month since. We believe that the small number of daily tests during April and May resulted in an artificially high positivity rate (relative to the rest of the time series) at the beginning of the time series. The estimated lag time between a change in positivity rates across Texas and hospitalization rates is 9.056 days, with a standard error of 3.808. The lag estimate and associated standard error appear to support the conclusions of Yang et al. (2020) who found a median of 9.5 (
Table 1
The posterior summaries of parameters for test positivity rates
| Node | Mean | SD | MC error | 2.50% | Median | 97.5% |
|---|---|---|---|---|---|---|
|
| 0.001458 | 1.18E-05 | ||||
|
| 3.01E-05 | 2.02E-07 | ||||
|
| 0.01717 | 6.70E-04 | ||||
|
| 0.4502 | 0.002138 | 0.8992 | |||
|
| 1.02 | 0.01937 | 8.59E-04 | 0.9803 | 1.023 | 1.055 |
|
| 0.715 | 0.01603 | 6.20E-04 | 0.6783 | 0.7156 | 0.744 |
|
| 0.002293 | 1.93E-05 |
Table 2
The posterior summaries of parameters for hospitalization rates
| Node | Mean | SD | MC error | 2.50% | Median | 97.5% |
|---|---|---|---|---|---|---|
|
| 0.00788 | 2.53E-04 | ||||
|
| 9.129 | 0.3412 | 0.01506 | 8.583 | 9.136 | 9.654 |
|
| 0.08602 | 3.18E-04 | 2.77E-06 | 0.0854 | 0.08602 | 0.08665 |
|
| 0.07568 | 0.004825 | 9.47E-05 | 0.06626 | 0.07566 | 0.08513 |
|
| 0.06259 | 0.002754 | ||||
|
| 1.95E-04 | 0.2884 | 0.001275 | 0.5799 | ||
|
| 0.421 | 0.06356 | 0.002798 | 0.3034 | 0.4173 | 0.5537 |
|
| 0.9366 | 0.008321 | 2.08E-04 | 0.9201 | 0.9367 | 0.9525 |
Figure 2.
Daily Hospitalization Rates Due to COVID-19 and Test Positivity Rates Combined.
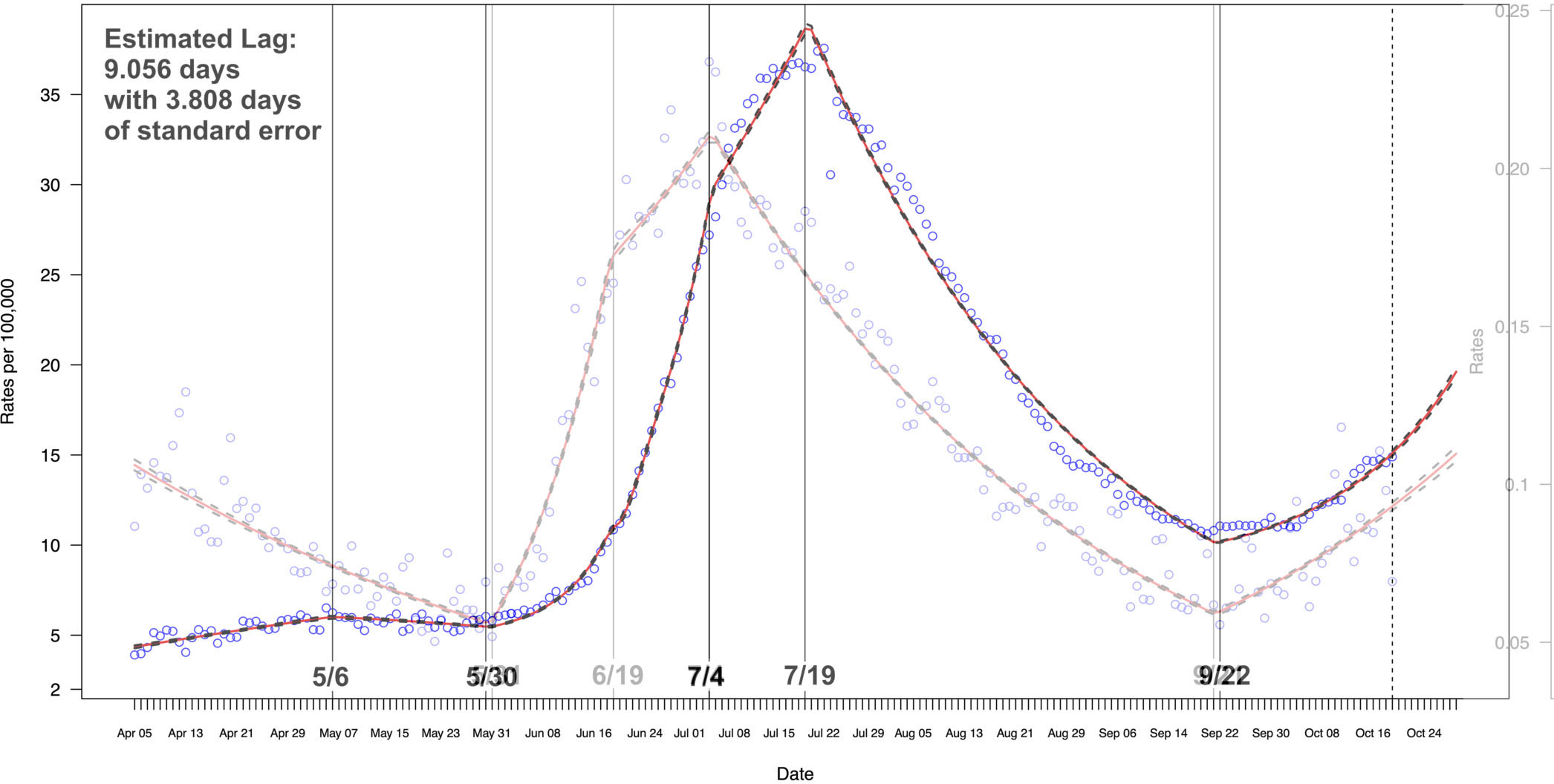
The mean, standard deviation, Monte Carlo errors, and associated 95% credible intervals for Hospitalization Rates model were presented in the table below.
4.Discussion
In this study, we adapted and applied the Bayesian joinpoint regression analysis to jointly model COVID-19 incidence rates and hospitalization trends in Texas. The analysis identified four distinct shifts in incidence trends and five shifts in hospitalization rates. The first stage estimation of incidence rates successfully informed the second stage estimation of hospitalization rates and provided an estimated lag time of 9.056 days (
Acknowledgments
The authors would like to thank the anonymous reviewer for the insightful comments that improved this paper.
References
[1] | ABC13 Eyewitness News ((2020) ). Texas colleges and universities’ fall 2020 plans. Available at https://abc13.com/college-back-to-school-plan-university-online-fall-2020/6350823. |
[2] | Berger, J. O., & Pericchi, L. R. ((2001) ). Objective bayesian methods for model selection: introduction and comparison. Lecture Notes-Monograph Series, 38: 135-207. |
[3] | Centers for Disease Control and Prevention ((2020) ). COVID-19 pandemic planning scenarios. Available at https://www.cdc.gov/coronavirus/2019-ncov/hcp/planning-scenarios.html. |
[4] | Ghosh, P., Basu, S., & Tiwari, R. C. ((2009) ). Bayesian analysis of cancer rates from SEER program using parametric and semiparametric joinpoint regression models. Journal of the American Statistical Association, 104: (486), 439-452. |
[5] | Kafle, R. C., Khanal, N., & Tsokos, C. P. ((2014) ). Bayesian age-stratified joinpoint regression model: An application to lung and brain cancer mortality. Journal of Applied Statistics, 41: (12), 2727-2742. |
[6] | Keskinocak, P., Orac, B. E., Baxter, A., Asplund, J., & Serbun, N. ((2020) ). Impact of social distancing on COVID 19 spread: State of Georgia case study. PLoS One, 15(10). |
[7] | Linton, N. M., Kobayashi, T., Yang, Y., Hayashi, K., Akhmetzhanov, A. R., Jung, S. M., Yuan, B., Kinoshita, R., & Nishiura, H. ((2020) ). Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: A statistical analysis of publicly available case data. Journal of Clinical Medicine, 9: (2), 538. |
[8] | Martinez-Beneito, M. A., García-Donato, G., & Salmerón, D. ((2011) ). A Bayesian joinpoint regression model with an unknown number of break-points. Annals of Applied Statistics, 5: (3), 2150-2168. |
[9] | Svitek, P. ((2020) ). Gov. Greg Abbott urges voluntary measures to curb coronavirus spread but says closing Texas will be “the last option”. The Texas Tribune. Available at https://www.texastribune.org/2020/06/22/texas-coronavirus-greg-abbott-press-conference/. |
[10] | Swaby, A., & Platoff, E. ((2020) ). Many Texas students will return to classrooms Tuesday. Little will be normal. The Texas Tribune. Available at https://www.texastribune.org/2020/09/08/texas-schools-reopening-in-person/. |
[11] | Texas Department of State Health Services ((2020) a). Accessible version (Excel). Available at https://dshs.texas.gov/news/updates.shtm. |
[12] | Texas Department of State Health Services ((2020) b). Opening the state of Texas. Available at https://www.dshs.state.tx.us/coronavirus/opentexas.aspx. |
[13] | Yang, X., Yu, Y., Xu, J., Shu, H., Xia, J., Liu, H., Wu, Y., Zhang, L., Yu, Z., Fang, M., Yu, T., Wang, Y., Pan, S., Zou, X., Yuan, S., & Shang, Y. ((2020) ). Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet Respiratory Medicine, 8: (5), 475-481. |




