Would wider adoption of reproducible research be beneficial for empirical software engineering research?
Abstract
Researchers have identified problems with the validity of software engineering research findings. In particular, it is often impossible to reproduce data analyses, due to lack of raw data, or sufficient summary statistics, or undefined analysis procedures. The aim of this paper is to raise awareness of the problems caused by unreproducible research in software engineering and to discuss the concept of reproducible research (RR) as a mechanism to address these problems. RR is the idea that the outcome of research is both a paper and its computational environment. We report some recent studies that have cast doubts on the reliability of research outcomes in software engineering. Then we discuss the use of RR as a means of addressing these problems. We discuss the use of RR in software engineering research and present the methodology we have used to adopt RR principles. We report a small working example of how to create reproducible research. We summarise advantages of and problems with adopting RR methods. We conclude that RR supports good scientific practice and would help to address some of the problems found in empirical software engineering research.
1Introduction
This paper reports some recent research results that have cast doubts about the reliability of current empirical software engineering research results. In the context of data mining studies, software engineering researchers have proposed reproducible research (RR) as a means to improve research practice (e.g. [36], and [15]). In this paper we ask the question “Would wider adoption of reproducible research be beneficial for empirical software engineering research involving human-centric experiments?”.
In Section 2, we discuss what we mean by reproducible research (RR) which is one of the methods being proposed to address problems with empirical research in software engineering data mining studies and other disciplines. We discuss the origin and scope of reproducible research, but also how it differs from, but supports, the concept of replication in human-centric software engineering studies. We report, in Section 3, problems found with recent empirical software engineering research. We also emphasize that the discussed problems are not unique to the software engineering domain. In Section 4, we examine how RR is currently being adopted in empirical software engineering research. To confirm the viability of RR, we identify, in Section 5, a set of free and open-source tools that we have been able to use in practice to produce reproducible research. In Section 6, we present an intentionally simple example of the RR process to help other researchers to understand how to construct reproducible research. We also highlight, in Section 7, the problems and benefits associated with RR from the viewpoint of software engineering researchers, as well as major initiatives related to RR. Finally, we present conclusions in Section 8. This is primarily a discussion paper. Our main contribution is to discuss the use of RR to address some of problems observed in software engineering experiments and to confirm the viability of RR with a small practical example of its use.
2Reproducible research: Origins and definition
Gandrud [12] attributes the term reproducible research to Professor Claerbout of Stanford University who, in 1990, imposed the standard of makefiles for all the figures and computational results published by the Stanford Exploration Project. Furthermore, in 2000, he and his students shared their experience of creating a reproducible research environment [38].
RR refers to the idea that the ultimate product of research is the paper plus its computational environment. That is, a reproducible research document incorporates the textual body of the paper (including any necessary supplementary materials, e.g., protocols or appendices) plus the data used by the study, and the analysis steps (algorithms) used to process the data, in the context of an open access environment that is used to compile these pieces of information into the resulting document. This triple is called the compendium by Gentleman and Lang [13]. Having access to this information, an independent researcher or data analyst can reproduce the results, verify the findings, and create new work based on the original research, for example, conduct alternative analyses of the same data, or replicate the original analysis on an updated or new data set. In addition, it should be easier to aggregate the outcomes of replicated studies using meta-analysis [23].
When reading the literature on RR, we noticed that some researchers (for example, Gandrud [12]) appear to use the terms reproducibility and replication interchangeably. However, in this paper we make a distinction between the concepts. Replication involves repeating an experiment with different participants or experimental materials to investigate whether previous experimental results are repeatable. However as Gomez et al. [14] point out many researchers talk about reproducibility in the context of replication. For example, Carver et al. talk about results being reproduced [6], and Runeson et al. [37] use the term reproduction to refer to a replication performed by independent researchers. In this paper, we take a restricted view of reproducible research defining it as the extent to which the report of a specific scientific study can be reproduced (in effect, compiled) from the reported text, data and analysis procedures, and thus validated by other researchers. Although we emphasise that reproducibility and replication are different things, it is also the case that research incorporating reproducibility is likely to be easier to replicate than research that does not.
RR is particularly important in the context of studies of computational algorithms where, as Vandewalle et al. [45] point out, details such as the “exact data set, initialization or termination procedure, and precise parameter values are often omitted” for reasons such as “a lack of space, a lack of self-discipline, or an apparent lack of interest to the readers”. In software engineering, this would apply to the data mining studies discussed by Robles and his colleagues (e.g. [15, 35, 36]), such as comparative studies of algorithms for test automation, comparative studies of cost estimation and of defect prediction, and any studies investigating the performance of evolutionary and machine learning algorithms. In this paper, we discuss, whether RR is also relevant to human-intensive experiments.
3Problems with empirical software engineering practice
Recent results in empirical software engineering have cast some doubts on the validity of our software engineering research results. For example, Shepperd et al. [39] analyzed the results of 42 papers reporting studies comparing methods for predicting fault-proneness. They found that the explanatory factor that accounted for the largest percentage of the differences among studies (i.e., 30%) was research group. In contrast prediction method, which was the main topic of research, accounted for only 1.3% of the variation among studies. They commented that “It matters more who does the work than what is done.” and “Until this can be satisfactorily addressed there seems little point in conducting further primary studies”. The papers overlapped in terms of the data sets used, and the defect prediction modelling methods used in primary papers. The fact that their results are inconsistent with respect to the impact of the fault prediction methods suggests significant reproducibility failures.
In the area of cost estimation, Kitchenham and Mendes pointed out that reported accuracy statistics for cost estimation studies claiming to use a specific method on a particular data set were inconsistent with the results they obtained using the specific method on the same data set [21]. More recently, Whigham et al. found that claims made in two recent cost estimation studies could not be confirmed by independent analyses [47]. RR emphasizes the need to specify, fully, any statistical analysis, in order to address problems such as these.
In the context of experiments and quasi-experiments, Vegas et al. [46] reviewed 39 papers using crossover designs (which are a form of repeated measures design) and found 58% of the papers did not use an analysis method consistent with the design, which could “compromise the validity of the findings”. Papers that used the invalid analysis are valueless scientifically, unless their raw data is available for re-analysis. RR practices require the publication of the raw data to address this problem.
In another recent study, Jørgensen et al. [19] suggested that the trustworthiness of software engineering experiments needs to be improved. They were particularly concerned about low power, researcher bias, and publication bias. Among their recommendations they include improving the reporting of study design, analysis and results, making data available, emphasising effect sizes and their confidence intervals, and undertaking more replications and meta-studies. The first two issues are directly supported by reproducible research requirements to make data and analysis available. The issues related to effect sizes and meta-analysis are supported by reproducible research, since if data is fully reported, subsequent studies can easily reanalyse the data to calculate effect sizes and perform meta-analysis.
Overall, these studies suggest that both our data intensive studies and human-centric experiments sometimes fail to provide reliable evidence to support technology adoption decisions. Reported issues also suggest that RR should be adopted more widely within the software engineering community.
Problems such as those discussed above are not unique to the software engineering domain. For example, in the context of drug trials, Osherovich reports that “an ‘unspoken rule’ among early stage VCs [Venture Capitals] is that at least 50% of published studies, even those in top-tier academic journals, can’t be repeated with the same conclusions by an industrial lab” [32]. In addition, Ioannidis and his colleagues reported that only 2 of 18 research papers published by Nature Genetics journal (one of the highest ranked journals in the world, with the impact factor about 30) could be fully reproduced [18]. The reasons for this included data sets and home made software disappearing, or the specification of data processing and analysis being incomplete. Probably the most striking summary of the research crisis in multiple disciplines is given by Ioannidis who (in his seminal paper with 3600+ citations) claims that “Most Research Findings Are False for Most Research Designs and for Most Fields” [17].
4Reproducibility in software engineering
Within the context of software engineering experiments, there has been discussion of laboratory packages, which were pioneered by Basili and his colleagues with the aim of assisting replications by providing additional detailed information about specific experiments, such as experimental materials, detailed instructions related to the experimental process and the methods used for data analysis (see [2], or [40]). However, laboratory packages were concerned with replication rather than reproducibility, and therefore did not emphasize the inclusion of data sets. Nonetheless, they could easily be extended to include the information needed for reproducibility.
Robles and his colleagues have discussed the importance of reproducibility from the view point of studies involving data mining from software repositories. Robles [35] undertook a systematic review of papers published in the former International Workshop on Mining Software Repositories (MSR) (2004–2006) and now Working Conference on MSR (2007–2009). He checked 171 papers for i) the public availability of the data used as case study, ii) the public availability of the processed dataset used by researchers and iii) the public availability of the tools and scripts. He found researchers mainly used publicly available data but the availability of the processed data used in specific studies was low. In the majority of papers, he could not find references to any tools even when authors said they had produced one. He concluded that there was a need for the community to address replicability in a formal way.
Robles and German [36] discuss best practices for supporting reproducibility, in particular providing a snapshot of the data as it was used in the study, versioning data sets, identifying the conditions under which the data can be used, making the data set available in a public repository, making tools available to others, licensing software, and providing the infrastructure to support tools via a forge. They also point out additional benefits of reproducibility to the data mining community such as providing worked examples for software engineering students and improved benchmarking.
González-Barahona and Robles [15] provide a method of assessing the reproducibility of a data mining study that is useful not only to other researchers but also to authors who can judge the main barriers to reproducing their study and reviewers who can assess whether a study will allow for easy reproduction.
In the context of software maintenance research, Dit et al. [10] have constructed a publicly available library of components and experiments aiming to improve the reproducibility and extensibility of software maintenance experiments.
In addition, Bowes et al. [5] have developed the SLuRp tool which provides the elements needed to support reproducibility for systematic reviews.
5Tools for reproducible research
We discuss the concept of Literate Programming behind RR and some of the tools that can be used to adopt RR, in Sections 5.1 and 5.2, respectively. Furthermore, we present a small working example of the RR approach in Section 6.
5.1Literate programming
Several researchers ([12, 26, 42]) have linked reproducible research to Knuth’s concept of Literate Programming [24]. They point out that if the output of research is not just a paper, but also the full computational environment, then researchers can use Knuth’s concept of Literate Programming [24] to achieve RR.
Literate Programming treats a program as a piece of literature addressed to human beings rather than a computer. The key assumptions are:
– A program should have plain language explanations interspersed with source code.
– The source code, data and plain language explanations are combined together.
– Results of program (or code chunks) are automatically included when document is created (so no exporting and/or importing is needed).
– After recompilation, changes are automatically incorporated if code or data sets change.
– Tools are available to make this simple to achieve.
5.2A reproducible research environment
We used the following freely available tools and formats to support our attempts to adopt reproducible research methods:
– The R programming language was used for all data analyses [34]. We also used the optional but useful R Studio integrated development environment which supports both, R and LATEX.
– The paper was written in LATEX and incorporated the R code using an R package called knitr [48]. Several other R packages were employed depending on particular requirements.
– Data sets and analytical procedures should be stored in a reliable manner and easily available to reviewers and readers. We decided to develop the reproducer R package [27] and made it available from CRAN – the official repository of R packages. Data sets analyzed in our three research papers [20, 22, 28] are encapsulated in the reproducer R package, while most of the figures and tables (particularly those which depend on data), as well as computational results, are built on the fly from data sets stored in the reproducer package and automatically exported into the manuscript rather than copied.
– All references were stored in the pure BibTeX format. R packages support generating BibTeX entries describing packages in a consistent way, including also the crucial information which package versions were used.
We used R language and environment for data analysis because we found it useful for several purposes. R is a mature language derived from S-plus. The R environment is open source and free to use. R is important for RR because the use of a statistical language and open source environment provide more traceability to the details of the statistical analysis than a closed source statistical package that includes various built-in defaults that are not accessible to check. In addition, typing an R script is more reproducible and easier to communicate than using the point-and-click user interface often adopted in other statistical packages. Last but not least, R provides not only an excellent support for quantitative analyses (including recent statistical methods and simulation), but also some support for qualitative analyses, e.g., the RQDA [16] and QCA packages [11]. Although we must make it clear that we have no first hand knowledge of applying RR to a qualitative study.
Other tools and formats can be used to achieve the goal of reproducible research, see, for example [25]. In addition, there are specialised tools that support RR in a specific context such as the Component Library developed by Dit et al. [10] to support software maintenance studies. However, in this paper, we concentrate on discussing the tools we ourselves have used.
The knitr package [48] provides the mechanism for linking R-code into basic LaTeX documents and can be easily accessed using R Studio. LaTeX, itself, is an ideal language for representing mathematical and statistical equations. We found xtable package [9] particularly useful for generating, on the fly, nicely styled tables in LaTeX or HTML from data structures produced using R. Another useful package, packrat [43], includes a collection of features for RR with R, e.g., the ability to install specific R package versions. This is important functionality because changes between packages can impede reproducibility. Therefore, we recommend recording the R session info, which makes it easy for future researchers to recreate what was done in the past and indentifies which versions of the R packages were used. The information from the session we used to create this research paper is shown in Output 7 in Section 6.
We used the BibTeX format to store references, because it allows us not only to easily import and populate BibTeX entries from the clipboard, files and digital libraries (e.g., ACM, IEEE, SpringerLink, Scopus), but also to automatically create BibTeX citations for R packages (which may include the data, analysis algorithms or both) inside a RR document, which makes reproducibility easier and less prone to typos. In our case references were managed using a reference management tool called BibDesk [30] but any other BibTeX-oriented reference manager could be used as well (e.g., JabRef).
6An example of the RR process
To assist the uptake of RR this section presents a small working example of the RR approach. We will use a real data set recently analysed by Madeyski and Jureczko [28] and available from the reproducer R package [27]. The analytic example presented in this section is based on empirical comparison of simple and advanced software defect prediction models performed on thirty-four (15 industrial and 19 open source) versions of software projects. The study investigated whether an advanced model, which includes both product metrics and the process metric NDC (Number of distinct committers), outperforms a simple model, which includes only product metrics. The performance of models was measured by the percentage of classes that must be tested in order to find 80% of the software defects.
It is worth mentioning that our example is deliberately simple so it can provide a starting point for novices. Researchers with more experience of RR concepts can view the reproducer R package [27] including data sets analyzed in our recently published research papers [20, 22, 28].
The steps needed to produce reproducible research follow indicating in each case the goal of the specific step:
1. Goal: Setup the basic RR environment.
Steps: Install R, LaTeX (e.g., TeX Live, MacTeX or Miktex) and RStudio (an integrated development environment which supports both, R and LaTeX)1.
2. Goal: Setup the convenient integrated development environment for RR.
Steps: Launch RStudio (navigate to the “Tools” menu and the “Global Options...” submenu, select the “Sweave” option and make sure the “Weave Rnw files using:” option is set to “knitr”, the “Typeset LaTeX into PDF using:” option is set to “pdfLaTeX”).
3. Goal: Setup the mechanism for linking the data analyses (and their the results) into text documents.
Steps: Click on “Console” window in RStudio, install and load the knitr R package by running:
install.packages (’knitr’,
dependencies=T, repos=
“http://cran.rstudio.com/”)
library (’knitr’) # Load
’knitr’
4. Goal: Setup the document containing text and analysis procedures that can be executed on data (analytic results, figures and tables can be produced on the fly from data).
Steps: Create an Example.Rnw file (in RStudio using the “File” → “New file” → “R Sweave” submenu option) containing text (with the LaTeX markup) and data analysis (with R code chunks, i.e., sections of R code). Each chunk can have its own options to configure how it is rendered. To reproduce the example, we recommend readers to open the file from http://madeyski.e-informatyka.pl/download/R/Example.Rnw, copy and paste it into the created Example.Rnw file in R Studio, instead of using copy and paste from this article in the PDF format.
The Example.Rnw file should then contain the following text:

5. Goal: Compile the.Rnw to.tex and.pdf. You may use RStudio, see Fig. 1.
Subsequent steps of data analysis, from the Example.Rnw file, and respective results (automatically embedded in this paper with the help of the knitr package) are briefly presented below2:
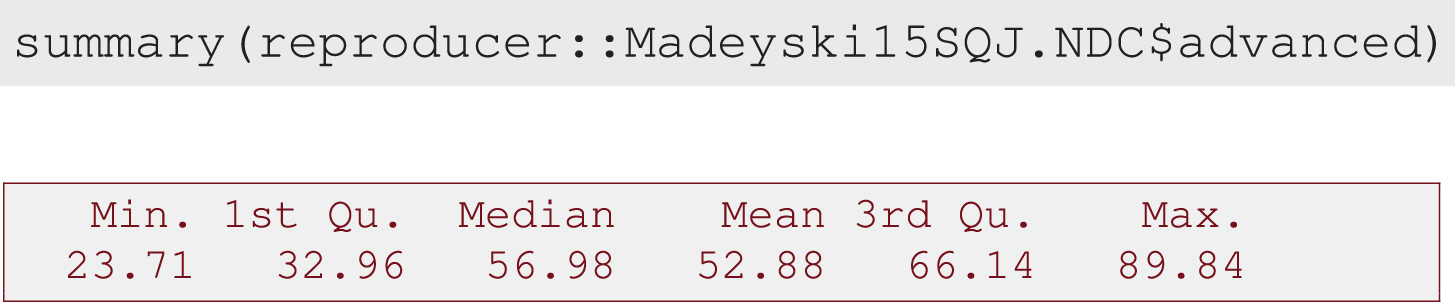
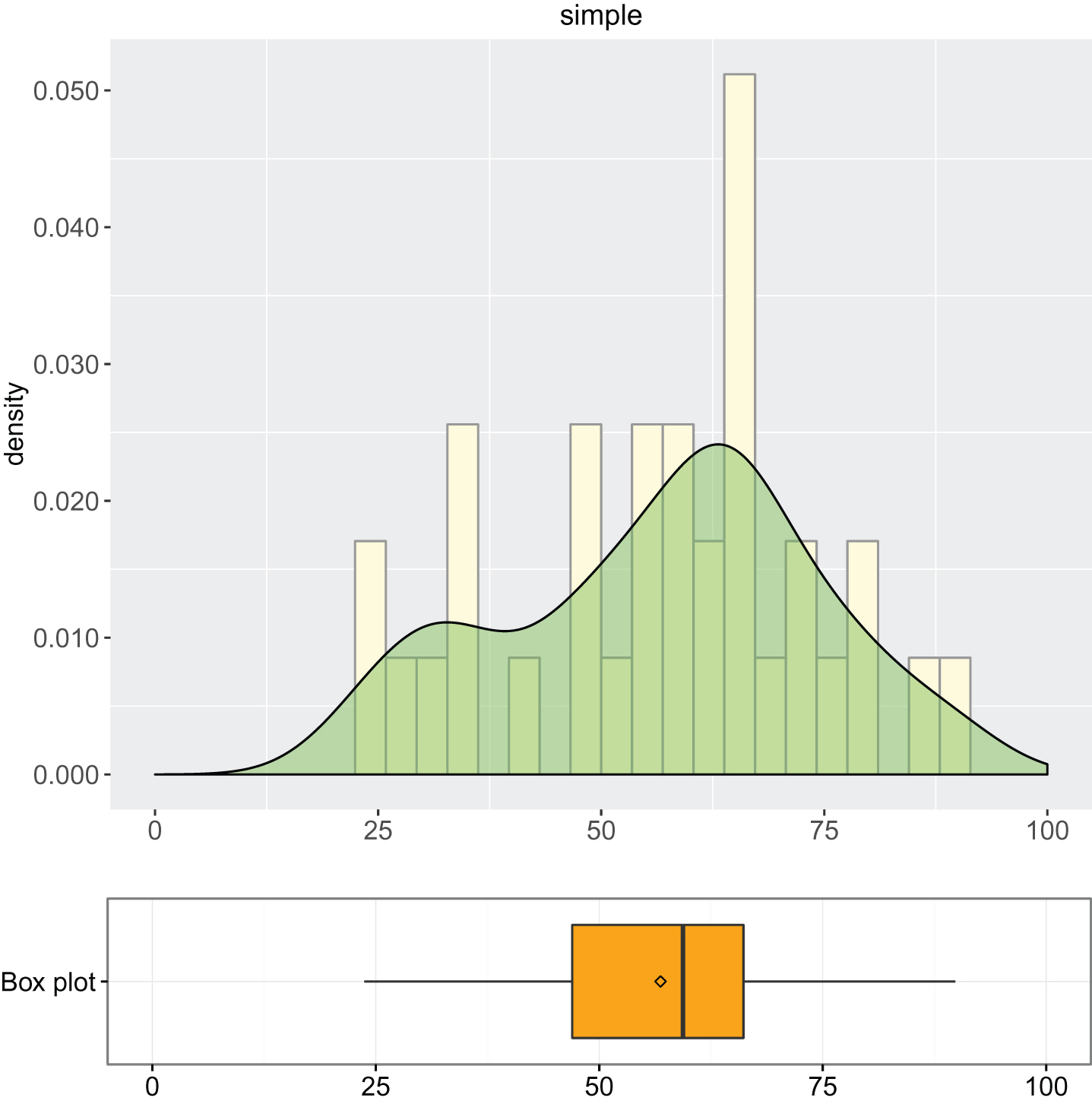
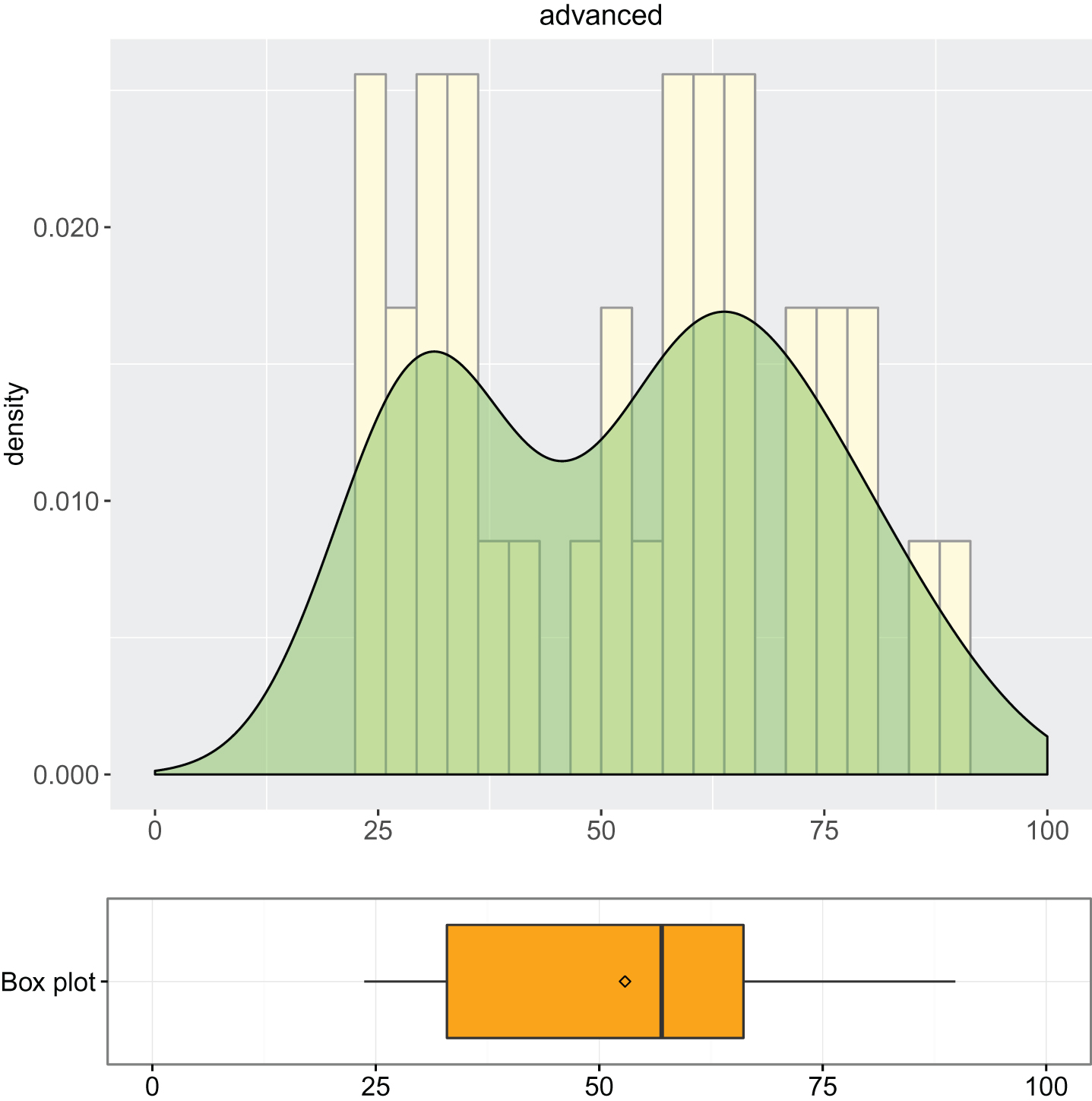
1. Descriptive analysis of simple and advanced models: summary of descriptive statistics (Output 1 and Output 2) and box plot combined with density curve laid out on histogram (Output 3, Output 4 and Output 5).
Output 1
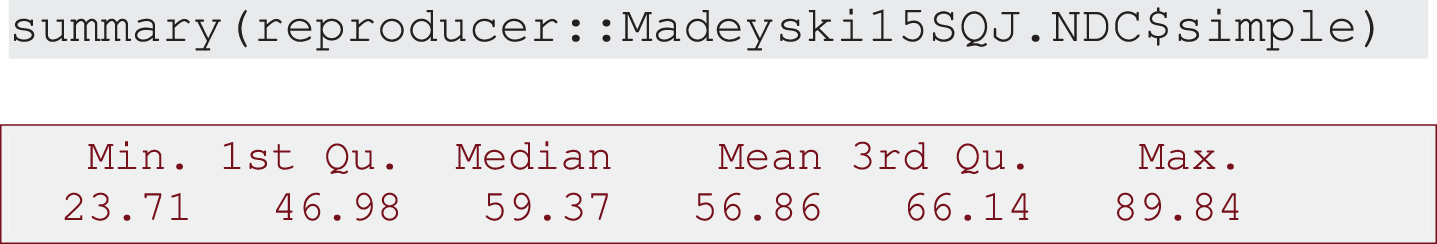
Output 2
Output 3

Output 4

Output 5

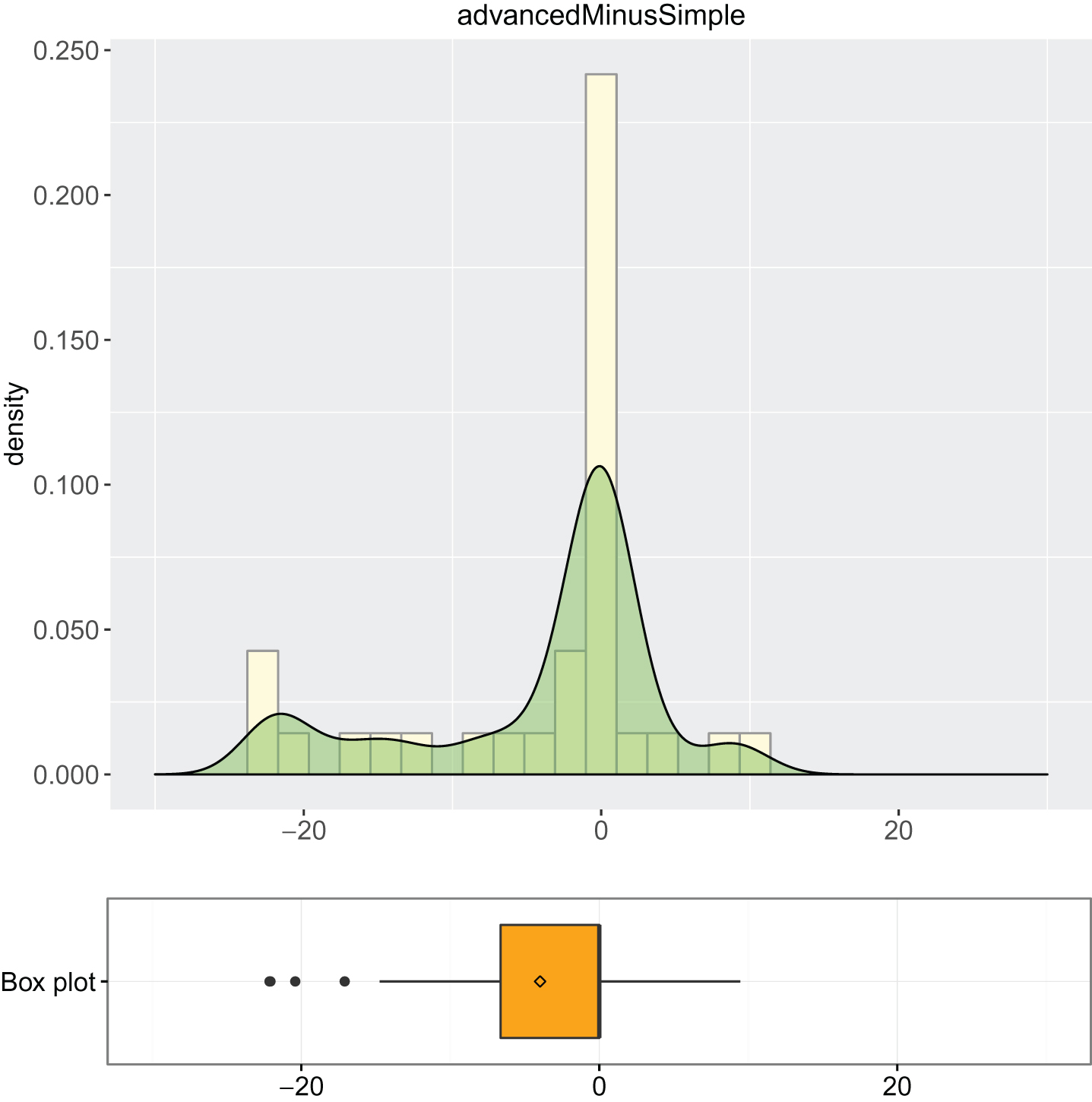
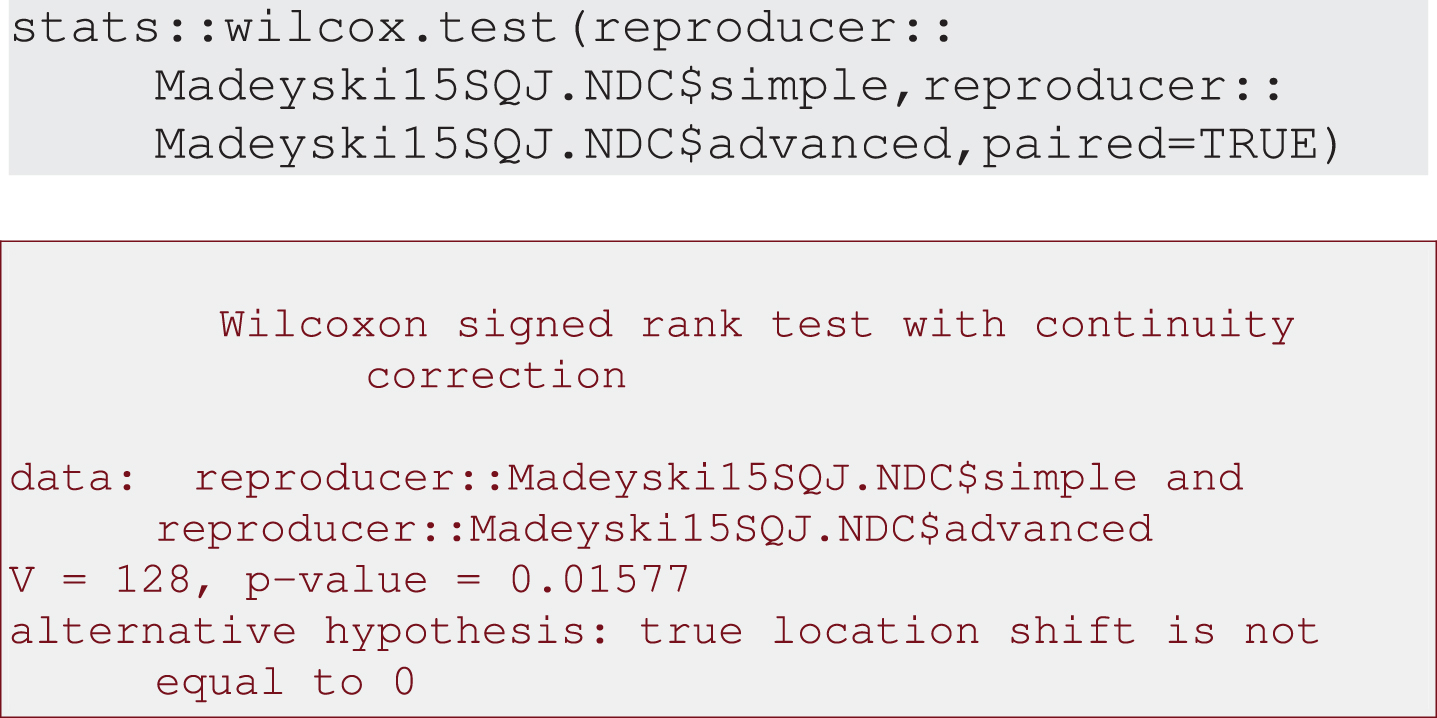
2. Inferential analysis using Wilcoxon test: as the data are non-normal, t-tests may not be appropriate and an alternative is Wilcoxon paired test, see Output 6.
Output 6
Since, as mentioned in Section 5.2, changes between packages can impede reproducibility, we recommend recording the R session info, which makes it easy for future researchers to recreate what was done in the past and which versions of the R packages were used. How to do it, as well as a result is shown inOutput 7.
Output 7
It is also worth mentioning that one can easily view the data set embeeded in the reproducer R package, seeOutput 8.
Output 8
The detailed description of the data set, as in case of other R packages, is available from CRAN(https://cran.r-project.org/web/packages/reproducer/reproducer.pdf).
Readers are invited to reproduce our small working example of reproducible research. All of our analyzes, as well as data, are encapsulated in the reproducer R package we created and made available from CRAN—the official repository of R packages [27]. Our simple working example (Example.Rnw file) is now available at http://madeyski.e-informatyka.pl/download/R/Example.Rnw.
We used the method proposed by González-Barahona and Robles [15] to assess reproducibility of our example, as shown in Table 1. The reproducer R package itself supports the availability, persistence and flexibility of the data set, whilst the use of R scripts (code chunks), which use functions embedded in R packages, helps to ensure that parameters of analysis are identified. In our case, the data set is held in the R package which is in effect the data source. The data used in our analysis (referred to as the Raw Data in [15]) is accessed by the R code chunks. Furthermore, all the tools we describe are freely available at no cost. An added benefit of using CRAN, is that it ensures that the data set variables and analysis package parameters are fully documented in the package. It is hard to overestimate the significance of such documentation. It should be noted that our example is quite simple because we do not need to extract the data set from a separate independent data source. When data are held in independent data sources, the tools we recommend do not address the issue of identifying and preserving the data source, however, the retrieval methodology and the raw data set can be specified in the R package.
7Discussion
In this section we discuss some of the pros and cons of reproducible research, as well as major RR initiatives.
7.1Advantages of reproducible research
Reproducible Research does not address all problems of the validity of experimental software engineering studies. It can only ensure that the data and analysis methods are available for inspection and that the results presented in the paper can be derived from the data and analysis procedures. However, if it were adopted, we hope that design and analysis errors, such as those reported by Vegas et al. [46] and Shepperd et al. [39] would be more likely to be uncovered, hopefully, prior to publication during the review process or soon after publication as the full details will be available to all interested readers. Furthermore, it would provide a valuable resource for training novice researchers. However, there are other advantages as well.
There is some research evidence that reproducibility improves the impact of research. For example, Piwowar et al. reported that from a set of 85 studies, the 48% with publicly available data received 85% of the total citations [33]. Data availability significantly increased citation rate (p = 0.006) independently of the impact factor of the source journal, date of publication, and author country of origin. Vandewalle et al. report a study that related the reproducibility of papers to the number of citations and conclude that “computational papers that do not have code and data available online have a low chance of being cited” [45].
Vandewalle et al. also described their own experiences of RR. They reported a gain in their own efficiency, because it was easier to pick up their work again, and positive feedback from colleagues and students who downloaded their code. In addition, they state that the availability of their code “allowed and simplified some collaborations and is a source of easily reusable demo material for students and visitors”.
For a research group, RR supports the preservation of group knowledge in terms of long-term conservation of experimental data together with supporting (e.g., statistical) analyses. This means that knowledge of current research is not lost when researchers move on, and it is easier for new members of a group to understand and build on previous research. It can also help researchers leverage their own research. For example, Bowes et al. report developing and using an environment to support complex systematic reviews that supports RR principles [5]. They have used this environment to support a number of different systematic reviews. In addition, funding agencies and journals are increasingly adopting open research policies and RR provides a means of complying with such policies.
Using tools such as R and RStudio as part of an RR project also supports the iterative nature of research where results are often revised and re-analysed by providing mechanisms to easily update tables and figures. Furthermore, having used an RR method to create a research paper, RStudio or slidify [44] allow us to produce interactive, reproducible conference and seminar presentations, generating figures and tables on the fly from our data using R. Thus, our presentations remain consistent with our papers and can be easily updated if data are changed or new analyses are required.
7.2Objections to reproducible research
There are two major objections related to RR. The first objection, discussed in Section 7.2.1, is the potential loss of intellectual property caused by making data freely available. The second one, examined in Section 7.2.2, is an additional effort imposed by RR.
7.2.1Potential loss of intellectual property
Sharing of data has attracted some strong disagreements among software engineering researchers. At the International Workshop on Empirical Software Engineering Issues held at Dagstuhl Castle, Germany in 2006 [3], participants discussed data sharing and concluded that there were areas of dissent among the software engineering community. Some people wanted licensing, others were opposed, and there was no agreement as to who owns the data. Subsequently, Basili et al. [4] published a proposal for data and artifact sharing agreements in software engineering research. This proposal concerned a framework for software engineering artifact agreement to “foster a market in making available and using such artifacts”. The limitation of the market-based viewpoint is that it fails to address two issues:
1. Ownership may not reside solely with the individual researchers but also with any research funding agency that supported the research.
2. Scientific ethics as well as research agencies advocate open sharing of results including data.
Stodden proposed an alternative approach called the Reproducible Research Standard (RRS) which aims to realign legal rights to fit scientific norms [41]. RRS is a legal tool for waiving as many rights as legally possible, worldwide. In particular, she suggests that authors:
– Release media components (text, figures) under CC BY, that is, the Creative Commons attribution license that does not have a Share Alike provision. This means that licensees may copy, distribute, display and perform the work and make derivative works based on it, only if they give the author or licensor the credits in the manner specified by the copyright holders.
– Release code components under a Berkeley Software Distribution (BSD) license that place few restrictions on re-use beyond attribution, creating an Intellectual Property framework resembling conventional scientific norms.
– Release data under the Science Commons Database Protocol3 because “raw data aren’t copyrightable” only “selection and arrangement” of data is copyrightable. Generally, data sets should be made available in recognized repositories for the field, if they exist. Otherwise, researchers may choose a repository for sharing, citing, analyzing, and preserving research data, which is open to all scientific data from all disciplines, e.g., Dataverse, OpenAIRE.
However, the approach we have adopted to make RR available is to create an R package [27], which is a free, open access way to share (via CRAN—the official R repository) both, data and code with accompanying detailed description of every field of the shared data sets, and every function and parameter of the shared code. As an alternative, Gandrud suggests using GitHub to share your research, pointing out that projects can can be kept private initially and then made public once the research results are published [12].
7.2.2Additional effort required by reproducible research
The second objection is that, in our experience, RR requires additional effort to write research papers. Some of the tools used to support RR (i.e., LaTeX, R and BibTeX) are mature tools that are already used by many software engineering researchers. However, tools such as RStudio and especially knitr that provide an environment to support RR may require time to become conversant with. Thus, although RR might be necessary for researchers aiming at internationally leading journals in critical disciplines such as medicine, some researchers might consider RR too much of an overhead for software engineering research.
There are other issues that make adoption of RR difficult for software engineers. In software engineering, it is often the case that data cannot be distributed due to confidentiality issues. However, there are tools that offer data anonymization functionality in a way expected by an industrial partner, e.g., DePress software measurement and prediction framework [29], developed as an open source project in close collaboration between Wroclaw University of Science and Technology and Capgemini software development company.
In addition, if data items need to be obtained from a variety of different sources and need to be integrated into a single data set, it may be very difficult to make the process of formatting the raw data into analyzable data completely reproducible.
7.3Reproducible research initiatives
Interest in reproducible research has led to two major scientific initiatives:
1. In psychology, the Open Science Collaboration aims to “to increase the alignment between scientific values and scientific practices” by open, large-scale, collaborative effort to estimate the reproducibility of psychological science [8, 31]. The recent result from this initiative has been published in Science in 2015 [7].
2. In medicine, Ioannidis and Goodman have established the Meta-Research Innovation Center at Stanford University (METRICS)4. This center aims to improve reproducibility by studying “how research is done, how it can be done better, and how to effectively promote and incentivize the use of best scientific practices”. In his recent video lecture5, Ioannidis proposed registration of data sets, protocols, analysis plans, and raw data. We expect results from this initiative provide further discussion of RR concepts.
These initiatives are likely to encourage funding agencies and journals to police their open science policies more rigorously.
8Conclusions
We have identified studies criticising the current software engineering practices. In our view these criticisms are serious enough that we need to consider carefully how we establish the validity of our research outcomes. Following ideas proposed in the data mining community and adopted in other empirical disciplines, we raise the question of whether it would be beneficial for researchers undertaking human-centric studies to adopt RR. In software engineering, particular research groups investigating specific topic areas are using RR principles, but there is little general agreement about whether the ideas should be more widely adopted. Critical issues are the extent to which researchers are willing to share their data and the time and effort needed to make data available for sharing. However, if we continue as we are, we run the risk of publishing more and more invalid and incorrect results with no systematic methods of correcting them.
RR, as discussed in this paper, concerns the extent to which the report of a specific study can be deemed trustworthy. It supports only the minimum level of validity that we should expect of research outcomes. It would, however, address problems currently being found in software engineering research by various leading researchers ([19, 39 and 46]). The example of reproducible research reported in this paper identifies free-to-use tools that are currently available to support RR. Our example shows how they can be integrated to adopt an RR approach. It should help other researchers to try out the RR approach for themselves. Other empirical software engineering researchers willing to share their data sets, and related analytic procedures, via the reproducer package may contact the first author—the maintainer of the package on CRAN.
Notes
2 Output 1–Output 8 include the R commands, as well as results.
References
[1] | Barr E. , Bird C. , Hyatt E. , Menzies T. and Robles G. , On the shoulders of giants. In Proceedings of the FSE/SDP Workshop on Future of Software Engineering Research, FoSER ’10, ACM, (2010) , pp. 23–28. |
[2] | Basili V.R. , Shull F. and Lanubile F. , Building knowledge through families of experiments, Transactions on Software Engineering 25: (4) ((1999) ), 456–473. |
[3] | Basili V.R. , Rombach D. , Schneider K. , Kitchenham B. , Pfhal D. and Selby R. , Empirical Software Engineering Issues: Critical Assessment and Future Directions LNCS 4336: , Springer, (2006) . |
[4] | Basili V.R. , Zelkowitz M.V. , Sjøberg D.I. , Johnson P. and Cowling A.J., Protocols in the use of empirical software engineering artifacts, Empirical Software Engineering 12: (1) ((2007) ), 107–119. |
[5] | Bowes D. , Hall T. and Beecham S. , Slurp: A tool to help large complex literature reviews. In Proceedings of the 2nd International workshop on Evidential Assessment of Software Technologies, EAST, (2012) , pp. 33–36. |
[6] | Carver J.C. , Juristo N. , Baldassarre M.T. and Vegas S. , Replications of software engineering experiments, Empirical Software Engineering 19: (2) ((2014) ), 267–276. |
[7] | Open Science Collaboration. Estimating the reproducibility of psychological science, Science 349: (6251) ((2015) ). |
[8] | Open Source Collaboration. An open, large-scale, collaborative effort to estimate the reproducibility of psychological science, Perspectives on Psychological Science 7: (6) ((2012) ), 657–660. |
[9] | Dahl D.B. , xtable: Export tables to LaTeX or HTML, (2016) . R package version 1.8–2. |
[10] | Dit B. , Moritz E. , Linares-Vásquez M. and Poshyvanyk D. , Supporting and accelerating reproducible research in software maintenance using tracelab component library. In Software Maintenance (ICSM), 2013 29th IEEE International Conference on, (2013) , pp. 330–339. |
[11] | Dusa A. and Thiem A. , Qualitative Comparative Analysis, (2014) . R Package Version 1.1–4. |
[12] | Gandrud C. , Reproducible Research with R and R Studio, CRC Press, (2013) . |
[13] | Gentleman R. and Temple Lang D. , Statistical analyses and reproducible research, Journal of Computational and Graphical Statistics 16: (1) ((2007) ), 1–23. |
[14] | Gómez O.S. , Juristo N. and Vegas S. , Understanding replication of experiments in software engineering: A classification, Information and Software Technology 56: ((2014) ), 1033–1048. |
[15] | González-Barahona J.M. and Robles G. , On the reproducibility of empirical software engineering studies based on data retrieved from development repositories, Empirical Software Engineering 17: ((2012) ), 75–89. |
[16] | Huang R. , RQDA: R-based Qualitative Data Analysis, (2014) . R package version 0.2–7. |
[17] | Ioannidis J.P.A. , Why most published research findings are false, PLoS Medicine 2: (8) ((2005) ), 696–701. |
[18] | Ioannidis J.P.A. , Allison D.B. , Ball C.A. , Coulibaly I. , Cui X. , Culhane A.C. , Falchi M. , Furlanello C. , Game L. , Jurman G. , Mangion J. , Mehta T. , Nitzberg M. , Page G.P. , Petretto E. and van Noort V. , Repeatability of published microarray gene expression analyses, Nature Genetics 41: ((2009) ), 149–155. |
[19] | Jørgensen M. , Dybå T. , Liestøl K. and Sjøberg D.I.K. , Incorrect results in software engineering experiments: How to improve research practices, Journal of Systems and Software 116: ((2016) ), 133–145. |
[20] | Jureczko M. and Madeyski L. , Cross–project defect prediction with respect to code ownership model: An empirical study, e-Informatica Software Engineering Journal 9: (1) ((2015) ), 21–35. |
[21] | Kitchenham B. and Mendes E. , Why comparative effort prediction studies may be invalid, In Proceedings of the 5th International Conference on Predictor Models in Software Engineering, PROMISE ’09, ACM, (2009) , pp. 4:1–4:5. |
[22] | Kitchenham B.A. , Madeyski L. , Budgen D. , Keung J. , Brereton P. , Charters S. , Gibbs S. and Pohthong A. , Robust statistical methods for empirical software engineering, Empirical Software Engineering ((2016) ). DOI: 10.1007/s10664-016-9437-5 (in press). |
[23] | Kitchenham B.A. and Madeyski L. , In Meta-analysis. Kitchenham B.A. , Budgen D. and Brereton P. , editors, Evidence-Based Software Engineering and Systematic Reviews, chapter 11, CRC Press, (2016) , pp. 133–154. |
[24] | Knuth D.E. , Literate programming, The Computer Journal 27: (2) ((1984) ), 97–111. |
[25] | Kuhn M. , Cran task view: Reproducible research. http://cran.r-project.org/web/views/ReproducibleResearch.html |
[26] | Leisch F. , Sweave, Part 1: Mixing R and LaTeX: A short introduction to the Sweave file format and corresponding R functions, R News 2/3: ((2002) ), 28–31. |
[27] | Madeyski L. , Reproducer: Reproduce Statistical Analyses and Meta-Analyses, (2016) . R package version 0.1.5 (http://CRAN.R-project.org/package=reproducer). |
[28] | Madeyski L. and Jureczko M. , Which process metrics can significantly improve defect prediction models? An empirical study, Software Quality Journal 23: (3) ((2015) ), 393–422. |
[29] | Madeyski L. and Majchrzak M. , Software measurement and defect prediction with dePress extensible framework, Foundations of Computing and Decision Sciences 39: (4) ((2014) ), 249–270. |
[30] | McCracken M. , Maxwell A. and Hofman C. , BibDesk. http://bibdesk.sourceforge.net/, (2015) . |
[31] | Nosek B.A. , Spies J.R. , Cohn M. , Bartmess E. , Lakens D. , Holman D. , Cohoon J. , Lewis M. , Gordon-McKeon S. , IJzerman H. , Grahe J. , Brandt M. , Carp J.M. and Giner-Sorolla R. , Open science collaboration, (2015) . |
[32] | Osherovich L. , Hedging against academic risk, Science–Business eXchange 4: (15) ((2011) ). |
[33] | Piwowar H.A. , Day R.S. and Fridsma D.B. , Sharing detailed research data is associated with increased citation rate, PLoS ONE 2: (3) ((2007) ). |
[34] | R Core Team, R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, (2015) . |
[35] | Robles G. , Replicating MSR: A study of the potential replicability of papers published in the Mining Software Repositories proceedings. In Mining Software Repositories (MSR), 2010 7th IEEE Working Conference on, (2010) , pp. 171–180. |
[36] | Robles G. and Germán D.M. , Beyond replication: An example of the potential benefits of replicability in the mining of software repositories community. In Proceedings 1st International Workshop on Replication in Empirical Software Engineering Research (RESER 2010), (2010) . |
[37] | Runeson P. , Stefik A. and Andrews A. , Variation factors in the design and analysis of replicated controlled experiments. Three (dis)similar studies on inspections versus unit testing, Empirical Software Engineering 19: ((2014) ), 1781–1808. |
[38] | Schwab M. , Karrenbach M. and Claerbout J. , Making scientific computations reproducible, Computing in Science and Engineering 2: (6) ((2000) ), 61–67. |
[39] | Shepperd M. , Bowes D. and Hall T. , Researcher bias: The use of machine learning in software defect prediction, IEEE Transactions on Software Engineering 40: (6) ((2014) ), 603–616. |
[40] | Shull F. , Basili V. , Carver J. , Maldonado J.C. , Travassos G.H. , Mendonça M. and Fabbri S., Replicating software engineering experiments: Addressing the tacit knowledge problem. In Proceedings of International Symposium on Empirical Software Engineering ISESE, (2002) , pp. 7–16. |
[41] | Stodden V. , The legal framework for reproducible scientific research licensing and copyright, Computing in Science and Engineering 11: (1) ((2009) ), 35–40. |
[42] | Stodden V. , Leisch F. and Peng R.D. editors. Implementing Reproducible Research. CRC Press, (2014) . |
[43] | Ushey K. , McPherson J. , Cheng J. and Allaire J.J. , Packrat: A Dependency Management System for Projects and their R Package Dependencies, (2015) . R package version 0.4.3. |
[44] | Vaidyanathan R. , slidify: Generate reproducible html5 slides from R markdown, (2012) . R package version 0.4.5. |
[45] | Vandewalle P. , Kovacevic J. and Vetterli M. , Reproducible research in signal processing, IEEE Signal Processing Magazine 26: (3) ((2009) ), 37–47. |
[46] | Vegas S. , Apa C. and Juristo N. , Crossover designs in software engineering experiments: Benefits and perils, IEEE Transactions on Software Engineering 42: (2) ((2016) ), 120–135. |
[47] | Whigham P.A. , Owen C. and MacDonell S. , A ine model for software effort estimation,20:1–20:, ACM Transactions on Software Engineering and Methodology 24: (3) ((2015) ), 11–basel. |
[48] | Xie Y. , knitr: A General-Purpose Package for Dynamic Report Generation in R, (2016) . R package version 1.12.3. |
Figures and Tables
Fig.1
Compilation of Example.Rnw in RStudio.
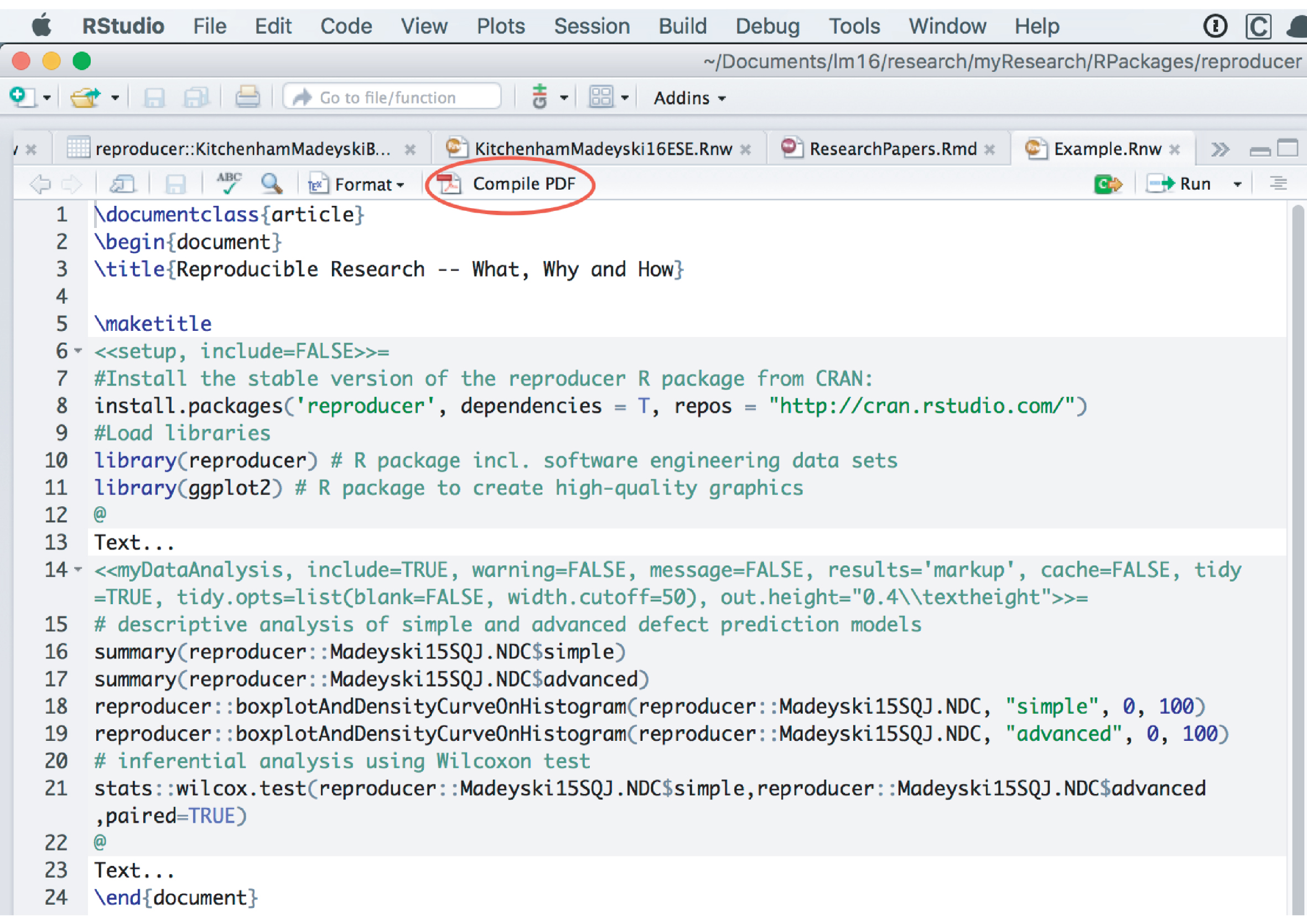
Table 1
Assessment of the Capability for Reproducibility of Our Tool Set
| Element | Assessment |
| Data Source | Usable |
| Retrieval Methodology | Usable |
| Raw Data Set | Usable |
| Extraction methodology | Usable |
| Available in Future | |
| Flexible | |
| Study Parameters | Usable |
| Analysis Methodology | Usable |
| Results Dataset | Usable |
| Flexible |




