DNAJB6b is Downregulated in Synucleinopathies
Abstract
Background:
α-synuclein (α-syn) aggregation contributes to the progression of multiple neurodegenerative diseases. We recently found that the isoform b of the co-chaperone DNAJB6 is a strong suppressor of α-syn aggregation in vivo and in vitro. However, nothing is known about the role of the endogenous isoform b of DNAJB6 (DNAJB6b) in health and disease, due to lack of specific antibodies.
Objective:
Here we generated a novel anti-DNAJB6b antibody to analyze the localization and expression of this isoform in cells, in tissue and in clinical material.
Methods:
To address this we used immunocytochemistry, immunohistochemistry, as well as a novel quantitative DNAJB6 specific ELISA method.
Results:
The endogenous protein is mainly expressed in the cytoplasm and in neurites in vitro, where it is found more in dendrites than in axons. We further verified in vivo that DNAJB6b is expressed in the dopaminergic neurons of the substantia nigra pars compacta (SNpc), which is a neuronal subpopulation highly sensitive to α-syn aggregation, that degenerate to a large extend in patients with Parkinson’s disease (PD) and multiple system atrophy (MSA). When we analyzed the expression levels of DNAJB6b in brain material from PD and MSA patients, we found a downregulation of DNAJB6b by use of ELISA based quantification. Interestingly, this was also true when analyzing tissue from patients with progressive supranuclear palsy, a taupathic atypical parkinsonian disorder. However, the total level of DNAJB6 was upregulated in these three diseases, which may indicate an upregulation of the other major isoform of DNAJB6, DNAJB6a.
Conclusion:
This study shows that DNAJB6b is downregulated in several different neurodegenerative diseases, which makes it an interesting target to further investigate in relation to amyloid protein aggregation and disease progression.
INTRODUCTION
Synucleinopathies refer to neurodegenerative diseases characterized by accumulation of misfolded alpha-synuclein (α-syn) in neurons, nerve fibers or glial cells. These include Parkinson’s disease (PD), dementia with Lewy bodies (DLB), multiple system atrophy (MSA), and other, multiple rare forms of synucleinopathies [1]. The most common synucleinopathy is PD which affects more than 2%of people above the age of 65 years old in the western world. The cardinal motor symptoms of PD are primarily associated with the selective loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) region of the brain. This neurodegenerative process correlates with the formation of large protein-rich cytoplasmic inclusions, known as Lewy bodies (LBs), in which aggregated α-syn is the main protein component [2–5]. α-syn is widely expressed in brain cells and mainly localized at the presynaptic terminals, where it is suggested to play a role in synaptic vesicle dynamics and dopaminergic neurotransmission [6, 7]. The SNCA gene, which encodes α-syn, is also linked to familial forms of PD caused by gene duplication/triplication and missense mutations that result in increased aggregation of the protein. It is therefore believed that α-syn aggregation plays a key role in PD pathogenesis [3, 8, 9].
The misfolding and aggregation of α-syn may be the result of an age-dependent impairment of the cellular mechanisms maintaining proteostasis in which molecular chaperones play a crucial role [10, 11]. 70 kDa heat shock proteins (HSP70) are protective against protein aggregation and a series of in vitro and in vivo studies have indeed demonstrated that Hsp70 can ameliorate α-syn aggregation and α-syn-induced neurotoxicity [12–14]. Hsp70 chaperones promote folding of unfolded or misfolded proteins, in which co-chaperones such as the DNAJ/Hsp40 family of proteins (hereafter named the DNAJ proteins), play a crucial role [15, 16]. The DNAJ proteins can bind to misfolded or unfolded polypeptides, and transfer these to the Hsp70 chaperones, which in turn promote either folding of the polypeptide into its native structure or degradation [17, 18]. There are more than 40 DNAJ proteins encoded in the human genome [15, 19]. Three of these DNAJ genes have been linked to rare forms of PD. These are the genes DNAJB2 [20], DNAJC6 [21, 22], and DNAJC13 [23, 24]. The mechanisms behind their roles in PD are still unknown. In addition, DNAJB1 has been shown to disaggregate α-syn fibrils in vitro [25]. One additional DNAJ member, DNAJB6, is expressed in neurons, and has been found to be present in LBs of PD patients [26]. In our previous cell-based studies we found that DNAJB6 is important for suppression of α-syn aggregation. In addition, we showed that DNAJB6 prevents α-syn aggregation in a HSP70 dependent manner [27]. Importantly, we found that DNAJB6 isoform “b” (hereafter DNAJB6b) was responsible for suppressing α-syn aggregation and that the “a” isoform (hereafter DNAJB6a) was not involved [27]. The a and b isoforms of DNAJB6 share the first 7 exons of the DNAJB6 gene (encoding 231 aa), but has 3 and 1 unique exons, in addition. This results in that DNAJB6a and DNAJB6b are 326 and 241 aa long, respectively. In support of the role of DNAJB6 as a suppressor of α-syn aggregation, using an unbiased α-syn FRET system to quantify α-syn aggregation, we also found that DNAJB6 prevents α-syn preformed fibrils (PFFs) induced α-syn aggregation [28]. This is important, as α-syn seeding by PFFs in cellular and animal models of the disease replicates many of the features seen in PD [29, 30]. It has been demonstrated by multiple research labs, that DNAJB6 suppresses aggregation of multiple amyloid proteins in cells apart from α-syn [31–33] and recently it was demonstrated that it suppresses disease progression in a Huntington’s disease (HD) mouse model [34]. These data suggest that DNAJB6 may be a dominant suppressor of amyloid protein aggregation in the brain. Since we had observed that it is only DNAJB6b, and not DNAJB6a, that suppresses α-syn aggregation, we wanted to explore specifically if DNAJB6b is dysregulated in PD and other neurodegenerative movement disorders. Here we show that DNAJB6b is downregulated in PD as well as the atypical movement disorders, MSA and progressive supranuclear palsy (PSP), whereas total DNAJB6 was found to be upregulated in those diseases.
MATERIALS AND METHODS
DNAJB6b antibody generation
Polyclonal rabbit anti-DNAJB6b antibody was generated by innovagen (Lund; Sweden) in the following manner: Synthetic peptide DNAJB6 (232–240): (NH2-) CKEQLLRLDNK (-COOH) was coupled to KLH protein and agarose bead matrix by SH-group of N-terminal cysteine. The peptide conjugate was used for immunizing one rabbit with 5 doses over a 12-week period. Serum was collected in week 6, week 9, and week 12. Serum sample 3 was used for antibody purification. The serum was purified on Protein G followed by affinity purification on the peptide-linked agarose matrix. Bound antibody was eluted by Glycine pH 2.7 into Tris-containing vials. Antibody fraction was then buffer exchanged into phosphate buffered saline (PBS) by gel filtration.
Immunohistochemistry, immunocytochemistry and fluorescence microscopy
Immunohistochemistry
Mice were anesthetized by an overdose of sodium pentobarbital and perfused first with 12 mL/min of 0.9%NaCl for 1 min, followed by 12 mL/min of ice-cold 4%paraformaldehyde (PFA in 0.1 M PBS, pH 7.4) for 5 min. Brains were removed from skulls and post-fixed in 4%PFA overnight and then transferred into 25%sucrose for cryoprotection at 4°C until sunk to the bottom of vials. Subsequently, the brains were cut into 40αm thick 8 series of coronal sections on a freezing microtome (Leica, Germany) and stored in an anti-freeze solution (0.5 M PBS, 30%glycerol, 30%ethylene glycol) at –20°C. The free-floating sections were washed thrice with PBS to discard to anti-freeze solution. Tyrosine hydroxylase (TH) and DNAJB6b staining required an antigen retrieval process carried out for 30 min at 80°C using Tris/EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 9.0), This step was followed by thrice washes in PBS. The sections were then incubated in 0.025%Triton X-100 (Sigma-Aldrich) in PBS containing 5%of normal serums (Vector Laboratories Inc, USA) matching the species used to raise the secondary antibody for that proper staining for 1 h. Immunofluorescence stainings were performed with sheep anti-TH (Abcam, cat# ab113, 1:2,000) and rabbit anti-DNAJB6b (Novagen, 1:50) antibodies. Primary antibodies were diluted in PBS and overnight incubated at 4°C on the shaker (IKA Vibrax XFR, Germany). The next day, the brain sections were washed thrice with PBS for 5 min, then 1 h incubated with donkey Alexa-488 anti-rabbit (cat# 711-545-152, Jackson ImmunoResearch, 1:300) and Cy5 labeled donkey anti-sheep (Cat# 713-175-147, Jackson ImmunoResearch, 1:300) secondary antibodies. The sections were mounted on positive charged superfrost plus glass slides (Thermo Scientific, USA) and cover-slipped using PVA/DABCO. The stainings were visualized on a Leica SP8 laser-scanning confocal microscope (Leica, Germany).
Immunocytochemistry
Cell lines: HEK293 cells, that were initially plated on poly-D-lysine coated cover slips, were fixed in for 20 min in cold 4%PFA, followed by 3x washes in PBS. Subsequently, cells were incubated in PBS + 0.5%Triton-X-100, for 5 min, washed 3 times and then blocked with 5%BSA in PBS for 1 h at room temperature (RT). Next, the cells were incubated with rabbit anti-DNAJB6b (Novagen) or rabbit anti-DNAJB6 (cat# 11707-1-AP, Proteintech) for 1 h, washed in PBS and then stained with secondary alexa488 labeled donkey anti-rabbit antibody (cat# 711-545-152, Jackson ImmunoResearch) as well as the F-actin stain phalloidin-atto550 (cat# 19083, Sigma-Aldrich) for 1 h at RT. Subsequently, to washes, DAPI containing mounting medium (Cat# H-1200, vector laboratories inc.) was added and the fixed cells on cover slips were stored at 4°C, until they were analyzed by fluorescence microscopy using a Nikon eclipse 80i microscope.
Primary neurons: Cultured neurons at 19-20 DIV were fixed in 4%paraformaldehyde (PFA) in PBS with 0.12 M sucrose for 20 min, at RT. Cells were then blocked in 0.1%saponin (Sigma-Aldrich), 1%bovine serum albumin (BSA; Sigma-Aldrich), and 2%normal goat serum (NGS; Thermo Fisher Scientific) in PBS for 1 h at RT. Cells were incubated in primary antibodies rabbit anti-DNAJB6b (Novagen) or rabbit anti-DNAJB6 (cat# 11707-1-AP, Proteintech) and sheep anti-MAP2 antibody ((cat# ab92434, Abcam) diluted in PBS+2%NGS overnight at 4°C. Cells were rinsed in PBS and incubated with anti rabbit alexa 488 and anti-sheep Cy5 secondary antibodies (Thermo Fischer) and in 2%NGS in PBS. Cells were rinsed in PBS and counterstained with DAPI (Sigma-Aldrich, 1:2,000). Imaging was performed with an inverted Olympus IX70 epifluorescence or an inverted Leica SP8 confocal microscope.
Neuronal cell culture
Primary neurons were cultured from the cortices and hippocampi of wt C57black6 mice (Jackson Labs, Maine, USA) embryos at embryonic day 16 (E16). Neurons were cultured as previously described [35]. Briefly, pregnant mice were deeply anesthetized using isoflurane (MSD Animal Health, Sweden) and sacrificed. Embryos were quickly removed, and biopsies were taken for genotyping. Brains were dissected under constant cooling with chilled (∼4°C) Hanks balanced salt solution (HBSS; Thermo Fisher Scientific). Cortices and hippocampi were retrieved and incubated in 0.05%trypsin (Thermo Fisher Scientific), followed by 2 washes with HBSS. Brain tissue was then triturated in 10%fetal bovine serum (FBS) supplemented Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific) using glass pipettes until neurons were dissociated. Neurons were seeded onto glass coverslips in 24-well plates (Sarstedt, Germany), coated with Poly-D-lysine (Sigma-Aldrich). Neurons were plated with 10%FBS and 1%penicillin-streptomycin in DMEM; following 3–5 h incubation, media was exchanged for complete Neurobasal solution, consisting of Neurobasal medium, B27 supplement, penicillin-streptomycin and L-glutamine (Thermo Fisher Scientific). All animal experiments were performed in accordance with the ethical guidelines and were approved by the Animal Ethical Committee at Lund University ethical permit number 5.8.18-05983/2019.
Western blot
Wild type C57BL/6J mice brain was dissected to cortex, striatum, olfactory bulbs, hippocampus and stored at –80°C until they were used. The brain samples were sonicated (Branson SLPe, USA) in buffer (1%SDS, 0.1 M Tris-HCl pH 7.4, and 1 mM Ethylenediaminetetraacetic acid (EDTA), with 1:100 protease inhibitor cocktail (cat# P8340, Sigma-Aldrich). The tissue debris was spun down at 20,000 rcf for 30 min at 4°C, and the supernatant was collected. The protein concentration was measured by BCA assay (Pierce BCA Protein Assy Kit, Thermo Scientific, USA). Tissue lysates mixed with 25%4 x Laemmli buffer containing 10%of 0.1 M Dithiothreitol (DTT) and boiled for 5 min at 96°C. 5αg of proteins from brain lysate samples were separated on 10%SDS-PAGE polyacrylamide gels, then the samples transferred onto Polyvinylidene fluorid (PVDF) membranes using the Trans-Blot Turbo Transfer System (Bio-Rad, USA). The membrane was blocked with 5%skimmed milk powder dissolved in PBS containing 0.05%Tween (PBS-T) for 1 h at RT, after which they were washed thrice for 5 min with PBS-T at RT. Subsequently, the membrane incubated in 2%skimmed milk in PBS-T with either rabbit anti-DNAJB6 primary antibody (cat# 11707-1-AP, Proteintech, 1:1,000) or rabbit anti-DNAJB6b (Novagen, 1:200) overnight shaking in 4°C. The next day, the membrane was washed thrice in PBS-T for 5 min and then incubated shaking at RT for 1 h in 3%skimmed milk in PBS-T with anti-rabbit secondary antibody (cat# P0448, Dako, 1:5,000). The membrane was washed then thrice for 5 min in PBS-T. Western Blotting Luminol Reagent (Santa Cruz Biotechnology, USA) was used for chemiluminescence reactions and protein bands were developed using a ChemiDoc™ XRS+Molecular Imager (Bio-Rad, USA) and the Image Lab software (Bio-Rad, USA). After stripping the membrane for 15 min at RT (Restore Buffer, Thermo Fisher Scientific, USA), the membrane was washed twice with PBS-T for 5 min and then blocked in 5%skimmed milk in PBS-T for 30 min before being incubated with a HRP coupled anti-β-actin primary antibody (cat# A3854, Sigma Aldrich, 1:10,000) and developed as described above.
Patient samples
Samples from 59 human brains were included in this part of the study. The brains had been donated generously from the Brain Bank at Bispebjerg-Frederiksberg Hospital (University Hospital of Copenhagen, DK, approved under the Danish Data Protection Agency, j.no.: BFH-2017-001, I-Suite no.: 05190). Demographic information is summarized in Table 1. Detailed neuropathological evaluations were performed with written content from donor or next of kin. All participants gave written consent or inclusion in accordance with the World Medical Association Declaration of Helsinki. All brains have undergone pathological examinations to verify diagnosis through the presence of LBs in PD patients, glial cytoplasmic inclusions (GCIs) in MSA patients and deposition of neurofibrillary tangles in PSP patients. All brains have been collected and handled in accordance with Danish Ethical Standards of Brain Banks and the Danish Health and Medicine Authorities. All brain samples were stored at –80°C prior to usage. The study is approved by the Regional Ethical Committee, the Capital Region of Denmark (ethical permission j. no.: H-16037525).
Table 1
Demographic information
| Group | Age | Sex (Male/ Female) | PMI (h) |
| Controls | 81.4 (11.2) [58–96] | 6/6 | 15.6 (16.8) [4–61] |
| PD | 80.0 (8.2) [66–95] | 10/3 | 28.2 (21.2) [11–86] |
| MSA | 63.7 (6.1) [56–75]*, # | 4/9 | 46.8 (25.5) [22–96]* |
| PSP | 71.9 (8.2) [56–87]§ | 9/4 | 31.9 (17.7) [12–74] |
| p-value | < 0.001 | 0.079 | < 0.001 |
PD, Parkinson’s disease; MSA, multiple system atrophy; PSP, progressive supranuclear palsy; PMI, postmortem interval. *significantly different from controls, p < 0.001. #significantly different from PD patients p < 0.01. §significantly different from controls (p < 0.05). Age, One-way ANOVA with Tukey post hoc test. Sex, Chi-squared test. PMI, Kruskall-Wallis test with Dunn’s post hoc test.
Tissue handling and protein extraction from patient material
Approximately 100 mg of brain tissue from the dorsomedial prefrontal cortex (dPFC) was added to 1 mL N-PER™ Neuronal Protein Extraction Reagent (cat# 87792, Life Technologies) with pre-added HALT™ Phosphatase Inhibitor Cocktail mix (cat# 788420, Life Technologies, 1:100) and HALT™ Protease Inhibitor Cocktail mix (cat# 87786, Life Technologies, 1:100). Brain tissue samples were homogenized using MagNA Lyser Green Beads tubes (cat# 03358941001, Roche Diagnostics) and homogenized twice on MagNA Lyser instrument (Roche Diagnostics, CH) for 25 s at 6,000 RPM followed by instant cooling using the MagNA Lyser Rotor Cooling Block (cat# 03358976001, Roche Diagnostics) for 90 s. Homogenates were spun for 1 min at 10,000×g at 4°C to reduce foam. Samples were aliquoted at stored at –80°C.
Measurement of DNAJB6 and DNAJB6b using ELISA
In-house sandwich ELISA was used to evaluate the absolute amounts of DNAJB6 and DNAJB6b from brain homogenates of PD, MSA, and PSP patients, compared to normal controls. In details: 96-well polystyrene MaxiSorp plates (cat# 144531, Nunc) were coated in 4°C Carbonate Buffer 0.1 M, pH 9.4 (cat# C3041, Sigma-Aldrich) overnight with rabbit anti-human DNAJB6 (cat# 11707-1-AP, Proteintech, [1αg/mL]) or DNAJB6b (Novagen, 7αg/mL). Recombinant DNAJB6b was a kind gift from Professor Cecilia Emanuelsson (Center for Molecular Protein Science, Division of Biochemistry and Structural biology, Lund University), which was produced as described in [36, 37]. The plates were emptied and blocked with PBS pH 7.4 containing 3%BSA fraction V (cat# 10735094001, Sigma-Aldrich) and 0.1%Tergitol™ solution (cat# NP40S, Sigma-Aldrich) for 2 h at RT. Plates were then washed five consecutive times in 300αl/well PBS + 0.1%-Tween-20 (cat# P1379, Sigma-Aldrich) using a WellWash instrument (Thermo Scientific, USA). Human brain homogenates from PD, MSA, and PSP patients as well as controls were diluted in PBS + 0.1%BSA solution (DNAJB6 1:20; DNAJB6b 1:1) and added to the plates in duplicates and incubated for 1 h at RT. After a washing step, the plates were incubated at RT for 2 h with 50αl of mouse anti-human DNAJB6 antibody (cat# H00010049-M01, Novus Biologicals, DNAJB6: 500 ng/mL, DNAJB6b: 2,000 ng/mL). After an additional washing step, 50αl of goat anti-mouse HRP-conjugated antibody (cat# ab98717, Abcam, DNAJB6: 1:10,000, DNAJB6b: 1:5,000) was added to each well and incubated for 2 h at RT. After a last washing step, 50αL of tetramethylbenzidine (TMB) Liquid Peroxidase Substrate (cat# T8665, Sigma-Aldrich) was added and stopped after 30 min incubation in dark at RT with 50αL 0.5 N H2SO4. The absorbance at 450 nm was measured on a Fisher Scientific MultiskanTM FC Microplate Reader (Thermo Fischer Scientific, USA). Standard curves were applied using under the same conditions as described above using the peptide DNAJB6b starting at 2,000 ng/mL and serial diluted at -3x. The ODs were interpolated using sigmoidal, 4PL, curves. For each assay standard curve assessment and spike-and-recovery was performed (Supplementary Figure 1). Demonstrating the preciseness and accuracy of our in-house ELISA setup, the coefficient of variabilities (CVs) in percentages were calculated to 3.01%for DNAJB6 and 2.97%for DNAJB6b, which was concluded sufficiently precise.
Immunohistochemistry on patient material
Samples of putamen from 6 human brains (2 healthy controls, 2 PD patients, 2 MSA patients) were included in this study. Tissue blocks that included putamen were excised from the fresh brains. Samples were fixed for minimum 48 h in 10%buffered formalin (cat# 1000.5000, CellPath). Samples were then embedded in paraffin on a Leica ASP300 S tissue processor (Leica, DE) before sectioning on a sliding microtome at 5–7αm. On the day of immunoreactions, slides were deparaffinized through heating for 45 min at 60°C followed by washing in xylene (#28973.294, VWR) for 2×5 min, 99%EtOH for 2×2.5 min, 96%EtOH for 2 min, 70%EtOH for 2 min, and running water for 5 min. Samples were demasked in boiling pH 9 TEG-buffer (cat# 862338, RegionH Apoteket) for 15 min followed by cooling for 20 min and washed in pH 7 PBS 2×5 min before blocking in 10%FCS in PBS. Slices were incubated overnight at RT, respectively, with either rabbit anti-human DNAJB6 (cat# 11707-1-AP, Proteintech, 0.02αg/mL) or DNAJB6b (Novagen, 0.28αg/mL). Endogenous peroxidases were blocked using 3%H2O2 (cat# 212892, Apotekernes A.m.b.a, DNK) for 8 min. After washing, samples were incubated with EnVision + System anti-rabbit (cat# K003, Dako) for 1 h. Reactions were developed for 10–15 min using a solution consisting 0.25%3,3′-diaminobenzidine tetrahydrochloride hydrate (cat# D5637, Sigma-Aldrich) and 0.15%H2O2 in PBS. Following washing in dH2O for 5 min, nuclei were stained in Mayer’s hematoxylin (cat# 860213, RegionH Apoteket) for 40 s, and then washed in running dH2O for 10 min. Finally, samples were dehydrated in 70%EtOH, 96%EtOH for 2 min, 99%EtOH for 2×2.5 min, and xylene for 2×5 min. Cover slides were mounted using Pertex Mounting Medium (cat# SEA-0100-00A, CellPath). Slides were studied using an Olympus BX60 microscope.
Statistics
Data analyses were performed using GraphPad Prism v. 8.01 (GraphPad Software Inc., USA). Demographic differences were tested using one-way ANOVA with Tukey post hoc test, Kruskal-Wallis with Dunn’s post hoc test, and the chi-squared test. Normality was assessed using D’Agostino Omnibus test. Data was log10-transformed if not passed for normality. Outliers were identified using ROUT test at 1%discovery rate. For group comparisons, one-way ANOVA with Tukey post hoc test for multiple testing. Differences were significant at p < 0.05.
RESULTS
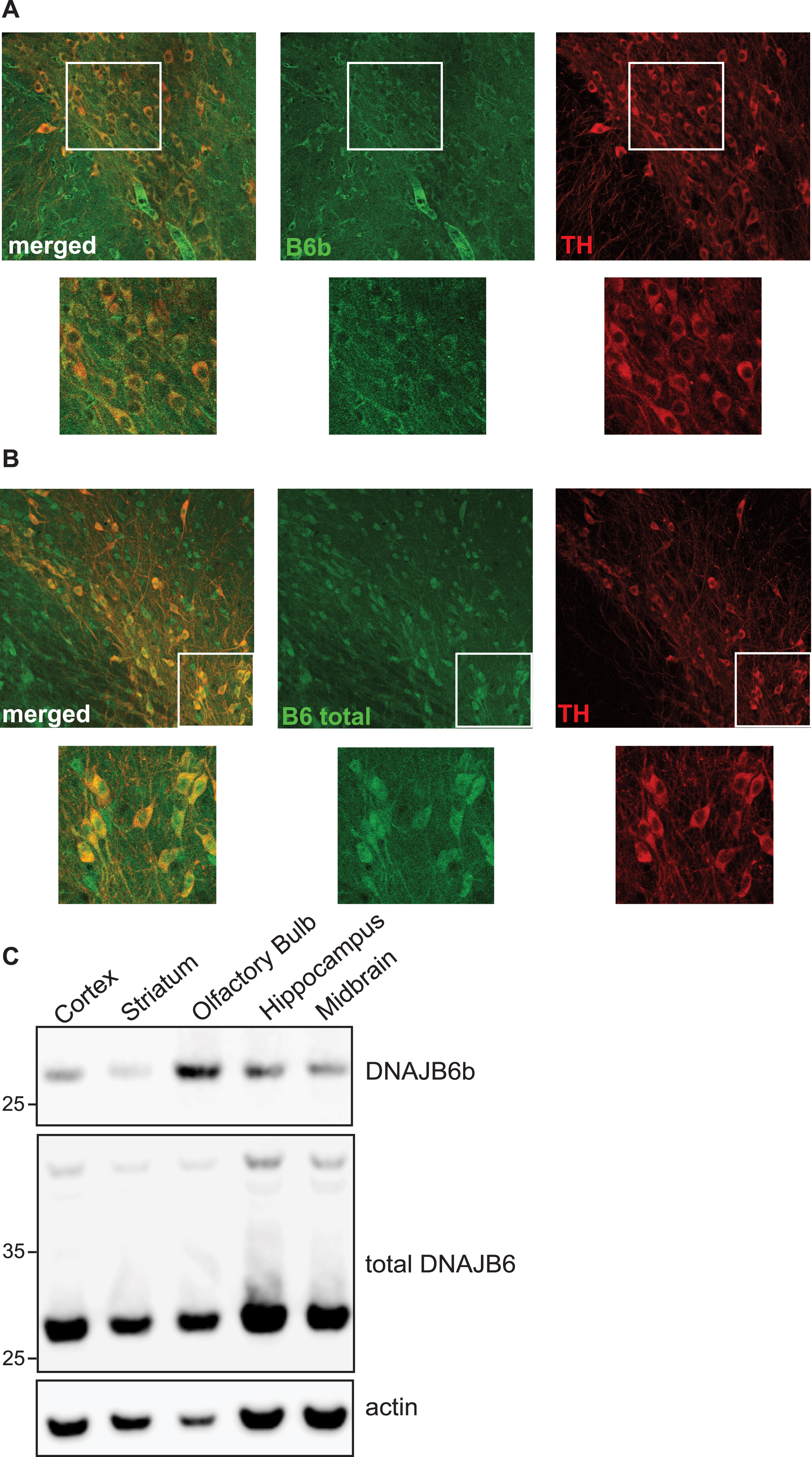
In order to assess the localization and expression of the DNAJB6b protein in cells and in tissues, we generated an antibody specific for DNAJB6b, which only has 10 amino acid residues at its C-terminus that are unique compared to the a-isoform (Fig. 1A). We tested this antibody in multiple cell lines using DNAJB6 KO cells previously generated, as a negative control [27] and found that the antibody was indeed specific for the DNAJB6b isoform (Fig. 1B). In addition, we also stained HEK293 wt cells or DNAJB6 KO HEK cells as a negative control and observed that the DNAJB6b isoform mainly localizes to the cytoplasm of cells, whereas as expected no staining could be seen in DNAJB6 KO cells, by immunocytochemistry (Fig. 1C). In comparison, staining of HEK293 wt cells with anti-(total) DNAJB6 antibody revealed a strong nuclear staining, which was expected as the DNAJB6a isoform carries a nucleus localization signal (Fig. 1A, D). Having determined the specificity of the antibody and the localization of the protein, we wanted to see how the DNAJB6b protein is localized in primary neurons, due to its potential important role as a suppressor of amyloid protein aggregation in neurons. We found that DNAJB6b was found to a large extend in the soma of neurons, but not in the nucleus (Fig. 2A). In addition, we observed that with regards to the projections from the primary neurons, DNAJB6b was found more in the dendrites than in the axons, as evaluated by staining with the anti-MAP2 antibody for dendritic localization (Fig. 2A, C). Indeed, the α-syn staining in proximity to DNAJB6b staining is presumably due to presynaptic terminals in proximity to dendrites (Fig. 2C), as it is well known that most of non-aggregated α-syn localize at the presynaptic terminals. In line with these results a staining for anti-(total)-DNAJB6 revealed similar localization results, albeit with slightly more staining of the nucleus, which we assume may be caused by the staining of DNAJB6a (Fig. 2B). The most common synucleinopathy, PD, is characterized by a loss of the dopaminergic neurons in the SNpc. Therefore, in order to know if DNAJB6b could potentially be protective in these neurons against α-syn aggregation, we wanted to know if the protein is expressed in these cells. Indeed, we observed that both DNAJB6b (Fig. 3A) and total DNAJB6 (Fig. 3B) is expressed in these cells as well as we observed that slightly more DNAJB6 was found in the nucleus, when we stained for total DNAJB6 (Fig. 3A, B). DNAJB6b and total DNAJB6 was furthermore found to be widely expressed in mouse brain, as revealed by western blotting analysis of mouse brain lysates (Fig. 3C).
Fig. 1
Expression of endogenous DNAJB6b as evaluated by western blot and immunocytochemistry. A) Illustration depicting the two major isoforms of DNAJB6 and their domains. nls, Nucleus Localization Signal. B) Expression of DNAJB6b in lysates from cell lines, analyzed by probing membranes with anti-DNAJB6b and HRP conjugated anti-rabbit antibodies. The membrane was probed with anti-total DNAJB6 and anti-actin as a control. C) DNAJB6 KO HEK 293 cells or wt HEK293 cells were probed with anti-DNAJB6b antibody, phalloidin-Alexa 547 as well as secondary anti-rabbit Alexa 488 coupled antibody. D) wt HEK293 cells were probed with anti- total DNAJB6 antibody, phalloidin-alexa 547 as well as secondary anti-rabbit alexa 488 coupled antibody.
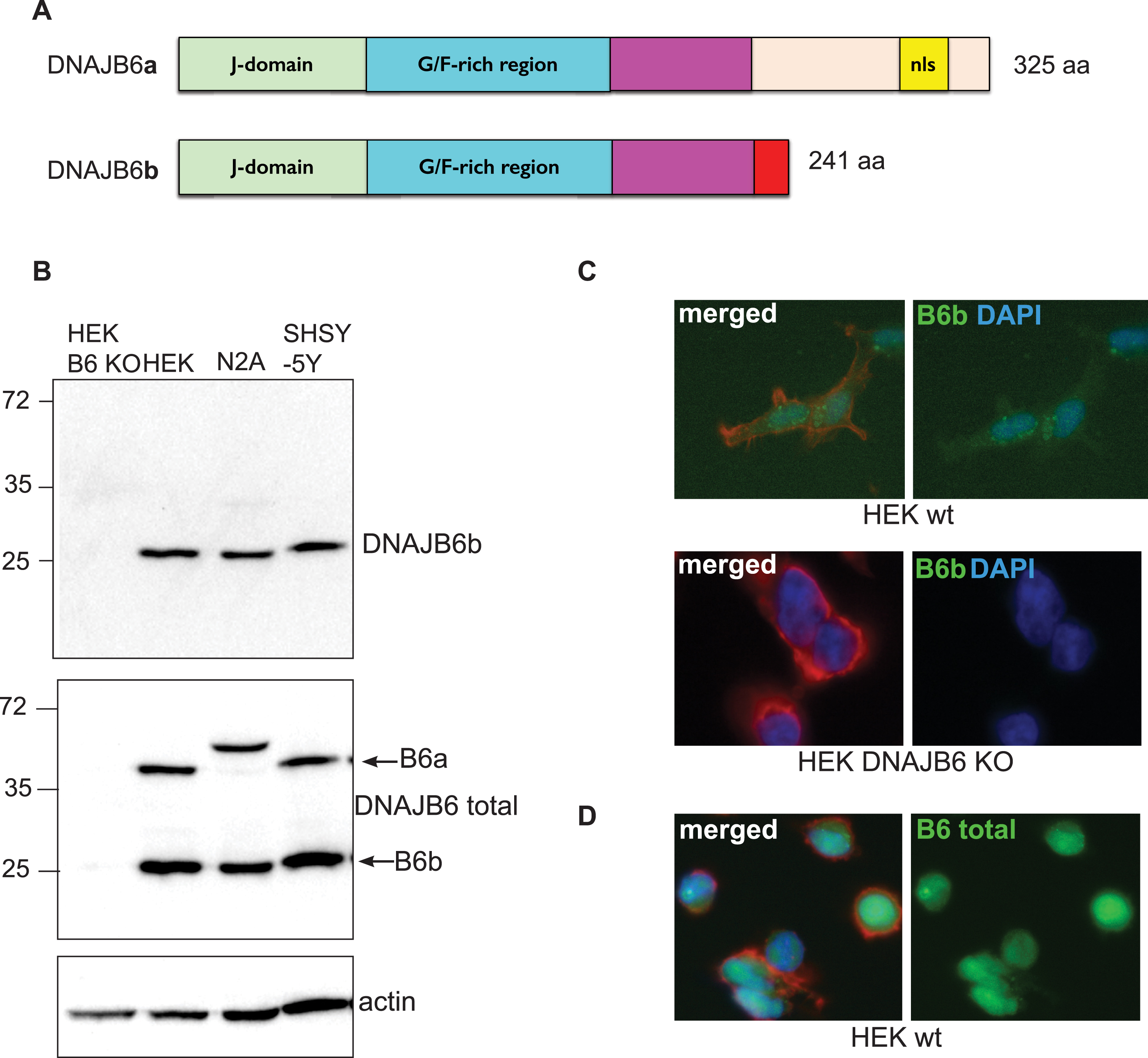
Fig. 2
DNAJB6b and total DNAJB6 are expressed in primary neurons. Fixed primary mouse neurons stained with anti-DNAJB6b (A) or anti-total DNAJB6 (B) as well as anti-MAP2, anti-α-syn and dapi and fluorescently labeled anti rabbit Alexa 488 and anti sheep Cy5 antibodies. C) Depicts a close up picture of a dendrite stained with antibodies against α-syn, MAP2, and DNAJB6b as well as secondary fluorescently labeled anti-rabbit and anti-sheep antibodies.
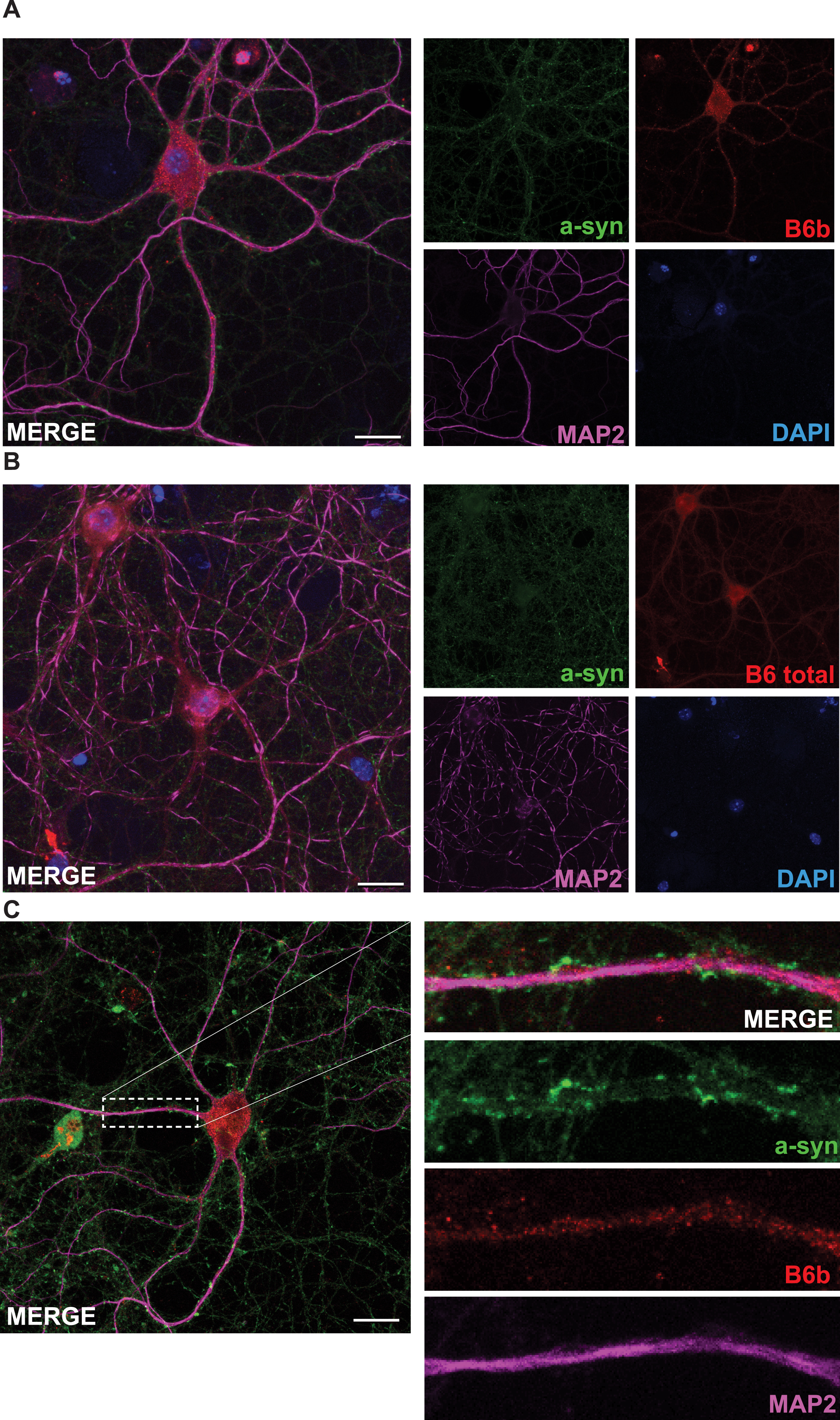
Fig. 3
DNAJB6b and total DNAJB6 are expressed in SNpc. 40 uM coronal sections from wt C57black6 mice containing SNpc, were probed with anti-DNAJB6b (A) or anti-total DNAJB6 (B) as well as anti-rabbit alexa 488 and anti-TH and well as anti sheep Cy5. C) Expression of DNAJB6b in brain lysates from different regions of mouse brain, as analyzed by probing membranes with anti-DNAJB6b, anti-total-DNAJB6 and HRP conjugated anti-rabbit antibody as well as HRP conjugated anti-actin antibody.
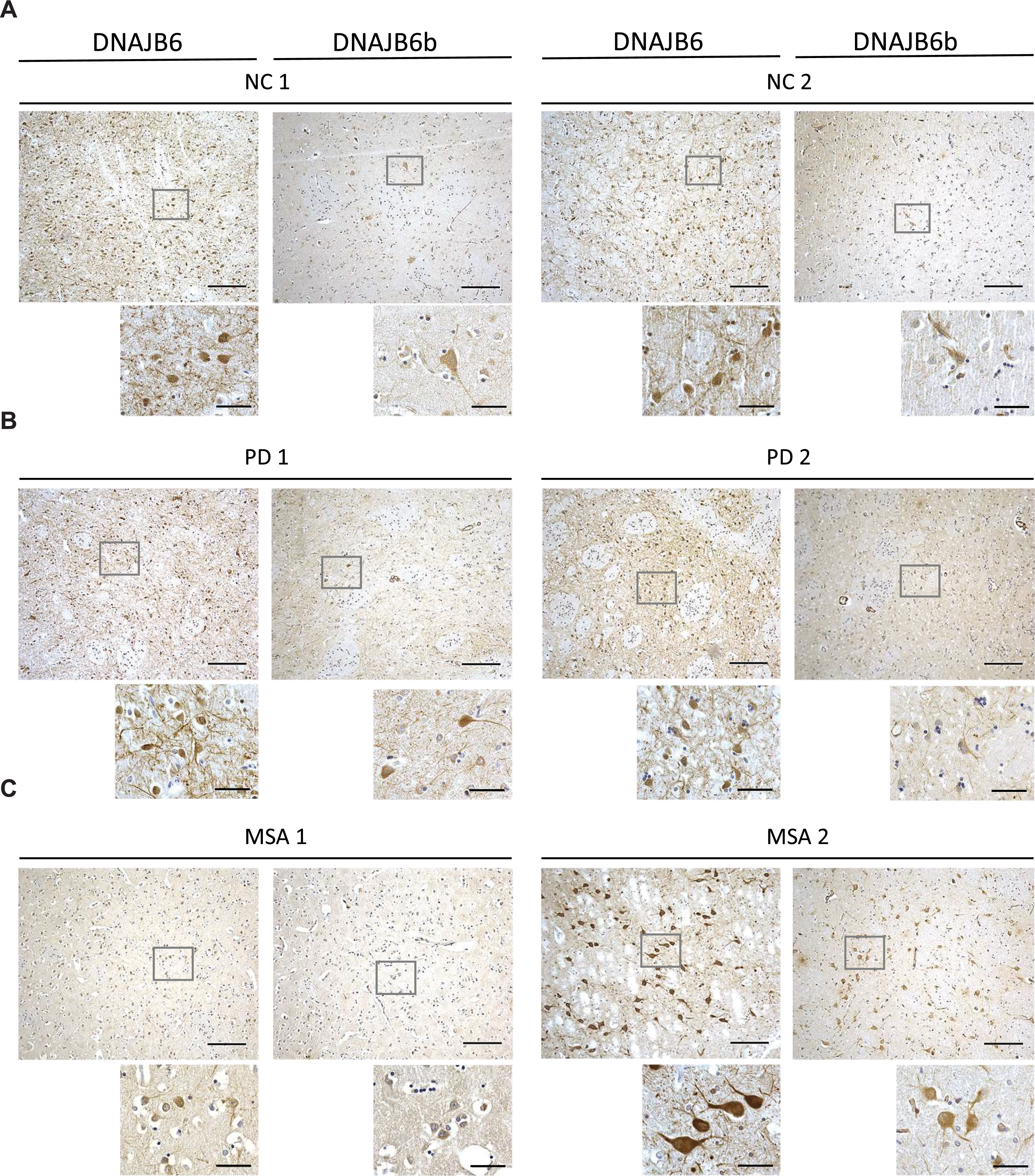
Due to the semi-quantitative nature of immunohistochemical staining we wanted to establish a more sensitive way to address if this protein could be dysregulated in a more disease environment. By use of recombinant DNAJB6b and commercially available anti-DNAJB6 antibodies, we established an ELISA based set up to measure DNAJB6 (Supplementary Figure 1A, C) and DNAJB6b (Supplementary Figure 1B, D) concentrations. We tested a cohort of postmortem brain material from patients with PD, MSA, PSP, and control subjects. Total levels of DNAJB6 in putamen samples were significantly different between the groups (F(3) = 6.2; p = 0.001) and found to be increased in PD (p = 0.048), MSA (p = 0.001), and PSP (p = 0.009) patients compared to normal controls (Fig. 4A). Inversely, the total levels of DNAJB6b were significantly different between groups (F(3) = 4.3; p = 0.009) and found to be reduced in PD (p = 0.048), MSA (p = 0.009), and PSP (p = 0.041) patients compared to normal controls (Fig. 4B). Further, representative immunohistochemical stainings from 2 normal controls, 2 PD, and 2 MSA patients (Fig. 5A-5C respectively) with anti-(total) DNAJB6 or anti-DNAJB6b antibodies, suggests that this dysregulation may largely be occurring in neurons rather than glial cells of patients. Western blot analysis of brain samples from control subjects (NC), PD, MSA, or PSP patients showed that the antibodies are specific for DNAJB6b and total DNAJB6 in human brain tissue samples as well (Supplementary Figure 2).
Fig. 4
Levels of brain DNAJB6 and DNAJB6b in PD, MSA, and PSP patients compared to normal controls. ELISA results of brain homogenate levels extracted from dorsomedial prefrontal cortex of DNAJB6 and DNAJB6b from 13 Parkinson’s disease (PD), 13 multiple system atrophy (MSA), and 13 progressive supranuclear ophthalmoplegia/palsy (Steele-Richardson-Olszewski disease) (PSP) patients compared to 12 normal controls (NC). Bar graph shows amount of proteins, DNAJB6 and DNAJB6b, in ng/mL. Data depicts mean±SEM. Data was analyzed using one-way ANOVA with Tukey post hoc test for multiple testing. Only significant p-values are shown. p-values below 0.05 were considered significant.
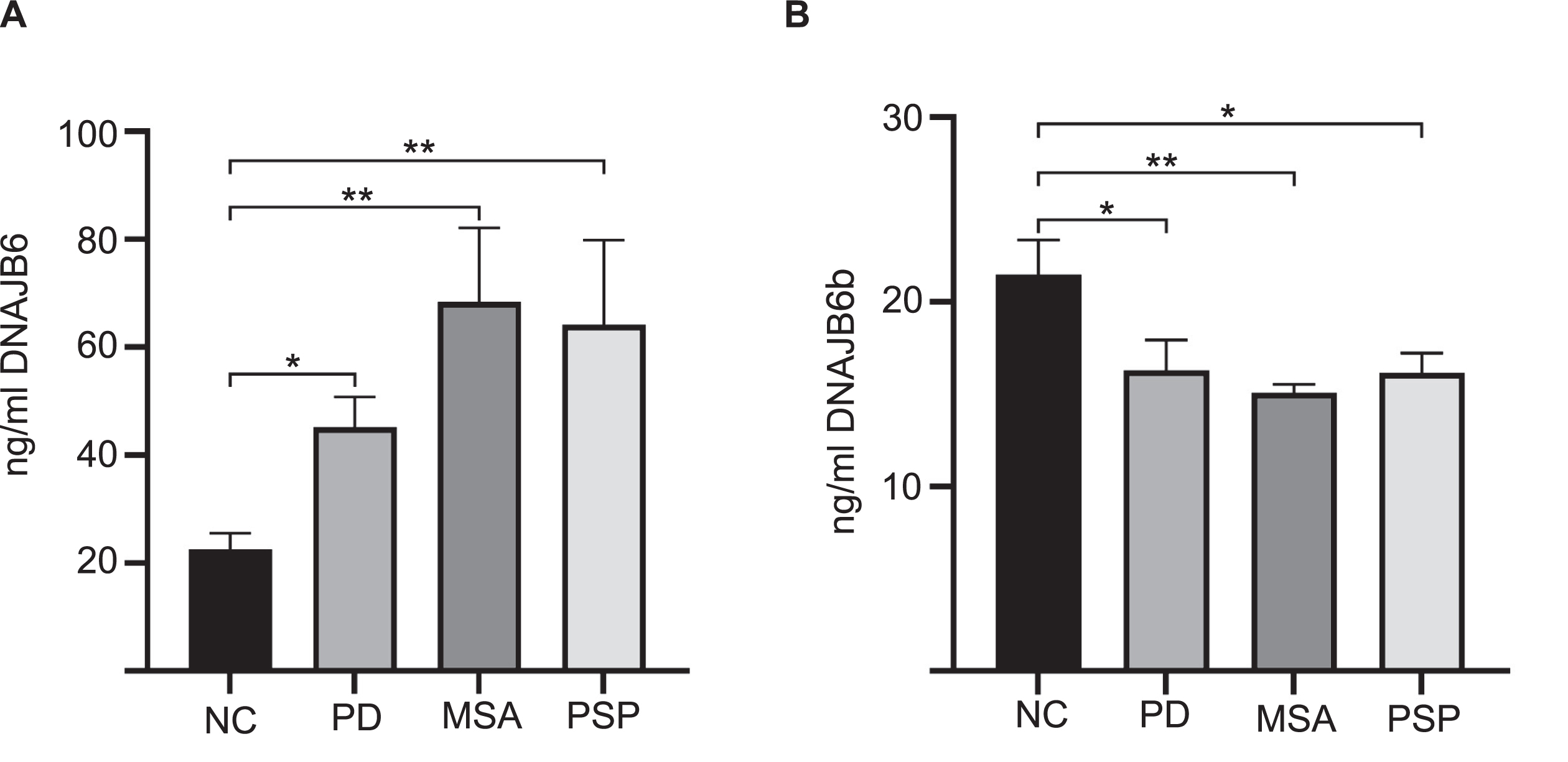
Fig. 5
Micrographs showing DNAJB6 and DNJB6B stained sections of the putamen from (A) 2 normal controls, (B) 2 PD patients, and (C) 2 MSA patients at ×10 (large pictures, scale bar = 200αm) and ×60 (small pictures, scale bar = 50αm) magnification.
DISCUSSION
Here we demonstrate that DNAJB6b is downregulated in PD, MSA, and PSP, whereas the total level of DNAJB6 protein was found to be upregulated in these diseases. One study has prior to this investigated the dysregulation of DNAJB6 in neurodegenerative diseases. In this study by Durrenberger et al., the total level of DNAJB6 was found to be upregulated in PD patients [26], which is in line with the results presented here. Durrenberger et al. further reported that the increase in DNAJB6 was due to glial increase. This is consistent with our results that MSA patients showed significantly higher amount of total DNAJB6, since α-syn aggregations are present in glial cells in GCIs in MSA and further suffers from high amount of gliosis [38]. However, our study is the first to describe the specific dysregulation of the DNAJB6b-isoform as well as total DNAJB6 in MSA and PSP patients (Figs. 4 and 5). Multiple studies have used plasmid induced overexpression of cDNA to address the localization of DNAJB6b isoform, and found that DNAJB6b is predominantly cytoplasmic, while DNAJB6a is localized in the nucleus [27, 39]. Our studies suggest that while DNAJB6b is indeed found in the cytoplasm in cell lines, primary neurons and dopaminergic neurons, it is also to some extend found inside the nucleus (Figs. 1–3). Why the suppression α-syn aggregation is specifically only caused by the b-isoform is still not known, but we strongly suspect that this is due to that DNAJB6b is found mainly in the cytoplasm whereas DNAJB6a is localized almost exclusively in the nucleus. The alternative splicing and the specific 10 amino acid residues unique to DNAJB6b are strongly conserved through evolution (Supplementary Fig. 3) suggesting that the isoforms most likely have important roles in cells, that are different from each other. Therefore, it is possible that the DNAJB6b and DNAJB6a found inside the nucleus of cells has other roles, which may be suppressing aggregation of nuclear localized proteins. Indeed, some amyloid proteins, including huntingtin, are found to a higher extend inside the nucleus, in contrast to α-syn, which is found at only at very sparse levels inside the nucleus. In addition, recent studies of biophysical properties suggest that the C-terminal domain of DNAJB6b, which is different from the C-terminal domain of DNAJB6a isoform, self-associates to form large oligomers through the C-terminal domain (CTD) which may be important for substrate binding and how it suppresses amyloid protein aggregation [40].
The finding that DNAJB6b is specifically downregulated in PD and MSA fits with that α-syn aggregates faster in the absence of DNAJB6b, and gives us a clue that this phenomenon that we have seen in cells [27, 28], in vitro as well as in an animal model of PD (Arkan et al., manuscript submitted), may have a clinical impact as well. In summary, these results highlight that drugs that aim to enhance expression or activity of DNAJB6b could be a promising target for neurodegenerative disease therapies.
ACKNOWLEDGMENTS
This study was supported by the Parkinson Foundation (Sweden), CH as well as the Stiftelsen Olle Enqvist Byggmästare (Sweden), SA and CH.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JPD-202512.
REFERENCES
[1] | Goedert M , Jakes R , Spillantini MG ((2017) ) The synucleinopathies: twenty years on. J Parkinsons Dis 7: , 69. |
[2] | Spillantini MG , Schmidt ML , Lee VM , Trojanowski JQ , Jakes R , Goedert M ((1997) ) Alpha-synuclein in Lewy bodies. Nature 388: , 839–840. |
[3] | Vekrellis K , Xilouri M , Emmanouilidou E , Rideout HJ , Stefanis L ((2011) ) Pathological roles of alpha-synuclein in neurological disorders. Lancet Neurol 10: , 1015–1025. |
[4] | Angot E , Steiner JA , Hansen C , Li JY , Brundin P ((2010) ) Are synucleinopathies prion-like disorders? Lancet Neurol 9: , 1128–1138. |
[5] | Hansen C , Li JY ((2012) ) Beyond alpha-synuclein transfer: pathology propagation in Parkinson’s disease. Trends Mol Med 18: , 248–255. |
[6] | Logan T , Bendor J , Toupin C , Thorn K , Edwards RH ((2017) ) alpha-Synuclein promotes dilation of the exocytotic fusion pore. Nat Neurosci 20: , 681–689. |
[7] | Burre J , Sharma M , Tsetsenis T , Buchman V , Etherton MR , Sudhof TC ((2010) ) Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329: , 1663–1667. |
[8] | Polymeropoulos MH , Lavedan C , Leroy E , Ide SE , Dehejia A , Dutra A , Pike B , Root H , Rubenstein J , Boyer R , Stenroos ES , Chandrasekharappa S , Athanassiadou A , Papapetropoulos T , Johnson WG , Lazzarini AM , Duvoisin RC , Di Iorio G , Golbe LI , Nussbaum RL ((1997) ) Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276: , 2045–2047. |
[9] | Goedert M , Spillantini MG , Del Tredici K , Braak H ((2013) ) 100 years of Lewy pathology. Nat Rev Neurol 9: , 13–24. |
[10] | Witt SN ((2013) ) Molecular chaperones, alpha-synuclein, and neurodegeneration. Mol Neurobiol 47: , 552–560. |
[11] | De Mattos EP , Wentink A , Nussbaum-Krammer C , Hansen C , Bergink S , Melki R , Kampinga HH ((2020) ) Protein quality control pathways at the crossroad of synucleinopathies. J Parkinsons Dis 10: , 369–382. |
[12] | Aprile FA , Sormanni P , Vendruscolo M ((2015) ) A rational design strategy for the selective activity enhancement of a molecular chaperone toward a target substrate. Biochemistry 54: , 5103–5112. |
[13] | Gao X , Carroni M , Nussbaum-Krammer C , Mogk A , Nillegoda NB , Szlachcic A , Guilbride DL , Saibil HR , Mayer MP , Bukau B ((2015) ) Human Hsp70 disaggregase reverses Parkinson’s-linked alpha-synuclein amyloid fibrils. Mol Cell 59: , 781–793. |
[14] | McLean PJ , Klucken J , Shin Y , Hyman BT ((2004) ) Geldanamycin induces Hsp70 and prevents alpha-synuclein aggregation and toxicity in vitro. Biochem Biophys Res Commun 321: , 665–669. |
[15] | Qiu XB , Shao YM , Miao S , Wang L ((2006) ) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 63: , 2560–2570. |
[16] | Mogk A , Bukau B , Kampinga HH ((2018) ) Cellular handling of protein aggregates by disaggregation machines. Mol Cell 69: , 214–226. |
[17] | Summers DW , Douglas PM , Ramos CH , Cyr DM ((2009) ) Polypeptide transfer from Hsp40 to Hsp70 molecular chaperones. Trends Biochem Sci 34: , 230–233. |
[18] | Summers DW , Wolfe KJ , Ren HY , Cyr DM ((2013) ) The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One 8: , e52099. |
[19] | Zarouchlioti C , Parfitt DA , Li W , Gittings LM , Cheetham ME ((2018) ) DNAJ proteins in neurodegeneration: essential and protective factors. Philos Trans R Soc Lond B Biol Sci 373: , 20160534. |
[20] | Sanchez E , Darvish H , Mesias R , Taghavi S , Firouzabadi SG , Walker RH , Tafakhori A , Paisan-Ruiz C ((2016) ) Identification of a large DNAJB2 deletion in a family with spinal muscular atrophy and parkinsonism. Hum Mutat 37: , 1180–1189. |
[21] | Olgiati S , Quadri M , Fang M , Rood JP , Saute JA , Chien HF , Bouwkamp CG , Graafland J , Minneboo M , Breedveld GJ , Zhang J , International Parkinsonism Genetics Network, Verheijen FW , Boon AJ , Kievit AJ , Jardim LB , Mandemakers W , Barbosa ER , Rieder CR , Leenders KL , Wang J , Bonifati V ((2016) ) DNAJC6 mutations associated with early-onset Parkinson’s disease. Ann Neurol 79: , 244–256. |
[22] | Elsayed LE , Drouet V , Usenko T , Mohammed IN , Hamed AA , Elseed MA , Salih MA , Koko ME , Mohamed AY , Siddig RA , Elbashir MI , Ibrahim ME , Durr A , Stevanin G , Lesage S , Ahmed AE , Brice A ((2016) ) A novel nonsense mutation in DNAJC6 expands the phenotype of autosomal-recessive juvenile-onset Parkinson’s disease. Ann Neurol 79: , 335–337. |
[23] | Yoshida S , Hasegawa T , Suzuki M , Sugeno N , Kobayashi J , Ueyama M , Fukuda M , Ido-Fujibayashi A , Sekiguchi K , Ezura M , Kikuchi A , Baba T , Takeda A , Mochizuki H , Nagai Y , Aoki M ((2018) ) Parkinson’s disease-linked DNAJC13 mutation aggravates alpha-synuclein-induced neurotoxicity through perturbation of endosomal trafficking. Hum Mol Genet 27: , 823–836. |
[24] | Vilarino-Guell C , Rajput A , Milnerwood AJ , Shah B , Szu-Tu C , Trinh J , Yu I , Encarnacion M , Munsie LN , Tapia L , Gustavsson EK , Chou P , Tatarnikov I , Evans DM , Pishotta FT , Volta M , Beccano-Kelly D , Thompson C , Lin MK , Sherman HE , Han HJ , Guenther BL , Wasserman WW , Bernard V , Ross CJ , Appel-Cresswell S , Stoessl AJ , Robinson CA , Dickson DW , Ross OA , Wszolek ZK , Aasly JO , Wu RM , Hentati F , Gibson RA , McPherson PS , Girard M , Rajput M , Rajput AH , Farrer MJ ((2014) ) DNAJC13 mutations in Parkinson disease. Hum Mol Genet 23: , 1794–1801. |
[25] | Wentink AS , Nillegoda NB , Feufel J , Ubartaite G , Schneider CP , De Los Rios P , Hennig J , Barducci A , Bukau B ((2020) ) Molecular dissection of amyloid disaggregation by human HSP70. Nature 587: , 483–488. |
[26] | Durrenberger PF , Filiou MD , Moran LB , Michael GJ , Novoselov S , Cheetham ME , Clark P , Pearce RK , Graeber MB ((2009) ) DnaJB6 is present in the core of Lewy bodies and is highly up-regulated in parkinsonian astrocytes. J Neurosci Res 87: , 238–245. |
[27] | Aprile FA , Kallstig E , Limorenko G , Vendruscolo M , Ron D , Hansen C ((2017) ) The molecular chaperones DNAJB6 and Hsp70 cooperate to suppress alpha-synuclein aggregation. Sci Rep 7: , 9039. |
[28] | Deshayes N , Arkan S , Hansen C ((2019) ) The molecular chaperone DNAJB6, but not DNAJB1, suppresses the seeded aggregation of alpha-synuclein in cells. Int J Mol Sci 20: , 4495. |
[29] | Luk KC , Kehm V , Carroll J , Zhang B , O’Brien P , Trojanowski JQ , Lee VM ((2012) ) Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338: , 949–953. |
[30] | Rey NL , Steiner JA , Maroof N , Luk KC , Madaj Z , Trojanowski JQ , Lee VM , Brundin P ((2016) ) Widespread transneuronal propagation of alpha-synucleinopathy triggered in olfactory bulb mimics prodromal Parkinson’s disease. J Exp Med 213: , 1759–1778. |
[31] | Rodríguez-González C , Lin S , Arkan S , Hansen C ((2020) ) Co-chaperones DNAJA1 and DNAJB6 are critical for regulation of polyglutamine aggregation. Sci Rep 10: , 8130. |
[32] | Gillis J , Schipper-Krom S , Juenemann K , Gruber A , Coolen S , van den Nieuwendijk R , van Veen H , Overkleeft H , Goedhart J , Kampinga HH , Reits EA ((2013) ) The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J Biol Chem 288: , 17225–17237. |
[33] | Hussein RM , Hashem RM , Rashed LA ((2015) ) Evaluation of the amyloid beta-GFP fusion protein as a model of amyloid beta peptides-mediated aggregation: a study of DNAJB6 chaperone. Front Mol Neurosci 8: , 40. |
[34] | Kakkar V , Mansson C , de Mattos EP , Bergink S , van der Zwaag M , van Waarde MA , Kloosterhuis NJ , Melki R , van Cruchten RT , Al-Karadaghi S , Arosio P , Dobson CM , Knowles TP , Bates GP , van Deursen JM , Linse S , van de Sluis B , Emanuelsson C , Kampinga HH ((2016) ) The S/T-rich motif in the DNAJB6 chaperone delays polyglutamine aggregation and the onset of disease in a mouse model. Mol Cell 62: , 272–283. |
[35] | Willen K , Sroka A , Takahashi RH , Gouras GK ((2017) ) Heterogeneous association of Alzheimer’s disease-linked amyloid-beta and amyloid-beta protein precursor with synapses. J Alzheimers Dis 60: , 511–524. |
[36] | Mansson C , Arosio P , Hussein R , Kampinga HH , Hashem RM , Boelens WC , Dobson CM , Knowles TP , Linse S , Emanuelsson C ((2014) ) Interaction of the molecular chaperone DNAJB6 with growing amyloid-beta 42 (Abeta42) aggregates leads to sub-stoichiometric inhibition of amyloid formation. J Biol Chem 289: , 31066–31076. |
[37] | Mansson C , van Cruchten RTP , Weininger U , Yang X , Cukalevski R , Arosio P , Dobson CM , Knowles T , Akke M , Linse S , Emanuelsson C ((2018) ) Conserved S/T residues of the human chaperone DNAJB6 are required for effective inhibition of Abeta42 amyloid fibril formation. Biochemistry 57: , 4891–4902. |
[38] | Salvesen L , Winge K , Brudek T , Agander TK , Lokkegaard A , Pakkenberg B ((2017) ) Neocortical neuronal loss in patients with multiple system atrophy: a stereological study. Cereb Cortex 27: , 400–410. |
[39] | Pei Y , Fu W , Yang E , Shen A , Chen YC , Gong H , Chen J , Huang J , Xiao G , Liu F ((2012) ) A Hsp40 chaperone protein interacts with and modulates the cellular distribution of the primase protein of human cytomegalovirus. PLoS Pathog 8: , e1002968. |
[40] | Karamanos TK , Tugarinov V , Clore GM ((2019) ) Unraveling the structure and dynamics of the human DNAJB6b chaperone by NMR reveals insights into Hsp40-mediated proteostasis. Proc Natl Acad Sci U S A 116: , 21529–21538. |




