Multiple Pathways of LRRK2-G2019S/Rab10 Interaction in Dopaminergic Neurons
Abstract
Background:
Inherited mutations in the LRRK2 protein are common causes of Parkinson’s disease, but the mechanisms by which increased kinase activity of mutant LRRK2 leads to pathological events remain to be determined. In vitro assays (heterologous cell culture, phospho-protein mass spectrometry) suggest that several Rab proteins might be directly phosphorylated by LRRK2-G2019S. An in vivo screen of Rab expression in dopaminergic neurons in young adult Drosophila demonstrated a strong genetic interaction between LRRK2-G2019S and Rab10.
Objective:
To determine if Rab10 is necessary for LRRK2-induced pathophysiological responses in the neurons that control movement, vision, circadian activity, and memory. These four systems were chosen because they are modulated by dopaminergic neurons in both humans and flies.
Methods:
LRRK2-G2019S was expressed in Drosophila dopaminergic neurons and the effects of Rab10 depletion on Proboscis Extension, retinal neurophysiology, circadian activity pattern (‘sleep’), and courtship memory determined in aged flies.
Results:
Rab10 loss-of-function rescued LRRK2-G2019S induced bradykinesia and retinal signaling deficits. Rab10 knock-down, however, did not rescue the marked sleep phenotype which results from dopaminergic LRRK2-G2019S. Courtship memory is not affected by LRRK2, but is markedly improved by Rab10 depletion. Anatomically, both LRRK2-G2019S and Rab10 are seen in the cytoplasm and at the synaptic endings of dopaminergic neurons.
Conclusion:
We conclude that, in Drosophila dopaminergic neurons, Rab10 is involved in some, but not all, LRRK2-induced behavioral deficits. Therefore, variations in Rab expression may contribute to susceptibility of different dopaminergic nuclei to neurodegeneration seen in people with Parkinson’s disease.
INTRODUCTION
Mutations in the LRRK2 gene are the most frequent genetic cause of late onset Parkinson’s disease. The G2019S mutation increases LRRK2 kinase activity [1], leading to a toxic cascade that kills dopaminergic neurons in the substantia nigra. This results in bradykinesia and sleep disturbances, while loss of dopaminergic amacrine cells in the retina contributes to visual deficits. However, the main steps between LRRK2 and these pathological outcomes remain to be determined. One of the first steps that has been suggested is that LRRK2-G2019S may phosphorylate a number of small GTPases (Rabs 3,5,8,10,29,35 & 43) [2]. Cell culture experiments have shown a particular effect on Rab10 [2–7] (see [8] for review), while a visual screen of Drosophila synaptic processing indicated a particular synergy between Rab10 and G2019S [9]. As a molecular switch, phosphorylation of a Rab protein could result in changed effector binding with a series of downstream consequences. However, the level of Rab10 is quite disparate, even among different types of dopaminergic neurons [9, 10] so that LRRK2 might not work in the same way in all dopaminergic neurons.
Since most genetic mouse models of Parkinson’s disease have very weak phenotypes, with lack of neuropathology [11], we turned to the fly where manipulation of Parkinson’s disease-related genes leads to marked phenotypes: bradykinesia [12], loss of dopaminergic neurons [13], tremor [14], retinal degeneration [15], and sleep disorder [16] (for review, see [17]). Parkinson’s disease-linked genes shown to have powerful effects in the fly include not just LRRK2, but also α-synuclein [12], parkin and PINK1 [18, 19], and DJ1 [20]. The advanced genetics, short lifespan, and high fecundity have allowed the identification of genes that are in the same pathway and thus show epistasis (e.g., parkin and PINK1 [18, 19]).
To determine how important Rab10 is in the pathological cascade initiated by LRRK2 activity, we now deploy fly genetics test the knock-down/knock-out of Rab10 in dopaminergic neurons in the intact organism. Flies use dopaminergic neurons in four physiologically measurable systems: movement, vision, sleep, and memory. We find the movement and visual deficits seen in flies expressing LRRK2-G2019S in dopaminergic neurons are rescued by the depletion of Rab10. However, the marked sleep phenotype caused by dopaminergic expression of LRRK2-G2019S is not affected by manipulation of Rab10. Conditioned courtship memory is not affected by LRRK2, though dopaminergic Rab10 reduction markedly improves this. We therefore find a role for Rab10 downstream of activated LRRK2 in some, but not all, dopaminergic neurons in the activation of LRRK2-associated physiological deficits in Drosophila.
MATERIALS AND METHODS
Flies
All flies tested were male Drosophila melanogaster, using TH-GAL4 (kind gift of Serge Birman) to manipulate dopaminergic neurons or nSyb-GAL4 (from Julie Simpson (corresponding Bloomington stock 51635)) for pan-neuronal expression. Gr5a-LexA was used to express the LexOp-ReachR channelrhodopsin in the sugar sensitive neurons independently from the GAL4-UAS manipulations. Flies were raised and crossed using standard Drosophila protocols [9]. The full list of fly lines is in Supplementary Table 1.
To investigate a role for Rab10 in dopaminergic neurons, downstream of activated LRRK2, we recombined the TH-GAL4 with UAS-G2019S and designated these ‘PD-mimic’ flies as THG2.
Western blots
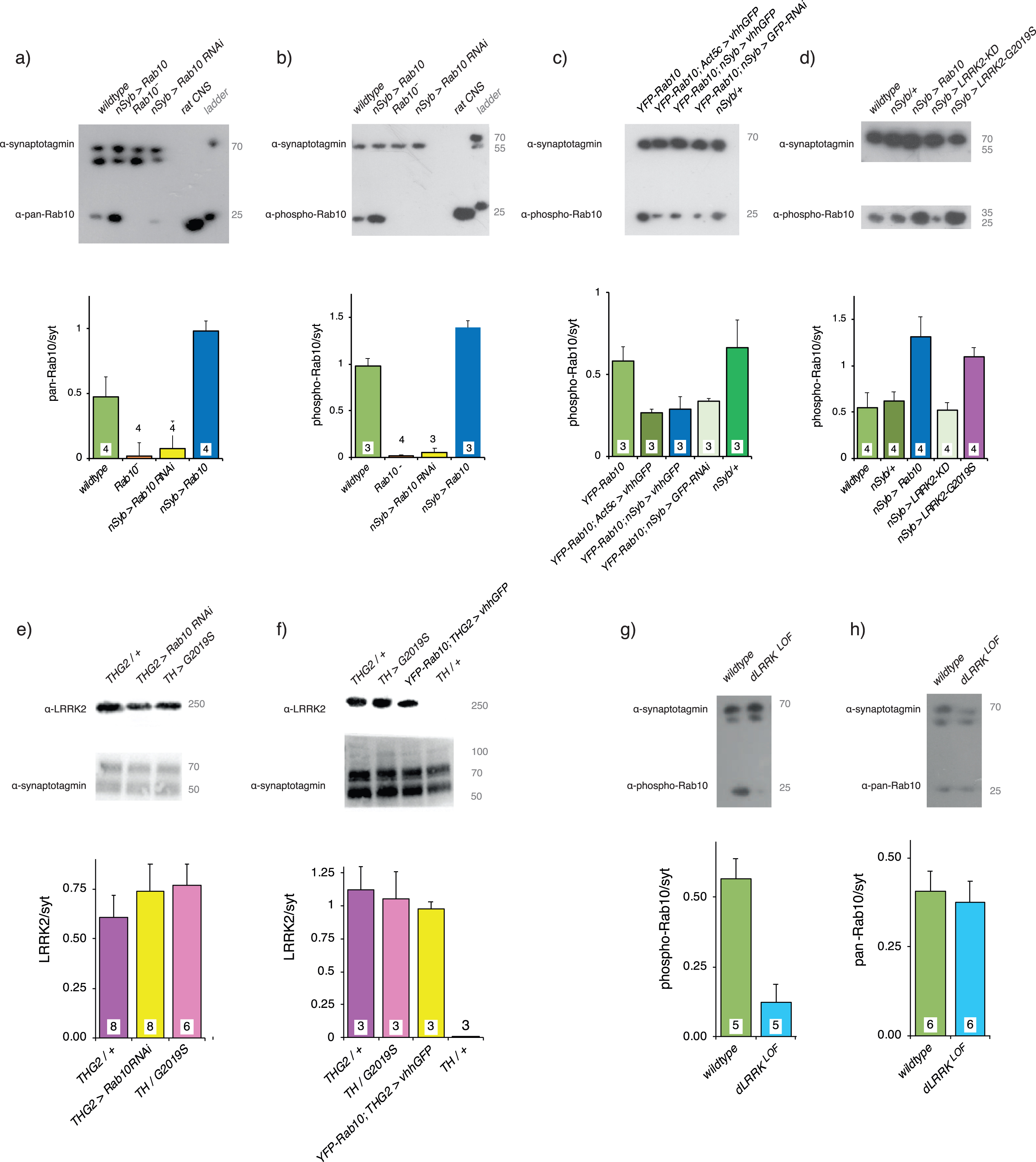
The levels of Rab10 and LRRK2-G2019S were assayed by western blot using standard protocols [9, 14]. Primary antibodies were as follows: α-pan-Rab10 (Nanotools, clone 605B11), α-phospho-Rab10 (Abcam, ab230261, 1:1000), and for LRRK2 (Neuromab, clone N241A/34). We used α-synaptotagmin 91 as a loading control [21]. Selectivity of Rab10 antibodies was confirmed by using a rat CNS extract [9]. YFP-Rab10 was assayed as described recently [9], except that sample preparation included boiling. This uncouples the YFP from Rab10, so that the MW was ∼25 kDa (Fig. 1), whereas unboiled samples run at ∼50 kDa [9]. Quantification of the blots was carried out in Fiji.
Fig. 1
Validation of Rab10 depletion and phosphorylation in vivo. a) A pan-hRab10 antibody detects dRab10 and shows that Rab10 is increased by neuronal (nSyb-GAL4) expression of UAS-Rab10, abolished in the Rab10− null, and substantially reduced by neuronal expression of Rab10RNAi. Antibody-specificity was confirmed by rat CNS binding. b) A phosopho-hRab10 antibody detects phospho-dRab10, which is increased by neuronal expression of UAS-Rab10, but undetectable with the Rab10− null or with neuronal expression of Rab10RNAi. c) Validation of deGradFP technique. In flies where endogenous Rab10 has been replaced by YFP-Rab10, global (Act5c-GAL4) or neuronal expression of the vhhGFP nanobody reduces the level of YFP-Rab10 by ∼50%. A slightly smaller reduction is achieved by neuronal expression of GFPRNAi. d) Neuronal expression of Rab10 increases pRab10, as does neuronal expression of LRRK2-G2019S. Neuronal expression of a kinase-dead LRRK2 (KD, LRRK2-G2019S-K1906M) has no effect on the phosphorylation of Rab10. e) Adding Rab10 RNAi expression does not reduce the level of LRRK2-G2019S. f) Adding expression of the GFP nanobody does not reduce the level of LRRK2-G2019S. Note that the THG2 and TH > G2019S samples in f are different to those in e. g) The dLRRKLOF mutations in the fly LRRK2 ortholog, dLRRK is a severe loss of function P-element insertion. This mutant reduces the phosphorylation of Rab10. h) dLRRKLOF, however, does not affect the pan-Rab10 level. Loading control: α-drosophila-synaptotagmin (α-syt), which recognizes at least 3 isoforms, depending on the blot time. Each panel contains a sample blot and quantification (using the upper α-syt band for each blot) below. Exact genotypes in Supplementary Table 2.
Bradykinesia
Bradykinesia was assessed using an optogenetic stimulus. Flies were fed retinal (1 mM) pipetted onto the surface of their food for 1 week in the dark at 29°C. They were restrained at 25°C for 3 h before the proboscis extension responses were observed with a Grasshopper 3 (Point Grey) camera mounted on a Zeiss Stemi microscope at 200 frames/s. A single flash was delivered from a ThorM470L3 LED, driven at 8 V for 7 ms. The stimulus was transcoded by LexOp-ReachR expressed in the Gr5a neurons. The area of the video occupied by the proboscis was automatically analyzed by python code https://github.com/biol75/PER, and graphed in Excel. Only those flies which showed smooth extension (Fig. 2 aiv) or smooth extension and relaxation (Fig. 2 aiii), with no erratic movements were used for full analysis. The Tukey - post-hoc test was applied in R.
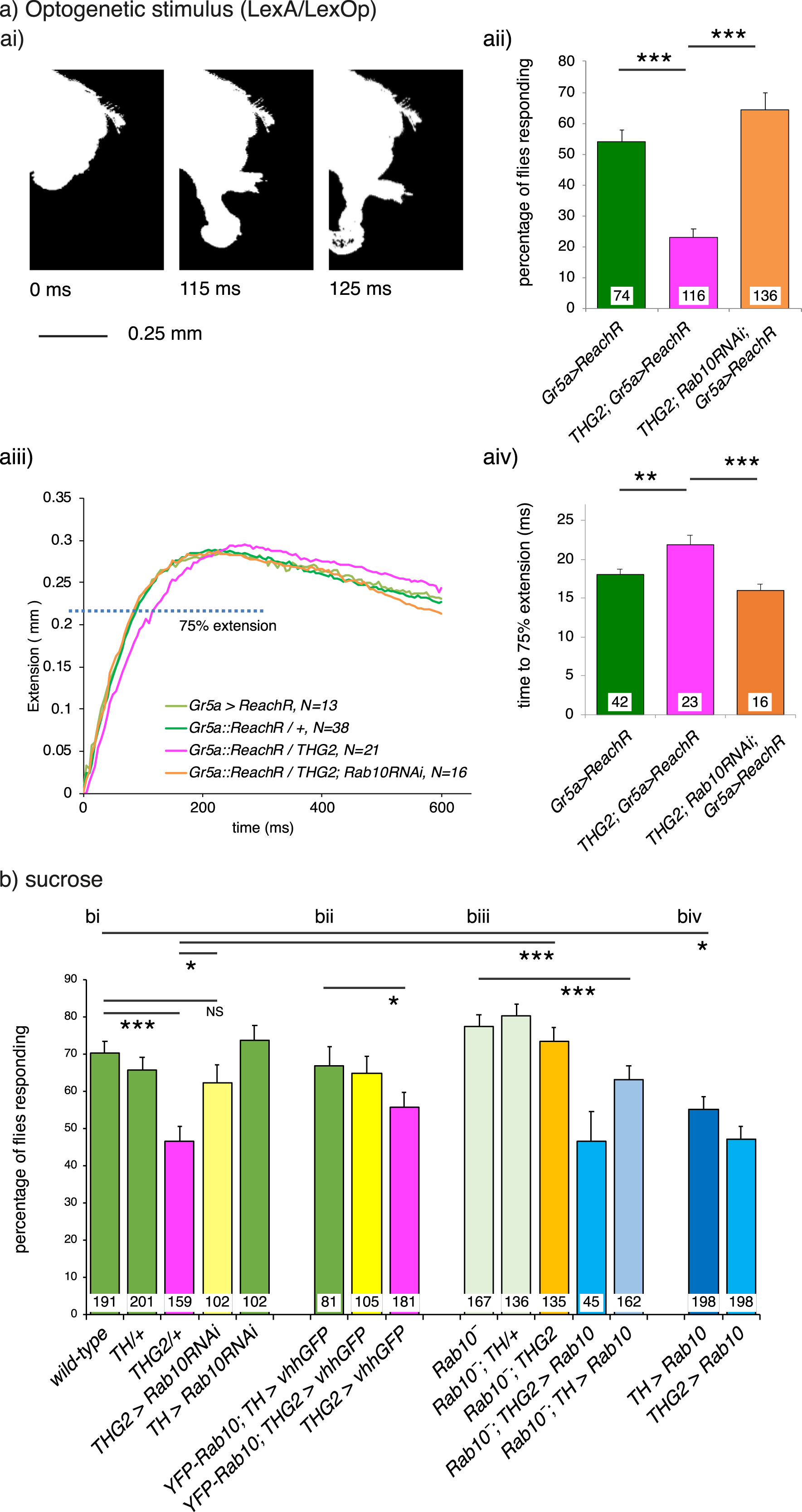
Fig. 2
Rab10 knock-down rescues LRRK2-G2019S -induced bradykinesia. a) Optogenetic stimulation of the Proboscis Extension Response (PER). ai) To reach for food or liquid, the fly extends its proboscis in response to an optogenetic stimulus to sensory neurons on the legs. The full extension response is shown in Supplementary Movie M1. aii) Expression of LRRK2-G2019S in dopaminergic neurons (THG2) reduces the proportion of flies that respond to a single flash of light, and this is rescued by Rab10 reduction with Rab10RNAi. aiii. Dopaminergic reduction of Rab10 rescues the bradykinesia (slower response) of flies expressing G2019S in their dopaminergic neurons. aiii). Raw traces; aiv). Mean data. To respond to the optogenetic stimuli all flies carry LexA/Op Gr5a> ReachR. b) Sucrose stimulation of the PER. bi) Flies expressing G2019S in their dopaminergic neurons (THG2, magenta bars) respond less frequently to sucrose than wild-type flies (green), i.e., they show akinesia. This is rescued in THG2 flies with dopaminergic reduction in Rab10 using Rab10RNAi (THG2 > Rab10RNAi). bii) Reduction of dopaminergic Rab10 with the deGradFP technique (Rab10 YFP; THG2 > vhhGFP, yellow bars) also rescues the G2019S-induced akinesia. biii) The Rab10 null (Rab10−, orange bar) also reverts the G2019S deficit, while expression of Rab10 in the null background again induces akinesia (Rab10−; TH > Rab10, light blue bar). biv) By itself, Rab10 overexpression phenocopies G2019S expression. Exact genotypes and full statistical data in Supplementary Table 3.
Akinesia
Akinesia was recorded from 1 week old flies, kept in the dark at 29°C. Flies were restrained as described [14] and starved at 29°C for 3 h before being offered a droplet of 100 mM sucrose three times. Each response was scored Yes/No and the median response for each fly used. The χ2-post-hoc test (Fifer) was done in R (3.3.3). Standard errors were estimated from the 95%confidence intervals generated by the Binomial prediction.
Visual assays
On the day of emergence, flies were placed in the dark or in disco-chambers at 29°C. One-week-old flies were prepared for SSVEP (steady state visual evoked potential) measurements as recently described [9]. Stimuli were generated and responses recorded by an Arduino Due system with FFTs and contrast sensitivity computed in Matlab. Dunnett’s post-hoc test was applied in R. Full code at https://github.com/wadelab/flyCode. DPP (deep-pseudo-pupil) images were captured with a Dino-Lite Camera and software, and images cropped, quantified, and converted to grey scale in Fiji. Both images in Fig. 3a ii were combined in Fiji before the color was removed and the contrast increased.
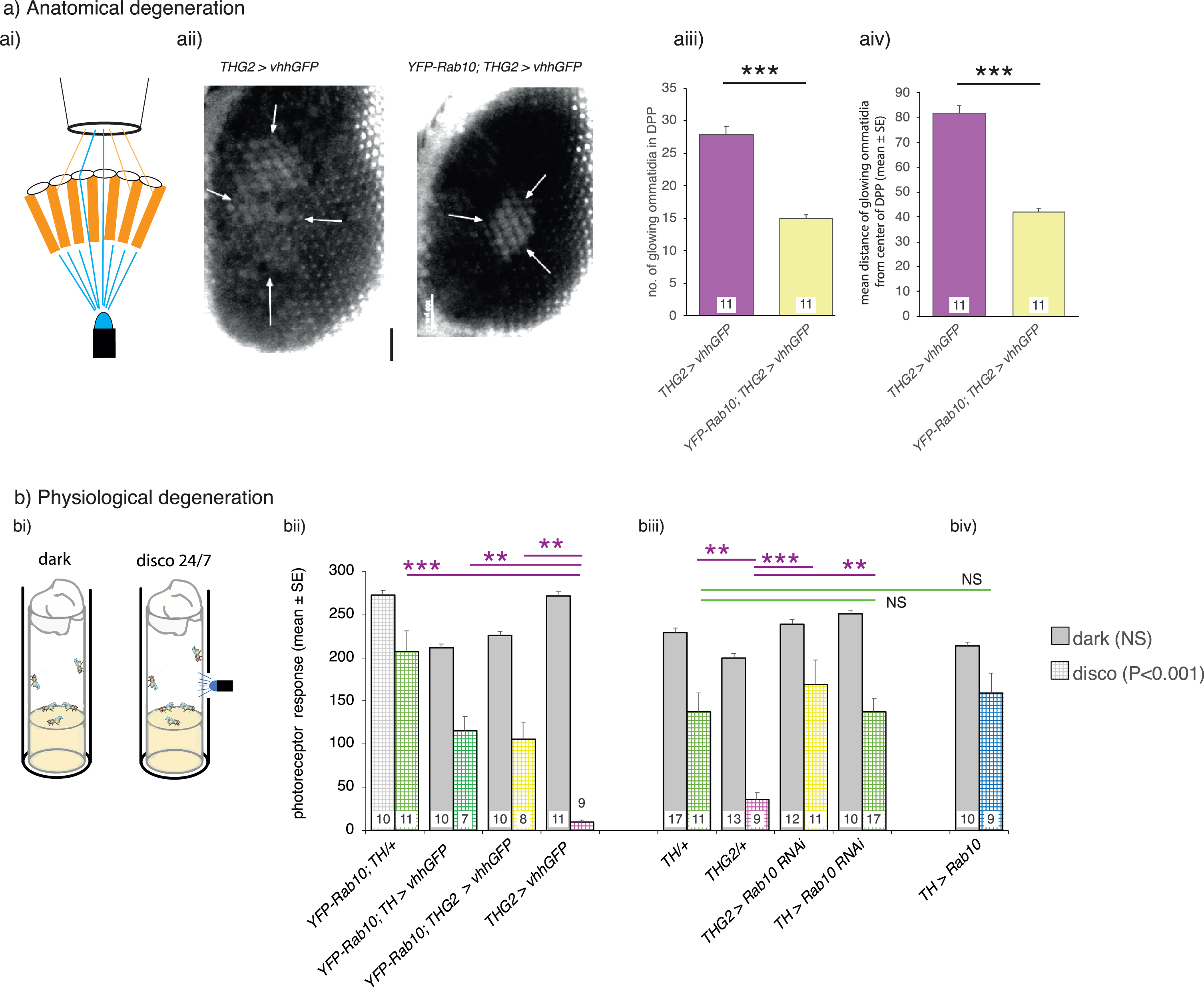
Fig. 3
Visual degeneration due to LRRK2-G2019S is rescued by dopaminergic knock-down of Rab10. a) Anatomically, dopaminergic Rab10 knock-down rescues the loss of eye structure. ai) Healthy flies have a marked DPP (Deep-Pseudo-Pupil) which can be seen when the eye is illuminated from underneath. Light passing directly though the eye is focused towards the observer, but light at an angle is blocked by the pigmentation in the ommatidia. aii-aiv). PD-mimic flies (THG2; vhhGFP) kept in the dark for 14 days lose their focused DPP as the eyes show an increased number of ommatidia transmitting light, spread over a wider area, with the loss of pigmentation indicating lysosomal dysfunction. Much less degeneration is seen when Rab10 is depleted using the deGradFP technique (YFP-Rab10; THG2; vhhGFP flies). b). Physiologically, dopaminergic Rab10 knock-down rescues neuronal vision. bi) Degeneration is accelerated by a mild visual stress achieved by keeping the flies in ‘disco-chambers’ where the light is turned on and off approximately every 2 seconds; these flies were compared with those kept in the dark. bii-iii). Flies kept in the dark have normal visual responses after 7 days. However, THG2 flies in disco chambers lose their physiological response after 7 days (hatched magenta bar, controls in green). This is rescued by deGradFP mediated knockdown of Rab10 protein (bi, Rab10 GFP; THG2; vhhGFP hatched yellow bar) or by dopaminergic knock-down of Rab10 by RNAi (biii, THG2 > Rab10RNAi yellow bar). biv) By itself, Rab10 overexpression has no effect. Exact genotypes and statistical data in Supplementary Table 4.
Circadian rhythms and sleep patterns
Circadian rhythms and sleep patterns were recorded as described recently [22] with a TriKinetics DAM system. Flies were placed in the monitor at 29°C on the day of eclosion, and locomotor activity was recorded in 1-min bins for 3 days under 12:12 h light/dark cycles followed by 7 to 10 days in constant darkness. Activity data was analyzed using the ActogramJ plugin for ImageJ and sleep data using the ShinyR-DAM code [23] using the branch at https://karolcichewicz.shinyapps.io/ShinyR-DAM_3_1_Beta/.
Conditioned courtship memory
Conditioned courtship memory was recorded using the well-developed conditioned courtship memory protocol for Drosophila [24, 25]. One-week-old males (kept at 29°C) were provided with a virgin female and the time spent in courting behaviors measured. Half of the males had recently (∼30 min before) spent an hour with a mated female; half were naïve. Males who fail to ‘remember’ their rejection will spend more time courting the virgin female than controls. The Courtship Index (CI) was calculated as the percentage of 10 min that a male spent courting; and the memory index (MI) by dividing the CI of each test fly by the mean CI of the naïve/sham flies of the same genotype. A score of 0 indicates highest memory performance possible and a score of≥1 indicates no memory. The distribution of MI was compared between genotypes using the Kolmogorov-Smirnov test. All naïve and trained groups contained 16–23 flies.
Immunocytochemistry
Immunocytochemistry was performed on 3–5-day-old flies expressing G2019S as described recently [14], using the LRRK2 mouse antibody (Neuromab, clone N241A/34). In some flies, mCD8-GFP was also expressed in the dopaminergic neurons. Each figure is representative of three preparations from at least two crosses.
Analysis of gene expression in the fly optic lobe
We interrogated the dataset generated by Davis et al. [10] for the cell types in the opticlobe which express Rab10 and its potential interactors, using the tool which they provided at http://www.opticlobe.com.
RESULTS
Validation of Rab10 knock-down and its phosphorylation by G2019S
We used three strategies for Rab10 reduction. 1) We used a CRISPR/Cas9-generated Rab10 null [26], in which Rab10 was severely reduced: both pan-Rab10 and phospho-Rab10 to less than 5%of wild-type (Fig. 1a,b). This line was not used in the visual assay because it carries a retinal RFP marker, which causes the eyes to fluoresce, and this would distort the disco-chamber data. 2) We used Rab10RNAi. This almost completely depleted phosphoRab10 (Fig. 1b), but a small amount (∼15%) of Rab10 was still present (Fig. 1a). 3) We used the deGradFP technique to knockout YFP-Rab10 protein with the vhhGFP nanobody, to target YFP -Rab10 to the proteasome. This was used in flies in which the wild-type Rab10 had been replaced by homologous recombination so that the YFP-Rab10 was expressed at wild-type levels. This reduced the amount of phospho-Rab10 to ∼50%of wild-type (Fig. 1c). In the GFP-Rab10 background, expression of a RNAi against GFP depleted some phospho-Rab10, but not as much as the nanobody, so we did not use this further.
The ability of LRRK2-G2019S to phosphorylate Rab10 in vivo was confirmed (Fig. 1d), by comparing nSyb > LRRK2-G2019S with the kinase-dead (KD) form (LRRK2-G2019S-K1906M). In the nSyb > LRRK2-G2019S flies, the level of pRab10 was similar to the effect of pan-neuronal expression of Rab10, being twice that of the wildtype or nSyb outcross or the nSyb > KD preparation.
In flies, the GAL4-UAS system (which we are deploying here) has proved a robust means of transgene expression. In several systems, it has been shown that adding extra UAS constructs does not dilute the phenotype produced by a particular transgene, e.g., [27]. We wanted to check if the level of LRRK2-G2019S protein remains the same when other transgenes are expressed at the same time. We confirmed that expressing Rab10RNAi or the vhhGFP nanobody did not affect the level of G2019S (Fig. 1e,f). No band was seen in wild-type flies [14], or those in which no G2019S was expressed (Fig. 1f).
We also tested if the fly homolog of LRRK2, dLRRK, phosphorylated Rab10. For this, we deployed a dLRRK loss of function mutant (dLRRKLOF) which contains a PiggyBac transposon, resulting in reduced protein expression [28]. The mutant has as much pan-Rab10 as the wild-type, but much less phospho-Rab10 (Fig. 1g,h). Here we note that in human and mouse, LRRK1 does not appear to phosphorylate Rab10.
A variety of physiological consequences of Rab10 knock-down in dopaminergic neurons expressing LRRK2-G2019S
We compared the ‘PD-mimic’ THG2 flies, in which only the dopaminergic neurons express LRRK2-G2019S with controls and with Rab10 knock-down in four types of behavioral/physiological phenotypes: bradykinesia in the proboscis extension response, neural signaling in the retina, sleep patterns and conditioned courtship memory. These four systems are controlled by different clusters of dopaminergic neurons.
Rab10 reduction rescues LRRK2-G2019S bradykinesia
The major features of Parkinson’s disease are movement deficits (bradykinesia), slowing or loss of movement and tremor. We therefore begin our analysis with the movement of PD-mimic THG2 flies. The Proboscis Extension Response (PER), a reaching movement used by Dipteran flies to obtain food, is particularly amenable to analysis [29] (Fig. 2a i, Supplementary Movie M1). When a fly walks across a surface and encounters a droplet of sugary solution, the sucrose-sensitive neurons on the legs are stimulated and this elicits a rapid extension of the proboscis. The PER is modulated by a single dopaminergic neuron, the Tyrosine Hydoxylase Ventral Unpaired Median (TH-VUM) neuron [30], which has a high level of Rab10 expression [9]. Expression of G2019S in this neuron results in movement deficits –akinesia (loss of the PER), slower response (bradykinesia) and tremor [14]. Further, the PER can be elicited using an optogenetic stimulus. In this paradigm, the sucrose-sensitive sensory neurons in the leg express a channelrhodopsin which is stimulated by a flash of light. Expression of a channelrhodopsin by the LexA/LexOp system in the sugar-sensitive ‘Gr5a’ neurons on the leg is independent of the expression of LRRK2-G2019S in the TH-VUM and other dopaminergic neurons by the GAL4-UAS system [31]. This precise control of the stimulus makes it possible to measure the time course of the reaching movement and separates the activation of the stimulus from manipulations using the GAL4 system in the dopaminergic neurons.
In our optogenetic setup, 60%of control flies respond to a single flash of light by smoothly extending their proboscis, whereas flies in which Tyrosine-Hydroxylase GAL4 was used to express LRRK2-G2019S (THG2) only respond 30%of the time, i.e., G2019S induces akinesia (Fig. 2a ii). Those THG2 flies that do respond, take 100 ms longer to achieve maximum extension, showing bradykinesia (Fig. 2a iii, iv). Knock-down of Rab10 using Rab10RNAi co-expressed with LRRK2-G2019S in the dopaminergic neurons, fully reverts both akinesia and the bradykinesia (Fig. 2a ii-iv).
We next tested if Rab10 knock-down would also rescue the deficits induced by G2019S in response to the natural stimulus, a sugar solution. Just over two-thirds of control flies respond to sucrose (70%, Fig. 2b i, green bars), whereas less than half of THG2 flies respond to sucrose (46 %, Fig. 2b i, magenta). This deficit is fully rescued when Rab10RNAi is co-expressed with THG2 (THG2 > Rab10RNAi Fig. 2b i, yellow). Dopaminergic expression of Rab10RNAi alone has no effect on the proportion of flies that respond to the sucrose solution (TH > Rab10RNAi, Fig. 2b i, green).
As RNAi may have off-target effects, we supplemented this with a nanobody-mediated-protein knockdown technique, deGradFP [32, 33]. In this, we deployed flies in which the wild-type Rab10 gene had been replaced by YFP- Rab10 by homologous recombination. Then we expressed an anti-GFP nanobody (vhhGFP) to deplete YFP-Rab10 protein in just the dopaminergic neurons using TH-GAL4. In this experiment, YFP- Rab10 flies with both G2019S and vhhGFP expression (Fig. 2b ii, yellow bar) responded identically to wild-types, or YFP- Rab10 controls in which dopaminergic neurons expressed just vhhGFP (Fig. 2b ii, YFP- Rab10; TH > vhhGFP green bar). This indicates a full rescue of the THG2 induced akinesia. We confirmed that expressing G2019S and vhhGFP in flies with wild-type Rab10 still resulted in akinesia (Fig. 2b ii, magenta bar).
While the Rab10 knockout is lethal in mammals [34], the fly Rab10 null (Rab10−) is viable [26]. We therefore tested if a global removal of Rab10 would also ameliorate the akinesia deficit in the THG2 flies. In the Rab10− background, the THG2 Proboscis Extension Response is fully rescued (Fig. 2b iii, orange), to the same level as wild-type controls (Fig. 2b i, green). In control experiments, the proportion of Rab10− flies that responded to sucrose was identical to wild-types; this was true for the homozygote stock and for an outcross to TH in males (Rab10 is on the X-chromosome) (Fig. 2b iii, pale green).
Rab10 is known to be phosphorylated by LRRK2 in vitro [2] and in the fly, in vivo (Fig. 1). Phosphorylation is likely to be part of the activation mechanisms of Rab10. If increasing phosphorylation of Rab10 is a key event in LRRK2-G2019S driven dysfunction in dopaminergic neurons, we would expect dopaminergic expression of Rab10 to phenocopy that of G2019S. We found that 55%of such TH > Rab10 flies responded to sucrose, in an almost identical manner to the THG2 flies (Fig. 2b iv) with pronounced akinesia. No further increase in akinesia is seen when both G2019S and Rab10 are expressed at the same time.
To confirm that akinesia was solely dependent on dopaminergic Rab10, we took the Rab10 null, and crossed it with flies expressing Rab10 in just the dopaminergic neurons. Such Rab10−; TH > Rab10 flies have akinesia when compared with the Rab10− flies (Fig. 2b iii). Similarly, the Rab10−; THG2 > Rab10 show even less response.
Thus, the G2019S-induced movement deficits in the PER are rescued by depleting Rab10 in the dopamine neurons either at the mRNA level using RNAi, or at the protein level using a nanobody, or with the global null. Increasing Rab10 in the dopaminergic neurons induces akinesia similar to the expression of LRRK2-G2019S.
Dopaminergic reduction in Rab10 rescues G2019S-induced visual deficits
People with Parkinson’s disease also show visual deficits including loss of dopaminergic neurons from the retina [35, 36]. Aged THG2 flies also show strong retinal degeneration with vacuoles throughout the optic lobe [15]. To demonstrate retinal degeneration, we deployed the ‘Deep-Pseudo-Pupil’ (DPP) assay [37]. When wild-type flies are illuminated from below, they show a DPP, with about 6–8 glowing ommatidia. In this, the normal retinal structure focuses light from directly below towards the observer through the center of the ommatidium, whereas light which is off-center is blocked by the pigment granules at the edge of the ommatidium (Fig. 3a i). We tested several types of genetic background for the THG2 ‘PD-mimic’ flies used in movement assays and found that the contrast between DPP and the red eye pigment was best when the THG2 was in the deGradFP background. Although the DPP was clear in these young flies, in flies aged for 14 days, the DPP is distorted in these THG2;vhhGFP flies (Fig. 3a ii-iv). The outline of the DPP is irregular, rather than a neat ellipse, and the number and dispersion of the glowing ommatidia was increased. However, when dopaminergic Rab10 is depleted in the THG2 deGradFP fly, the DPP is much less disrupted (YFP-Rab10; THG2 > vhhGFP Fig. 3a ii-iv). There are fewer light-transmitting ommatidia and they are all adjacent. Thus, dopaminergic Rab10 reduction is sufficient to rescue the G2019S-induced neurodegeneration. We conclude that expression of G2019S leads to degeneration of the retina, including disruption of the pigment granules and regular hexagonal patterning.
In people with Parkinson’s disease, the retinal degeneration of dopaminergic amacrine cells is accompanied by changes in contrast detection, e.g., [38]. We therefore tested if the anatomical degeneration of the fly eye is linked to a loss of visual physiological contrast sensitivity. To do this, we used the SSVEP (steady state visual evoked potential) method which is automated to distinguish the photoreceptor contrast sensitivity from the synaptic response of the second order lamina neurons. At 7 days, all dark-reared control and THG2 flies show a strong SSVEP photoreceptor signal, indicating that the eyes are functioning normally. There is no effect of the manipulation of Rab10 (Fig. 3b, solid bars). On the other hand, when exposed to a mild visual stress to accelerate neurodegeneration (by being kept in the disco chamber for 1 week [15]), the THG2 flies lose almost all their visual response—it is reduced to 18%of the wild-type dark level (Fig. 3b ii,iii, hatched magenta bars; controls in green), indicating loss of photoreceptor function. As with the anatomical measure, the contrast sensitivity of the eye was rescued using the deGradFP manipulation (YFP-Rab10; THG2 > vhhGFP, Fig. 3b ii, yellow bar). With co-expression of Rab10RNAi, this THG2 deficit is also fully rescued (Fig. 3b iii, yellow bar).
Thus, both electrophysiological response and structural observation of the DPP indicate that Rab10 knock-down rescues the effect of dopaminergic LRRK2-G2019S.
LRRK2-induced daily activity deficits are not affected by Rab10
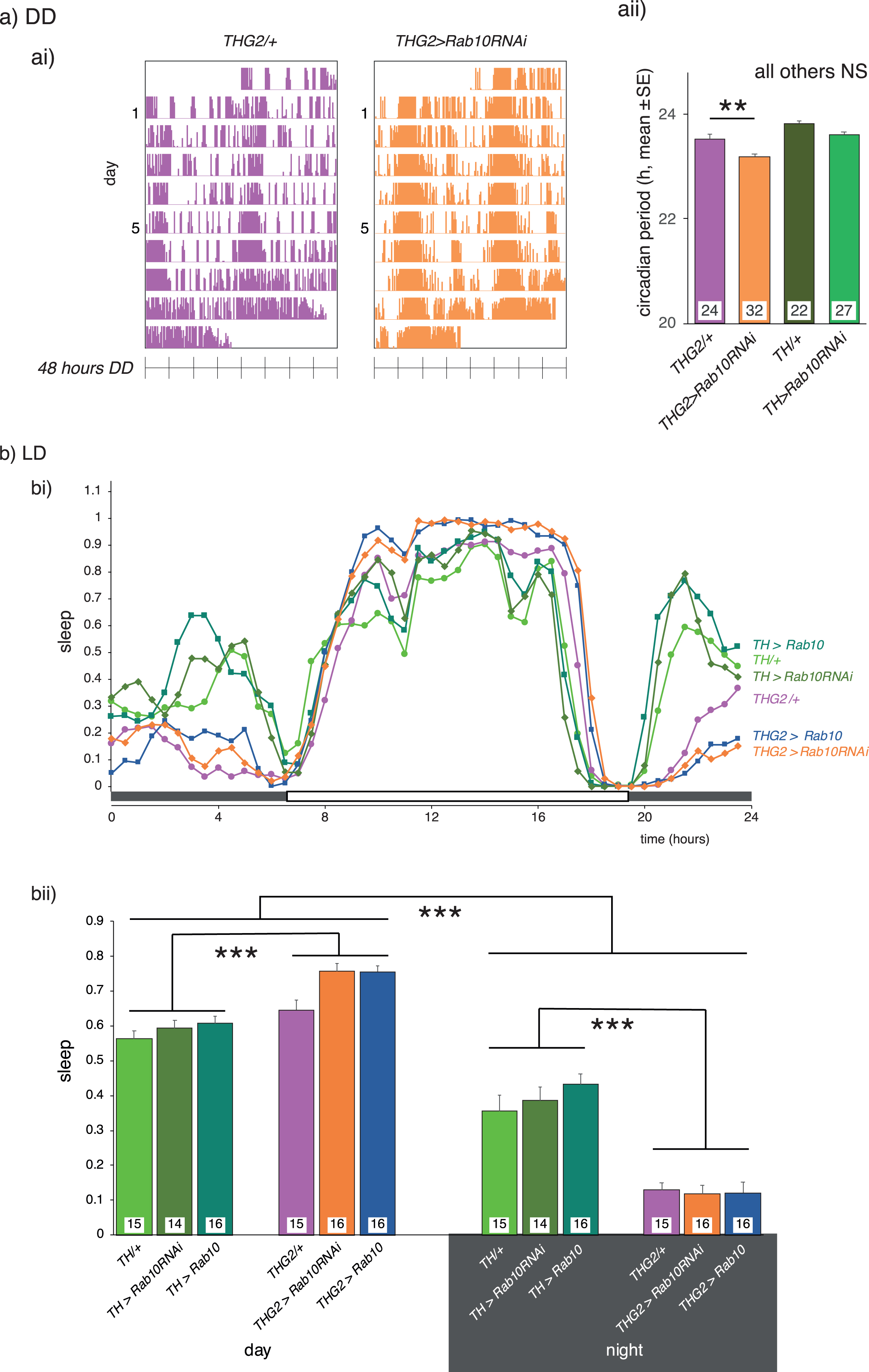
A third behavior modulated by dopaminergic neurons is the daily sleep-wake rhythm, both in human [39] and fly [40] (see [41] for review). The dopaminergic neurons that innervate the fly mushroom bodies are key players in the daily activity rhythm. Locomotor activity measured over several days provides a measure of circadian rhythm and sleep-wake behavior (Fig. 4a). Periods of inactivity longer than 5 min defined as ‘sleep’ (see [42]). In 12 h:12 h light on:off cycles, all flies tested show a daily sleep-wake rhythm, sleeping more in the light than in the dark, with THG2 flies sleeping 15%more than the controls during the ‘day’ (Fig. 4b). At the light/dark transition, all flies have high, persistent activity, and little sleep. THG2 flies continue their activity during the dark, spending less than 50%of the time ‘asleep’ in the dark that control flies managed (TH/+, Fig. 4b). Neither reduction nor increased expression of Rab10 in dopaminergic neurons affected the sleep/wake cycle of either the control or LRRK2-G2019S flies. Following light-dark entrainment (LD), in the dark (DD), the fly locomotor activity pattern persists with a ∼24 h cycle providing a measure of intrinsic circadian rhythms. The period of the circadian rhythm was not affected by LRRK2-G2019S or Rab10 manipulation, though a small reduction was seen when both transgenes were expressed (Fig. 4a). This is not unexpected as the circadian period is not affected by dopamine manipulations [43].
Fig. 4
Rab10 knock-down does not rescue sleep deficits induced by dopaminergic expression of LRRK2-G2019S. A. There is no effect of LRRK2-G2019S or Rab10 expression on circadian period in continuous darkness (DD), and only a small reduction in circadian period when both genes are expressed. ai) Raw actograms; aii) mean period from days 6-9 in DD. Our control flies, in which no transgene was expressed is TH/+. b) In Light/dark (LD), THG2 flies show increased sleep during the day, and reduced sleep at night. Neither reduced nor increased expression of Rab10 affects the daily pattern of sleep (bi), with summary data in bii. Sleep is defined as periods of inactivity longer than 5 min. Exact genotypes and statistical results in Supplementary Table 5.
Thus, the normal sleep-wake cycle is disrupted by dopaminergic LRRK2-G2019S, but not by Rab10 manipulation, even though Rab10 in the context of LRRK2-G2019S is seen to regulate the PER and retina.
Improved Memory due to Rab10 depletion is insensitive to LRRK2-G2019S
Dopaminergic circuits also affect memory in both human [44] and fly [45, 46]. We deployed the conditioned courtship short-term memory protocol, which is dependent on the well-known dopaminergic neurons that project to the mushroom bodies (MB) [47] to assess the effects of Rab10 and G2019S. In this assay, males are allowed to court mated females, which reject their advances. When presented with more receptive virgin females, they tend to court less as they ‘remember’ their rejection. We found that dopaminergic knock-down of Rab10 substantially reduced the memory index, with just over half of the TH > Rab10RNAi flies having a memory index less than 0.05, i.e. they have excellent memory. In contrast, only 10%of control flies had a memory index less than 0.05 (Fig. 5). When G2019S was expressed, no change in the distributions was seen, and Rab10 depletion still improved memory (Fig. 5).
Fig. 5
Memory depends on dopaminergic Rab10 but not LRRK2-G2019S. A) Depletion of Rab10 in the control background (TH > Rab10RNAi) increases the proportion of flies with low memory index (MI) compared to flies with no transgene expression (TH/+). Expression of LRRK2-G2019S has no effect on performance; either for the THG2/+v TH/+control flies, nor for the flies with Rab10RNAi. a) Raw scores, b) summary data. Exact genotypes and statistical results in Supplementary Table 6.
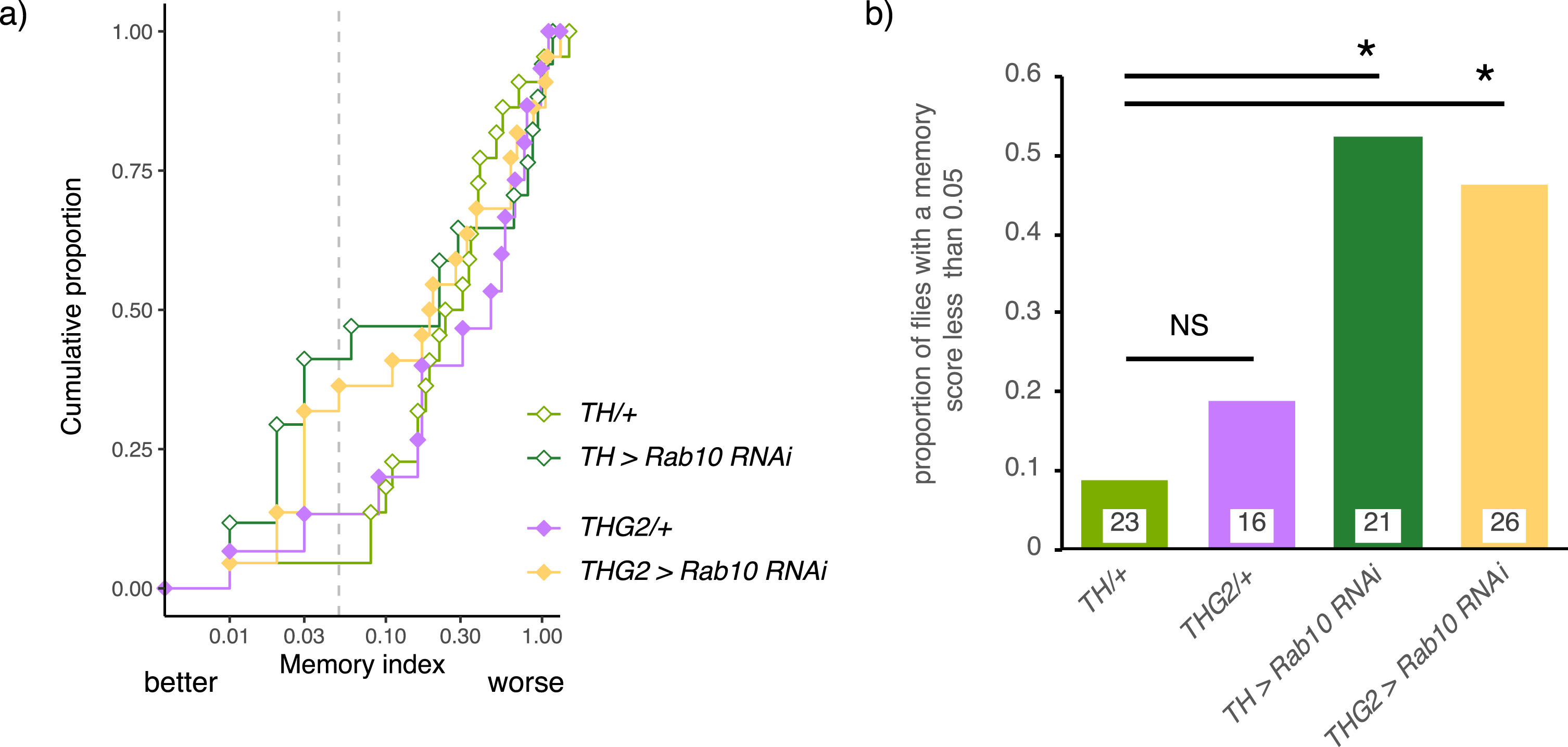
We conclude that, in the conditioned courtship assay, Rab10 is present and has a key role in the dopaminergic neurons, but that the behavior is not affected by dopaminergic expression of G2019S.
Cytoplasmic location of G2019S and Rab10
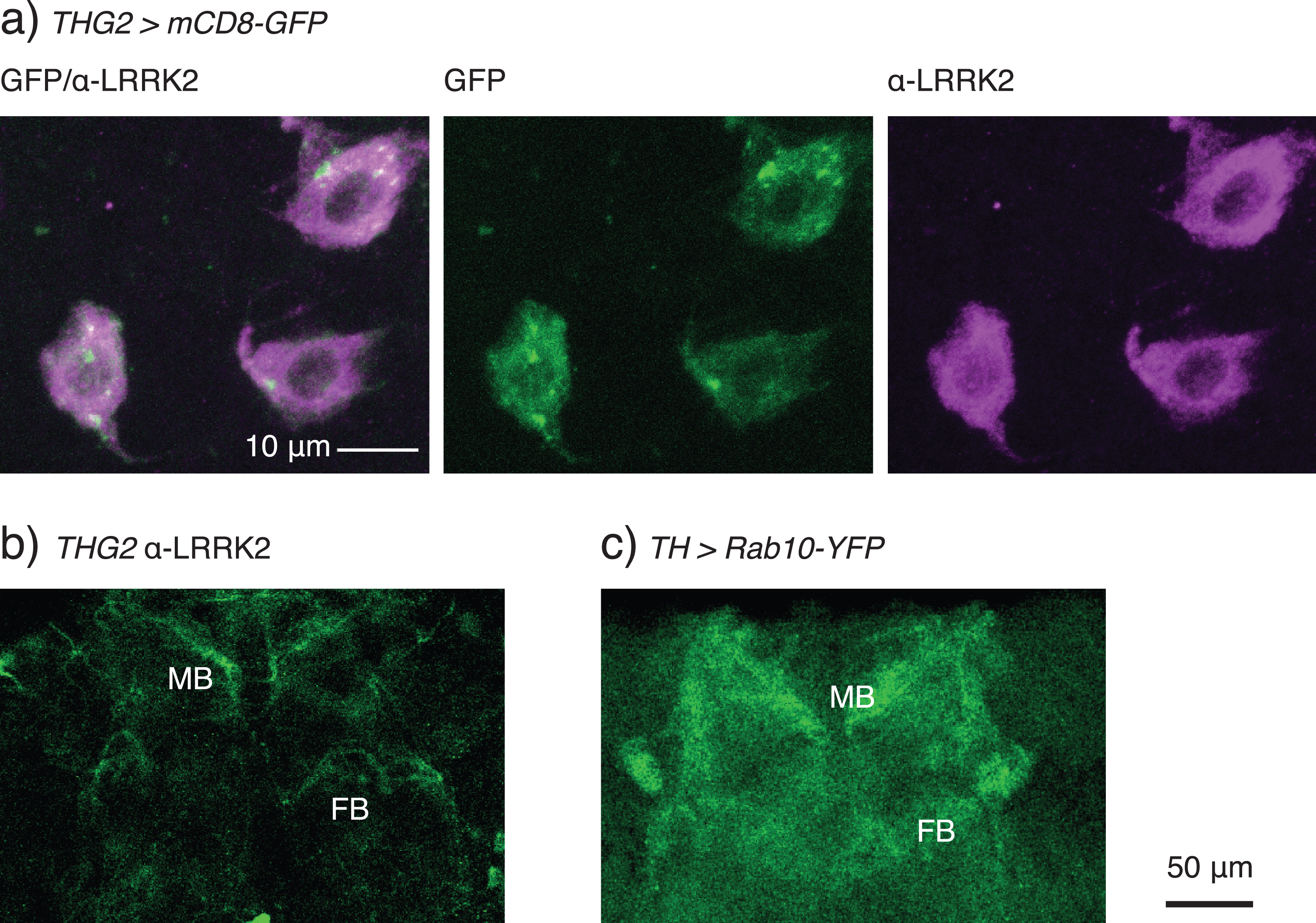
A variety of distributions for LRRK2 has been reported (microtubule [48], TGN [49], mitochondria [50], lysosome [51], or on synaptic vesicles [52]), but the location of LRRK2 has not, to our knowledge, been reported in fly neurons. In comparison, the fly ortholog, dLRRK, is focused at the Golgi-membranes [28]. We therefore tested where ectopically expressed LRRK2-G2019S protein is detected in dopaminergic neurons. Staining was seen more strongly in the cytoplasm than in the nucleus (Fig. 6a). Additionally, there is weak staining along the axons and of the synaptic endings, including the mushroom bodies and fan-shaped body (Fig. 6b). A similar pattern of staining is seen with dopaminergic expression of hLRRK2- wild-type (data not shown). Rab 10 expression (TH > Rab10-YFP flies, Fig. 6c) shows a staining in the cytoplasm, with the strongest fluorescence at the synaptic terminals, noticeably in the neuropil of the mushroom bodies and fan-shaped body. The similar distribution allows for plenty of interaction sites, which would be necessary for phosphorylation.
Fig. 6
LRRK2-G2019S is found in the cytoplasm of the cell soma and in the synaptic endings of dopaminergic neurons. a) Confocal stack through the TH-VUM and DADN neurons in the ventral part of the brain of a THG2 > mCD8-GFP fly. Note that LRRK2 protein is found more in the cytoplasm than the nucleus, and that there are areas of the cytoplasm with less LRRK2 staining. mCD8-GFP expression is used to mark dopaminergic neurons. The confocal stack spans the whole depth of these dopaminergic neurons. b) LRRK2 is found in the synaptic endings in the mushroom bodies (MB) and weakly in the fan-shaped body (FB). Single confocal section with α-LRRK2 antibody, cropped to select the neuropil. c) Dopaminergically expressed Rab10-YFP is found in the same synaptic neuropils. Stack of two confocal images, cropped to include just the neuropil. Images representative of at least 3 preparations. Exact genotypes in Supplementary Table 7.
Expression of Rab10 and its potential interactors in PAM neurons
The gene expression pattern in the opticlobe and in three types of dopaminergic PAM neurons has been reported [10]. We interrogated this dataset to examine the expression of a Rab10 GEF, two GAPs and three effectors (Table 1). While two of the Rab10 effectors (Ehbp1 and Rilpl (CG11448)) are expressed in every cell type, the other genes are not universally recorded. In fact, the neurons in which the genes are expressed differ substantially. This is indicated by analysis of the PAM neurons: PAM1 and PAM3 omit Crag and Mical, whereas PAM4 does not express plx or Mical. We also note that dLRRK is not expressed in the PAM neurons.
Table 1
Distribution of Rab10 and its potential effectors in the fly optic lobe dataset (http://www.opticlobe.com, Davis et al., 2020 [10])
| Rab10 | Crag | plx | Evi5 | Ehbp1 | Rilpl (CG11448) | Mical | dLRRK | ||
| Role with Rab10 | GEF | GAP | GAP | effector | effector | effector | |||
| Dopaminergic | PAM 1 | Y | N | Y | Y | Y | Y | N | N |
| PAM neurons | PAM 3 | Y | N | Y | Y | Y | Y | N | N |
| PAM 4 | Y | Y | N | Y | Y | Y | N | N | |
| %of cell types in which the gene was detected | 77 | 3 | 80 | 73 | 100 | 100 | 9 | 74 |
DISCUSSION
Having previously demonstrated a strong genetic interaction, in vivo, between LRRK2 and Rab10 [9], we sought to determine whether manipulation of Rab10 affected the physiological deficits associated with flies expressing LRRK2-G2019S. We have identified a requirement for Rab10 in mediating the movement and visual deficits induced by dopaminergic expression of LRRK2-G2019S. In contrast, in two other dopaminergic physiological systems, there was no interaction: in one (the circadian sleep/wake cycle) LRRK2-G2019S has a marked phenotype independent of Rab10; in the other (conditioned courtship memory) Rab10 has a distinct role, but there is no LRRK2-G2019S phenotype. As such, we have begun to identify the individual dopaminergic circuits in the fly that are most sensitive to LRRK2-Rab10 interplay (Fig. 7).
Fig. 7
Multiple pathways of LRRK2-G2019S / Rab10 interaction in dopaminergic neurons. Rab10 depletion ameliorates the proboscis extension bradykinesia and loss of synaptic signaling in the retina induced by LRRK2-G2019S expression (magenta arrows/orange crosses). Rab10 manipulation does not affect the ‘sleep’ phenotype from LRRK2-G2019S (magenta arrow). Reduction of Rab10 facilitates conditioned courtship memory, but LRRK2 has no effect (yellow arrow). All manipulations of Rab10 and G2019S in dopaminergic neurons, shown in the outline of the brain (filled cells have high levels of Rab10 [9]). We conclude that Rab10 and LRRK2 interact in some, but not all dopaminergic neurons. This may underlie differences in the susceptibility of different human striatal dopaminergic cells to Parkinson’s disease and explain why differences in the age at which symptoms are first noted.
![Multiple pathways of LRRK2-G2019S / Rab10 interaction in dopaminergic neurons. Rab10 depletion ameliorates the proboscis extension bradykinesia and loss of synaptic signaling in the retina induced by LRRK2-G2019S expression (magenta arrows/orange crosses). Rab10 manipulation does not affect the ‘sleep’ phenotype from LRRK2-G2019S (magenta arrow). Reduction of Rab10 facilitates conditioned courtship memory, but LRRK2 has no effect (yellow arrow). All manipulations of Rab10 and G2019S in dopaminergic neurons, shown in the outline of the brain (filled cells have high levels of Rab10 [9]). We conclude that Rab10 and LRRK2 interact in some, but not all dopaminergic neurons. This may underlie differences in the susceptibility of different human striatal dopaminergic cells to Parkinson’s disease and explain why differences in the age at which symptoms are first noted.](https://content.iospress.com:443/media/jpd/2021/11-4/jpd-11-4-jpd202421/jpd-11-jpd202421-g007.jpg)
Rab10 knock-down rescues movement and visual deficits from dopaminergic LRRK2-G2019S expression
Movement and visual deficits induced by LRRK2-G2019S expression have been linked to low dopamine release in the TH-VUM and some PPL/MC neurons that control the PER and vision respectively [14, 15]. We focus here on how the LRRK2-Rab10 interaction might lead to a potential loss of dopamine release. First, Rab10 is strongly expressed in these neurons [9] and so could play a role in controlling dopamine traffic or exocytosis. LRRK2-G2019S expression in the fly CNS increases the phosphorylation of Rab10 approximately two-fold. Knock-down by Rab10RNAi reduces the level of pRab10 below the level at which it is detected, though some Rab10 is still present. Additionally, protein knock-down using deGradFP lowers pRab10 and rescues the proboscis extension/visual deficits, though the reduction in Rab10 reported by Western Blot was not so strong as with Rab10RNAi. Thus, the amelioration, in vivo of G2019S-induced movement and visual deficits by Rab10RNAi is in accord with data from cell culture that Rab10 is a key target of LRRK2 [2–7]. Our data additionally shows, at least for a subset of Drosophila dopaminergic neurons, Rab10 is likely a relevant physiological substrate for LRRK2-G2019S.
This genetic interaction between Rab10 and LRRK2-G2019S does not imply that the physical interaction is direct, though the anatomical evidence shows that both proteins spread along the axons to the synaptic endings and are likely in proximity. Rab proteins often work in functionally linked chains—for example Rab29 and Rab10 both interact with LRRK2 [53, 54]. Our data does not exclude that the LRRK2-G2019S deficits also involve other Rabs.
The cellular consequences of phosphorylation of Rab10 are not yet fully clear, but it has been suggested that this may switch the effectors to which Rab10 binds [8, 54, 55]. In dividing cells in culture phosphorylation of Rab10 by LRRK2 reduces ciliogenesis [56, 57]. However, the relevance of this to the terminally differentiated adult Drosophila CNS neurons is not clear. Both LRRK2 and Rab10 have been linked to mitochondrial damage resolution, through the Rab10 effector OPTN (optineurin) [50] though Drosophila do not have an evident OPTN homolog (this function could be fulfilled by an as yet unidentified protein). Another role for LRRK2—control of vesicle transport at the TGN—has support from HEK cells [49] and fly neurons. In the fly retina, a Rab10 interaction has been reported occurring at the TGN with its GEF Crag and effector Ehbp1 [58], affecting vesicle budding. Interestingly, the fly ortholog of LRRK2, which is dLRRK, is linked to the golgin protein, Lava lamp, at Golgi outposts in neurites [28]. An effect on vesicle transport at the TGN, along with consequent endosomal disruption [51, 53, 59, 60] may provide an explanation for a lack of dopamine release.
Sleep/wake cycles and conditioned courtship memory do not depend on an interaction between Rab10 and LRRK2-G2019S
In stark contrast, in the same animals, dysfunction in other dopamine-mediated behaviors does not require Rab10. Rab10RNAi did not ameliorate the change in daily sleep defect induced by LRRK2-G2019S, and overexpression of Rab10 did not phenocopy G2019S- induced sleep defects. While Rab10RNAi expression affected the conditioned courtship memory, indicating the presence of Rab10 and its importance in these dopaminergic neurons, LRRK2-G2019S had no phenotype. Thus, Rab10 seems to be present, but not affected by LRRK2-G2019S.
Dopaminergic neurons display varied levels of Rab10 even within a particular cluster [9]. Consequently, it is possible that the Rab10 GEFs, GAPs and effectors may also differ between the subsets of the dopaminergic neurons in the PAM, PPL1 and PPM3 neurons, the clusters that regulate the sleep-wake cycle [40, 61, 62]—indeed the three types of PAM neurons express a different subset of Rab10 interactors. Further, dLRRK is expressed in the dopaminergic neurons in the optic lobe, but not in the PAM neurons. The variation in gene expression may explain why some labs have identified other potential LRRK2 interactors—for example EndophilinA, in Drosophila larval glutamatergic neuromuscular synapses [63].
CONCLUSION
Our key finding is that that LRRK2-G2019S may signal by several pathways, even within the dopaminergic neuron population. We conclude that an uneven distribution of Rabs, their effectors and binding partners may contribute to the differences in the rate of dopaminergic neuron degeneration in the human striatum. Multiple LRRK2 pathways may also explain why the range of Parkinson’s disease symptoms initiate at different ages.
ACKNOWLEDGMENTS
We are grateful for the gifts of flies from Kristin Scott, Zhuohua Zhang, Wanli Smith, Cheng-Ting Chien, Julie Simpson, Serge Birman, Robin Hiesinger, and Stefan Heidmann. We also thank the York Biology Technology Facility, Bloomington Drosophila Supply Center and Flybase for their provision. Sean Sweeney, Chris MacDonald, and Amy Cording kindly read a draft manuscript. We are particularly grateful to Parkinson’s UK and to their volunteers for support (K-1704, G-1804).
This work was supported by Parkinson’s UK, (K-1704, G-1804).
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JPD-202421.
REFERENCES
[1] | Greggio E , Cookson MR ((2009) ) Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: Three questions. ASN Neuro 1: , AN20090007. |
[2] | Steger M , Diez F , Dhekne HS , Lis P , Nirujogi RS , Karayel O , Tonelli F , Martinez TN , Lorentzen E , Pfeffer SR , Alessi DR , Mann M ((2017) ) Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. Elife 6: , e31012. |
[3] | Fan Y , Howden AJM , Sarhan AR , Lis P , Ito G , Martinez TN , Brockmann K , Gasser T , Alessi DR , Sammler EM ((2018) ) Interrogating Parkinson’s disease LRRK2 kinase pathway activity by assessing Rab10 phosphorylation in human neutrophils. Biochem J 475: , 23–44. |
[4] | Jeong GR , Jang E-H , Bae JR , Jun S , Kang HC , Park C-H , Shin J-H , Yamamoto Y , Tanaka-Yamamoto K , Dawson VL , Dawson TM , Hur E-M , Lee BD ((2018) ) Dysregulated phosphorylation of Rab GTPases by LRRK2 induces neurodegeneration. Mol Neurodegener 13: , 8. |
[5] | Thirstrup K , Dächsel JC , Oppermann FS , Williamson DS , Smith GP , Fog K , Christensen KV ((2017) ) Selective LRRK2 kinase inhibition reduces phosphorylation of endogenous Rab10 and Rab12 in human peripheral mononuclear blood cells. Sci Rep 7: , 10300. |
[6] | Liu Z , Bryant N , Kumaran R , Beilina A , Abeliovich A , Cookson MR , West AB ((2018) ) LRRK2 phosphorylates membrane-bound Rabs and is activated by GTP-bound Rab7L1 to promote recruitment to the trans-Golgi network. Hum Mol Genet 27: , 385–395. |
[7] | Kelly K , Wang S , Boddu R , Liu Z , Moukha-Chafiq O , Augelli-Szafran C , West AB ((2018) ) The G2019S mutation in LRRK2 imparts resiliency to kinase inhibition. Exp Neurol 309: , 1–13. |
[8] | Alessi DR , Sammler E ((2018) ) LRRK2 kinase in Parkinson’s disease. Science 360: , 36–37. |
[9] | Petridi S , Middleton CA , Ugbode C , Fellgett A , Covill L , Elliott CJH ((2020) ) In vivo visual screen for dopaminergic rab< ->LRRK2-G2019S interactions in drosophila discriminates Rab10 from Rab3. G3 (Bethesda) 10: , 1903–1914. |
[10] | Davis FP , Nern A , Picard S , Reiser MB , Rubin GM , Eddy SR , Henry GL ((2020) ) A genetic, genomic, and computational resource for exploring neural circuit function. Elife 9: , e50901. |
[11] | Daniel G , Moore DJ ((2014) ) Modeling LRRK2 pathobiology in Parkinson’s disease: From yeast to rodents. Curr Top Behav Neurosci 22: , 331–368. |
[12] | Feany MB , Bender WW ((2000) ) A Drosophila model of Parkinson’s disease. Nature 404: , 394–398. |
[13] | Liu Z , Wang X , Yu Y , Li X , Wang T , Jiang H , Ren Q , Jiao Y , Sawa A , Moran T , Ross CA , Montell C , Smith WW ((2008) ) A Drosophila model for LRRK2-linked parkinsonism. Proc Natl Acad Sci U S A 105: , 2693–2698. |
[14] | Cording AC , Shiaelis N , Petridi S , Middleton CA , Wilson LG , Elliott CJH ((2017) ) Targeted kinase inhibition relieves slowness and tremor in a Drosophila model of LRRK2 Parkinson’s. NPJ Parkinsons Dis 3: , 34. |
[15] | Hindle SJ , Afsari F , Stark M , Middleton CA , Evans GJO , Sweeney ST , Elliott CJH ((2013) ) Dopaminergic expression of the Parkinsonian gene LRRK2-G2019S leads to non-autonomous visual neurodegeneration, accelerated by increased neural demands for energy. Hum Mol Genet 22: , 2129–2140. |
[16] | Julienne H , Buhl E , Leslie DS , Hodge JJL ((2017) ) Drosophila PINK1 and parkin loss-of-function mutants display a range of non-motor Parkinson’s disease phenotypes. Neurobiol Dis 104: , 15–23. |
[17] | Hewitt VL , Whitworth AJ (2017) Mechanisms of Parkinson’s disease. In Current Topics in Developmental Biology, pp. 173-200. |
[18] | Park J , Lee SB , Lee S , Kim Y , Song S , Kim S , Bae E , Kim J , Shong M , Kim J-M , Chung J ((2006) ) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441: , 1157–1161. |
[19] | Clark IE , Dodson MW , Jiang C , Cao JH , Huh JR , Seol JH , Yoo SJ , Hay BA , Guo M ((2006) ) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441: , 1162–1166. |
[20] | Oswald MCW , Brooks PS , Zwart MF , Mukherjee A , West RJH , Giachello CNG , Morarach K , Baines RA , Sweeney ST , Landgraf M ((2018) ) Reactive oxygen species regulate activity- dependent neuronal plasticity in Drosophila. Elife 7: , e39393. |
[21] | West RJH , Lu Y , Marie B , Gao F-BF-B , Sweeney ST ((2015) ) Rab8, POSH, and TAK1 regulate synaptic growth in a Drosophila model of frontotemporal dementia. J Cell Biol 208: , 931–947. |
[22] | Nippe OM , Wade AR , Elliott CJH , Chawla S ((2017) ) Circadian rhythms in visual responsiveness in the behaviorally arrhythmic Drosophila clock mutant Clk Jrk. J Biol Rhythms 32: , 583–592. |
[23] | Cichewicz K , Hirsh J ((2018) ) ShinyR-DAM: A program analyzing Drosophila activity, sleep and circadian rhythms. Commun Biol 1: , 25. |
[24] | Griffith LC , Ejima A ((2009) ) Courtship learning in Drosophila melanogaster: Diverse plasticity of a reproductive behavior. Learn Mem 16: , 743–750. |
[25] | Koemans TS , Oppitz C , Donders RAT , van Bokhoven H , Schenck A , Keleman K , Kramer JM ((2017) ) Drosophila courtship conditioning as a measure of learning and memory. J Vis Exp e55808. |
[26] | Kohrs F , Daumann I , Pavlović B , Jin E , Kiral F , Lin S , Port F , Wolfenberg H , Mathejczyk T , Linneweber G , Chan C , Boutros M , Hiesinger P ((2021) ) Systematic functional analysis of rab GTPases reveals limits of neuronal robustness to environmental challenges in flies.. eLife 10: . https://doi.org/10.7554/eLife.59594. |
[27] | Tariq Ahmad S , Sweeney ST , Lee JA , Sweeney NT , Gaoa FB ((2009) ) Genetic screen identifies serpin5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc Natl Acad Sci U S A 106: , 12168–12173. |
[28] | Lin C-H , Li H , Lee Y-N , Cheng Y-J , Wu R-M , Chien C-T ((2015) ) Lrrk regulates the dynamic profile of dendritic Golgi outposts through the golgin Lava lamp. J Cell Biol 210: , 471–483. |
[29] | Dethier VG ((1976) ) The hungry fly: A physiological study of the behavior associated with feeding, Harvard U Press. |
[30] | Marella S , Mann K , Scott K ((2012) ) Dopaminergic modulation of sucrose acceptance behavior in Drosophila. Neuron 73: , 941–950. |
[31] | Zhou Y , Cao LH , Sui XW , Guo XQ , Luo DG ((2019) ) Mechanosensory circuits coordinate two opposing motor actions in Drosophila feeding. Sci Adv 5: , eaaw5141. |
[32] | Urban E , Nagarkar-Jaiswal S , Lehner CF , Heidmann SK ((2014) ) The cohesin subunit Rad21 is required for synaptonemal complex maintenance, but not sister chromatid cohesion, during Drosophila female meiosis. PLoS Genet 10: , e1004540. |
[33] | Caussinus E , Kanca O , Affolter M ((2012) ) Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol 19: , 117–121. |
[34] | Lv P , Sheng Y , Zhao Z , Zhao W , Gu L , Xu T , Song E ((2015) ) Targeted disruption of Rab10 causes early embryonic lethality. Protein Cell 6: , 463–467. |
[35] | Harnois C , Di Paolo T ((1990) ) Decreased dopamine in the retinas of patients with Parkinson’s disease. Invest Ophthalmol Vis Sci 31: , 2473. |
[36] | Ortuño-Lizarán I , Sánchez-Sáez X , Lax P , Serrano GE , Beach TG , Adler CH , Cuenca N ((2020) ) Dopaminergic retinal cell loss and visual dysfunction in Parkinson’s disease. Ann Neurol 88: , 893–906. |
[37] | Franceschini N , Kirschfeld K ((1971) ) Les phénomènes de pseudopupille dans l’œil composé deDrosophila. Kybernetik 9: , 159–182. |
[38] | Gupta HV , Zhang N , Driver-Dunckley E , Mehta SH , Beach TG , Adler CH ((2019) ) Contrast acuity with different colors in Parkinson’s disease. Mov Disord Clin Pract 6: , 672–677. |
[39] | Monti JM , Monti D ((2007) ) The involvement of dopamine in the modulation of sleep and waking. Sleep Med Rev 11: , 113–133. |
[40] | Sitaraman D , Aso Y , Rubin GM , Nitabach MN ((2015) ) Control of sleep by dopaminergic inputs to the Drosophila mushroom body. Front Neural Circuits 9: , 73. |
[41] | Dubowy C , Sehgal A ((2017) ) Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205: , 1373–1397. |
[42] | Helfrich-Förster C ((2018) ) Sleep in insects. Annu Rev Entomol 63: , 69–86. |
[43] | Hirsh J , Riemensperger T , Coulom H , Iche M , Coupar J , Birman S ((2010) ) Roles of dopamine in circadian rhythmicity and extreme light sensitivity of circadian entrainment. Curr Biol 20: , 209–214. |
[44] | Shohamy D , Adcock RA ((2010) ) Dopamine and adaptive memory. Trends Cogn Sci 14: , 464–472. |
[45] | Cognigni P , Felsenberg J , Waddell S ((2018) ) Do the right thing: Neural network mechanisms of memory formation, expression and update in Drosophila. Curr Opin Neurobiol 49: , 51–58. |
[46] | Takemura SY , Aso Y , Hige T , Wong A , Lu Z , Xu CS , Rivlin PK , Hess H , Zhao T , Parag T , Berg S , Huang G , Katz W , Olbris DJ , Plaza S , Umayam L , Aniceto R , Chang LA , Lauchie S , Ogundeyi O , Ordish C , Shinomiya A , Sigmund C , Takemura S , Tran J , Turner GC , Rubin GM , Scheffer LK ((2017) ) A connectome of a learning and memory center in the adult Drosophila brain. Elife 6: , e26975. |
[47] | Montague SA , Baker BS ((2016) ) Memory elicited by courtship conditioning requires mushroom body neuronal subsets similar to those utilized in appetitive memory. PLoS One 11: , e0164516. |
[48] | Watanabe R , Buschauer R , Böhning J , Audagnotto M , Lasker K , Wen Lu T , Boassa D , Taylor SS , Villa E ((2020) ) The in situ structure of Parkinson’s disease-linked LRRK2. Biophys J 118: , 486a. |
[49] | Beilina A , Bonet-Ponce L , Kumaran R , Kordich JJ , Ishida M , Mamais A , Kaganovich A , Saez-Atienzar S , Gershlick DC , Roosen DA , Pellegrini L , Malkov V , Fell MJ , Harvey K , Bonifacino JS , Moore DJ , Cookson MR ((2020) ) The Parkinson’s disease protein LRRK2 interacts with the GARP complex to promote retrograde transport to the trans-Golgi network. Cell Rep 31: , 107614. |
[50] | Wauters F , Cornelissen T , Imberechts D , Martin S , Koentjoro B , Sue C , Vangheluwe P , Vandenberghe W ((2019) ) LRRK2 mutations impair depolarization-induced mitophagy through inhibition of mitochondrial accumulation of RAB10. Autophagy 16: , 203–222. |
[51] | Schapansky J , Khasnavis S , DeAndrade MP , Nardozzi JD , Falkson SR , Boyd JD , Sanderson JB , Bartels T , Melrose HL , LaVoie MJ ((2018) ) Familial knockin mutation of LRRK2 causes lysosomal dysfunction and accumulation of endogenous insoluble α-synuclein in neurons. Neurobiol Dis 111: , 26–35. |
[52] | Pischedda F , Piccoli G ((2021) ) LRRK2 at the pre-synaptic site: A 16-years perspective. J Neurochem 157: , 297–311. |
[53] | Eguchi T , Kuwahara T , Sakurai M , Komori T , Fujimoto T , Ito G , Yoshimura S , Harada A , Fukuda M , Koike M , Iwatsubo T ((2018) ) LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc Natl Acad Sci U S A 115: , E9115–E9124. |
[54] | Rivero-ríos P , Romo-lozano M , Fernández B , Fdez E ((2020) ) Distinct roles for RAB10 and RAB29 in pathogenic LRRK2-mediated endolysosomal trafficking alterations. Cells 9: , 1719. |
[55] | Liu Z , Xu E , Zhao HT , Cole T , West AB ((2020) ) LRRK2 and Rab10 coordinate macropinocytosis to mediate immunological responses in phagocytes. EMBO J 39: , e104862. |
[56] | Dhekne HS , Yanatori I , Gomez RC , Tonelli F , Diez F , Schüle B , Steger M , Alessi DR , Pfeffer SR ((2018) ) A pathway for Parkinson’s disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. Elife 7: , e40202. |
[57] | Lara Ordónez AJ , Fernández B , Fdez E , Romo-Lozano M , Madero-Pérez J , Lobbestael E , Baekelandt V , Aiastui A , López De Munaín A , Melrose HL , Civiero L , Hilfiker S ((2019) ) RAB8, RAB10 and RILPL1 contribute to both LRRK2 kinase-mediated centrosomal cohesion and ciliogenesis deficits. Hum Mol Genet 28: , 3552–3568. |
[58] | Nakamura Y , Ochi Y , Satoh T , Satoh AK ((2020) ) Crag/Rab10/Ehbp1 regulate the basolateral transport of Na+K+ATPase in Drosophila photoreceptors. J Cell Sci 133: , jcs.238790. |
[59] | Kuwahara T , Inoue K , D’Agati VD , Fujimoto T , Eguchi T , Saha S , Wolozin B , Iwatsubo T , Abeliovich A , Paisan-Ruiz C , Zimprich A , Satake W , Simon-Sanchez J , Zhang FR , Umeno J , Lewis PA , Manzoni C , Greenman C , Hassin-Baer S , MacLeod DA , Pihlstrom L , Bultema JJ , Pietro Di SM , Ma J , Plesken H , Treisman JE , Edelman-Novemsky I , Ren M , Hermann GJ , Grill B , MacLeod D , Plowey ED , Cherra SJ , Liu YJ , Chu CT , Parisiadou L , Chan D , Citro A , Cordy JM , Shen GC , Wolozin B , Beilina A , Tong Y , Herzig MC , Hinkle KM , Tong Y , Fuji RN , Newell-Litwa K , Seong E , Burmeister M , Faundez V , Dell’Angelica EC , Wolozin B , Gabel C , Ferree A , Guillily M , Ebata A , Kuwahara T , Sakaguchi-Nakashima A , Meir JY , Jin Y , Matsumoto K , Hisamoto N , Currie E , Dell’Angelica EC , Shotelersuk V , Aguilar RC , Gahl WA , Bonifacino JS , Peden AA , Shotelersuk V , Dell’Angelica EC , Hartnell L , Bonifacino JS , Gahl WA , Zhen L , Swank RT , Reddington M , Howlett O , Novak EK , Seong E , Biskup S , Steger M , Dell’Angelica EC , Ooi CE , Bonifacino JS , Faundez V V. , Kelly RB , Azevedo C , Burton A , Ruiz-Mateos E , Marsh M , Saiardi A , Martin I , West AB , Smith WW , Guttentag SH , Brenner S , Saha S , Kuwahara T , Tonegawa R , Ito G , Mitani S , Iwatsubo T , Ito G , Inoue K , Inoue K ((2016) ) LRRK2 and RAB7L1 coordinately regulate axonal morphology and lysosome integrity in diverse cellular contexts. Sci Rep 6: , 29945. |
[60] | Rivero-Ríos P , Romo-Lozano M , Fasiczka R , Naaldijk Y , Hilfiker S ((2020) ) LRRK2-related Parkinson’s disease due to altered endolysosomal biology with variable Lewy dody pathology: A hypothesis. Front Neurosci 14: , 556. |
[61] | Driscoll M , Coleman V , McLaughlin M , Nguyen A , Sitaraman D (2020) Dopamine neurons promote arousal and wakefulness via Dop1R receptor in the Drosophila mushroom body. bioRxiv, 2020.04.29.069229. |
[62] | Potdar S , Sheeba V ((2018) ) Wakefulness is promoted during day time by PDFR signalling to dopaminergic neurons in Drosophila melanogaster. eneuro. eneuro 5: , ENEURO.0129-18.2018. |
[63] | Soukup SF , Kuenen S , Vanhauwaert R , Manetsberger J , Hernández-Díaz S , Swerts J , Schoovaerts N , Vilain S , Gounko N V. , Vints K , Geens A , De Strooper B , Verstreken P ((2016) ) A LRRK2-dependent endophilinA phosphoswitch is critical for macroautophagy at presynaptic terminals. Neuron 92: , 829–844. |




