A high-throughput marker-assisted selection system combining rapid DNA extraction high-resolution melting and simple sequence repeat analysis: Strawberry as a model for fruit crops
Abstract
BACKGROUND:
The strawberry is an important commodity as well as a model plant species in the Rosaceae. DNA marker availability for various traits in octoploid cultivated strawberry has increased intensely in recent years. To date, laborious and expensive DNA extraction procedures have been required to achieve sufficient DNA quality in this recalcitrant species. When combined with gel-based marker systems, current genotyping methods are becoming a bottleneck for marker-assisted selection (MAS) in large strawberry breeding populations.
OBJECTIVE:
The main objective of this work was to develop a high-throughput marker system that combines 1) rapid DNA extraction and 2) rapid, cost-effective, and accurate genotyping, even when using crude strawberry DNA extracts. To this end, we aimed to develop high-throughput high-resolution melting (HRM) and SSR assays for selection at the FaFAD1 locus conferring peach-like aroma (γ-decalactone).
METHODS:
Eight cultivars and four advanced selections of strawberry (Fragaria×ananassa Duchesne) were used in this study. A rapid DNA extraction method from strawberry leaf discs was developed by modifying a previously published NaOH-based method. Three high-throughput HRM markers were developed for detecting the presence/absence of FaFAD1, and HRM analysis was performed using a Roche LightCycler 480.
RESULTS:
Rapid NaOH-based DNA extraction was successful for accurate and repeatable marker screening when diluted 5-fold prior to PCR and when non-acetylated BSA and PVP were added to the PCR reaction. Three HRM markers were successfully developed and used to detect the presence/absence of FaFAD1, with equivalent results to those obtained using the previously published gel-based marker.
CONCLUSIONS:
This high-throughput genotyping system has been successfully employed to screen approximately 6,000 seedlings in the University of Florida strawberry breeding program, and the method is being expanded for MAS at additional loci. The system should be adaptable to other berry crops for which fast DNA extraction, and low-cost and accurate genotyping are needed for large breeding populations.
1Introduction
The cultivated strawberry (Fragaria×ananassa Duchesne) is an allo-octoploid (2n = 8x = 56) belonging to the Rosaceae. The genome of diploid strawberry (Fragaria vesca) has recently been sequenced, rearrangement of scaffold improved, and genome scanning with sub-genome specificity is now routine [1–4]. Thus, molecular marker techniques can be now widely applied for marker-assisted selection (MAS) in strawberry breeding [5–10]. To successfully apply MAS, it is necessary to rapidly and accurately screen a large number of individuals for multiple target traits. Traditional cetyl trimethylammonium bromide (CTAB) DNA extraction and gel-based genotyping are becoming a bottleneck for MAS in strawberry and in other berry and fruit crops. It is, therefore, critical to develop a high-throughput genotyping system that is faster and more cost-effective while still maintaining accuracy.
For effective MAS in strawberry and other Rosaceae fruit crops, a rapid DNA extraction method is necessary. Alkaline (NaOH) DNA extraction has been successfully used for many important crops [11–16]. Most notably, Xin et al. [14] reported a rapid extraction method that was successfully used for PCR amplifications in various plant species, and compatible with the use of fluorescently-labeled primers for PCR fragment analysis. Although rapid DNA extraction has been successfully adapted in other plant breeding programs, this method has never been reported in strawberry, which is particularly recalcitrant to DNA extraction. Strawberry leaf tissues produce high levels of secondary metabolites such as polyphenols and polysaccharides, which inhibit the activity of DNA polymerases and restriction enzymes [17, 18].
High-resolution melting (HRM) analysis is a powerful and cost-effective method to detect mutations and polymorphisms. This genotyping technique has been widely used for genetic studies in humans for over a decade, and recently applied for numerous plant breeding programs, because of its simplicity, accuracy, reproducibility, and low-cost [19]. Since HRM can distinguish multiple SNPs in PCR amplicons containing heteroduplex molecules, it can identify multiple alleles in polyploids and is emerging as a powerful tool for polyploid genetics [20, 21]. Therefore, this method is ideal for high-throughput genotyping, mapping genes, and DNA testing through MAS in plant breeding, especially for strawberry.
The γ-decalactone (peachy aroma) gene, FaFAD1, is one the most important traits in University of Florida strawberry breeding program. It has been described that strawberry varieties without FaFAD1 do not produce γ-decalactone. The functional presence/absence (PA) polymorphism of FaFAD1 has been reported in γ-decalactone producing and non-producing octoploid strawberry varieties [8, 23].
Here we describe the development of a rapid NaOH-based DNA extraction system in strawberry, combined with a new high-resolution melting (HRM) analysis for detecting the presence/absence of the FaFAD1 locus.
2Materials and methods
2.1Plant materials
A total of 12 UF strawberry cultivars and advanced selections were used in this study, ‘Florida Elyana’, Sweet Sensation® ‘Florida 127’, ‘Florida Radiance’, ‘Sweet Charlie’, Winterstar™, FL10-121, 11.28-34, 11.31-54, ‘Strawberry Festival’, ‘Florida Ninety’, ‘Winter Dawn’, and FL10-46. They are known as γ-decalactone producer or nonproducer based on the metabolite analysis (18 and unpublished data).
2.2Rapid DNA extraction
A set of NaOH-based, rapid DNA extractions were conducted following the methods described by Xin et al. [17] with modifications. Four leaf discs (each disc 3 mm in diameter) from 14-day-old strawberry seedlings (or young leaves from 2–3 months old plants) were collected into each well of a 96-well plate using a handheld hole puncher. With the plate on ice, buffer A (100 mM NaOH, 2% Tween 20) and buffer B [100 mM Tris-HCl (pH 8), 2 mM EDTA] were prepared. Buffer A (50 μl) was added to each well, and the plate was sealed with a foil lid (Extreme weather aluminum foil tape, 2.83 inch, HomeDepot or Lowe’s, USA) before centrifuging at 4,000 rpm for 2 min. The plate was then heated at 95°C for 10 min using an Eppendorf Mastercycler® ep gradient S Thermal Cycler (Eppendorf AG, Germany). Buffer B (50 μl) was subsequently added to each well, and the plate was again centrifuged at 4,000 rpm for 2 min and kept at 4°C for further use. To prevent possible cross-contamination, the plate was sealed with aluminum sealing foil and centrifuged at 4,000 rpm for 1 min before opening.
2.3Gel-based PCR detection of FaFAD1
PCR was performed in a 20 μl volume containing 1×Standard Taq Reaction Buffer (New England BioLabs, MA, USA), 0.2 mM dNTPs (New England BioLabs, MA, USA), 0.25 μM of each forward and reverse primer, 1.25 U Taq DNA polymerase (New England BioLabs, MA, USA), 0.1% bovine serum albumin (acetylated or non-acetylated BSA) (Sigma-Aldrich, MO, USA), 1% polyvinylpyrrolidone (PVP, MW 40,000) (Fisher Scientific, PA, USA), and 1.0 μl DNA solution (NaOH-crude extract from one to five-fold dilution, or CTAB-purified DNA). For large numbers of samples, PCR was performed in a 10 μl volume in 384 well plates with the following protocol; 5 μl of 2×PCR super mix (AccuStart II PCR Tough Mix, QuantaBio), 0.25 μM of each forward and reverse primer, 1.0 μl DNA solution (NaOH-crude extract from one to five-fold dilution, or CTAB-purified DNA). As the positive control, the gel-based marker for FaFAD1 described by Chambers et al. [8] was used to determine the presence or absence of FaFAD1 gene for all varieties tested in this study. The reactions were pre-incubated at 95°C for 3 min for initial denaturation, followed by 33 cycles of denaturation at 95°C for 20 sec, annealing at 62°C for 20 sec, and extension at 72°C for 15 sec, and completed with the final extension at 72°C for 10 min. All amplified PCR products were analyzed by electrophoresis in 1.5 % agarose gels stained with GelRed (Biotium, CA) and visualized under a UV transilluminator.
2.4Development of high-throughput high-resolution melting markers for FaFAD1
Three HRM markers, UFGDHRM2, UFGDHRM4 and UFGDHRM5, were developed to detect FaFAD1 in strawberry (Fig. 2). All primer sets were designed from the publically available FaFAD1 sequence (GenBank accession #KF887973) using IDT’s PrimerQuest Software (www.idtdna.com). The primer set for UFGDHRM2 (Forward: CGGTAGAGTACAGAAGCAGTGA, Reverse: GGCCGTTGTACTTGGCATTTA) binds the second exon of FaFAD1, and produces 154 base pair (bp) amplicons. The UFGDHRM4 marker (Forward: ACCTCGATCATAGCTACACTCTTT, Reverse: TAGAGTGAATGGCGGAGTCG AAAC) targets the first exon of FaFAD1 and produces 152 bp amplicons. UFGDHRM5 targets the promoter region of FaFAD1 (Forward: CCTCGATCATAGCTACACTCTTTC, Reverse: AGCCTTTGACGTGTCCTTATT) (Fig. 2).
2.5HRM and SSR marker analysis for FaFAD1
Both PCR and HRM analysis were carried out using a LightCycler® 480 system II (Roche Life Science, Switzerland). PCR amplifications were performed in a 10 μl reaction with 0.5 μl of template DNA using LightScanner Master Mix (BioFire, UT, USA) or AccuStart II PCR Tough Mix (Quantabio, MA, USA) in LightCycler® 480 Multiwell 96-well plates. The HRM PCR mixture was pre-incubated at 95°C for 3 min for initial denaturation, followed by 33 cycles of denaturation at 95°C for 20 sec, annealing at 62°C for 20 sec, and extension at 72°C for 15 sec. After PCR amplification, the samples were heated at 95°C for 1 min and then cooled at 4°C for 1 min. Melting curves were obtained by melting over the desired range (70–95°C) at a rate of 25 acquisitions per 1°C.
For SSR marker analysis, the primer information for detecting FaFAD1 was obtained from the previous study [18]. The PCR amplification was conducted in a 20 ul reaction with 0.5 μl of template DNA, 2 μl of 10×PCR buffer, 0.4 μl of dNTP (10 mM), 0.4 μl of forward (5 μM), 0.8 μl of reverse primer (5 μM), 0.8 μl of M13-FAM primer (5 μM), 1 unit of Taq polymerase (NEB, MA, USA): initial denaturation – 3 min at 94°C; 33 cycles – 30 s at 94°C, 40 s at 60°C, 40 s at 72°C; 10 cycles – 30 s at 94°C, 40 s at 53°C, 40 s at 72°C; final extension – 10 min at 72°C; 4°C forever. PCR samples were diluted 5-fold and fragment analysis was performed on ABI3730 at Interdisciplinary Center for Biotechnology (ICBR), University of Florida.
3Results and discussion
The rapid DNA extraction method developed in this study was first tested using the gel-based marker developed by Chambers et al. [8] for FaFAD1, which encodes a fatty acid desaturase essential for the biosynthesis of the peach-like aroma compound γ-decalactone (see also Sanchez-Sevilla et al. [22]). As shown in Fig. 1, PCR using crude DNA extract (no dilution or 2-fold dilution) failed to amplify FaFAD1 in cultivars and advanced selection that are known to produce γ-decalactone, such as ‘Florida Elyana’, Sweet Sensation® ‘Florida 127’, ‘Florida Radiance’, ‘Sweet Charlie’, Winterstar™ ‘FL 05-107’, FL10-121, 11.28-34, and 11.31-54. After further dilution of the crude DNA extracts from four leaf discs (5-fold dilution), FaFAD1 was strongly detected in all accessions producing γ-decalactone, producing results nearly equivalent to the CTAB-purified DNA samples (Fig. 1). Other individuals that do not produce γ-decalactone (i.e. ‘Strawberry Festival’, ‘Florida Ninety’, ‘Winter Dawn’) and FL10-46 did not generate a product upon PCR amplification of FaFAD1. We also found that the crude DNA extract from one leaf disc instead of four discs resulted in successful FaFAD1 amplification in some cases (not shown). The positive effect of dilution illustrates the inhibitory effects of secondary metabolites in strawberry leaves on PCR.
It has been reported that a PCR mixture with 0.1% BSA and 1% PVP effectively reduced the inhibition of DNA polymerase by secondary metabolites [10]. The NaOH-based rapid DNA extracts from plants such as Arabidopsis, tobacco, sorghum, and moss can be directly used for PCR amplification without BSA and PVP. However, in the present study we found that for strawberry leaf tissue, PCR amplification from crude DNA failed without BSA and PVP, again indicating high levels of PCR inhibitors in strawberry leaf tissue. We conducted PCR amplifications with two different types of bovine serum albumins (BSA), acetylated and non-acetylated. Amplification of FaFAD1 was successful only in the presence of the non-acetylated form (Fig. 1). Acetylated BSA has been shown to inhibit PCR amplification [23].
High resolution melting (HRM) is a high-throughput genotyping method that detects genetic variation in PCR amplicons without the use of gels or sequencing. Thus, HRM can provide higher throughput than most other methods while lowering per-sample costs. Since the null allele of FaFAD1 is a deletion of the entire gene, three HRM markers behaved in a dominant fashion (presence vs. absence). The upper melt curve (blue) indicates the presence of FaFAD1 and the lower curve (red) indicates the absence of FaFAD1. In each case, the HRM curve results agreed between CTAB-DNA and crude DNA templates (5-fold dilution) (Fig. 2). In addition, we tested the high-throughput rapid DNA extraction method for SSR-based genotyping. The SSR marker for FaFAD1 [18] was tested with NaOH-based crude DNA. As shown in Fig. 3, the unique fragment (205 bp) was only present in γ-decalactone producing varieties, 11.28-34, ‘Florida Elyana’, Sweet Sensation® ‘Florida 127’, ‘Florida Radiance’, ‘Sweet Charlie’, and Winterstar™ ‘FL 05-107’. Taken together, our findings demonstrate that high-throughput HRM analysis and SSR genotyping can be performed directly from strawberry leaf tissues without conventional DNA extraction, remarkably.
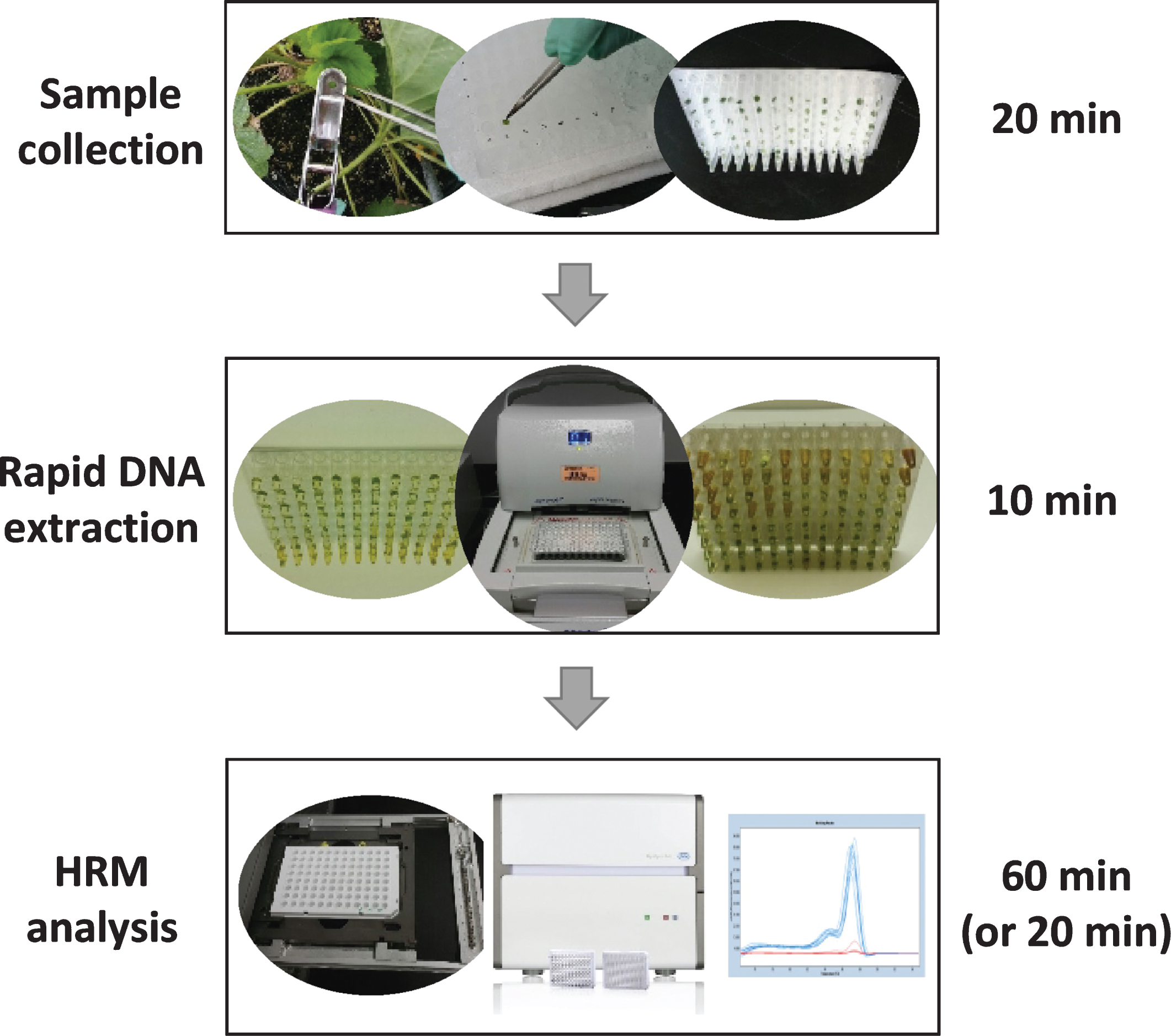
In the present study, we developed a high-throughput genotyping system that can be effectively utilized for marker-assisted breeding in strawberry. The entire procedure from sample collection and rapid DNA extraction to genotyping for a 96-well PCR plate takes about two hours (Fig. 4). The use of HRM eliminates the need for gel electrophoresis, and is accurate and repeatable. The estimated material and reagent cost is about USD 0.1 per sample (5 μl for single PCR reaction) when use a 384 well PCR plate, and one trained person can reasonably complete the whole procedure from rapid DNA extraction to genotyping about 1,000 samples in a day. PCR can be done in any PCR thermocycler with subsequent scanning for 20 min in a LightCycler 480 system II.
In general, molecular beacons, SNP microarrays, genotype-by-sequencing (GBS), and high resolution melting are the most common techniques for SNP genotyping. The Affymetrix IStraw90 Axiom Array and GBS are excellent tools for whole-genome (or targeted region) SNP genotyping, but it costs very high, at least USD 50 per sample (Axiom Array), for strawberry, and both are not suitable for seedling selection applications. Both probe-based (or TaqMan-based) and HRM-based assays can be performed by special instruments designed for SNP genotyping, such as the LightCycler 480 system II that costs little more than regular quantitative real time PCR systems. However, the cost for each probe is expensive (approximately USD 200 per probe; generally two probes are needed for each marker), especially for large breeding population, and also requires unique PCR reagents designed for the assay. For probe-based SNP genotyping including Kompetitive Allele Specific PCR (KASP), the specific SNP at the target region must be confirmed by sequencing prior to designing allele specific probes. However, HRM is post-PCR analysis, and thus does not require advance sequencing of the target region. HRM markers (about $6 per marker) can detect any known and/or unknown SNP or Indels present in a targeted region. The marker development for the KASP assay can be time-consuming because the whole development process must be completed by the company (LGC group, England). More importantly, this assay requires the use of only the company’s proprietary KASP primer mix for SNP genotyping (about USD 0.3 per reaction) and also is not suitable for multiplex PCR, which limits to modify the marker and/or PCR protocol needed for the future analysis. In the 2015 and 2016 breeding cycles, the University of Florida strawberry breeding program screened approximately 6,000 and 20,000 seedlings for the presence/absence of FaFAD1 using this system, and the breeding program is currently expanding the system to other trait loci.
Our future goals are to develop co-dominant HRM assays for the FaFAD1 locus and for other traits such as disease resistance. These would be particularly suitable for systems utilizing crude DNA extracts, since scoring would be based on the shape of the melt curve rather than the presence or absence of the curve. This provides a distinct advantage over gel-based detection since quantitative changes in amplification are less likely to influence scoring.
4Conclusions
This report details the development of two novel MAS components; 1) the modification of an NaOH-based crude DNA extraction method enabling successful PCR amplification in strawberry, and 2) the development of three HRM markers to replace gel-based detection of the FaFAD1 locus in octoploid strawberry. The combination of these two elements resulted in a rapid and cost-effective MAS system in strawberry. The system and marker set are immediately applicable in strawberry breeding programs, as our previous study [8] indicates that FaFAD1 should segregate in most cultivated strawberry germplasm. We anticipate that, with minor modifications, the system should be highly transferable to other berry and fruit crop breeding programs, including those species for which DNA extraction is highly problematic.
Acknowledgments
The authors thank the Florida Strawberry Growers Association (FSGA) for financial support and Nahla Bassil and the rest of the RosBREED community for helpful technical discussions about DNA tests in strawberry breeding.
References
[1] | Bassil NV , Davis TM , Zhang H , Ficklin S , Mittmann M , Webster T , et al. Development and preliminary evaluation of a 90 K Axiom(R) SNP array for the allo-octoploid cultivated strawberry Fragaria x ananassa. BMC Genomics. (2015) ;16: :155. |
[2] | Shulaev V , Sargent DJ , Crowhurst RN , Mockler TC , Folkerts O , Delcher AL , et al. The genome of woodland strawberry (Fragaria vesca). Nature Genetics. (2011) ;43: (2):109–16. |
[3] | Tennessen JA , Govindarajulu R , Ashman TL , Liston A . Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biology and Evolution. (2014) ;6: (12):3295–313. |
[4] | Darwish O , Shahan R , Liu Z , Slovin JP , Alkharouf NW . Re-annotation of the woodland strawberry (Fragaria vesca) genome. BMC Genomics. (2015) ;16: :29. |
[5] | Zorrilla-Fontanesi Y , Cabeza A , Dominguez P , Medina JJ , Valpuesta V , Denoyes-Rothan B , et al. Quantitative trait loci and underlying candidate genes controlling agronomical and fruit quality traits in octoploid strawberry (Fragaria x ananassa). TAG Theoretical and Applied Genetics Theoretische und Angewandte Genetik. (2011) ;123: (5):755–78. |
[6] | Sanchez-Sevilla JF , Horvath A , Botella MA , Gaston A , Folta K , Kilian A , et al. Diversity Arrays Technology (DArT) Marker Platforms for Diversity Analysis and Linkage Mapping in a Complex Crop, the Octoploid Cultivated Strawberry (Fragaria x ananassa). PloS One. (2015) ;10: (12):e0144960. |
[7] | Roach JA , Verma S , Peres NA , Jamieson AR , van de Weg WE , Bink MC , et al. FaRXf A locus conferring resistance to angular leaf spot caused by Xanthomonas fragariae in octoploid strawberry. TAG Theoretical and Applied Genetics Theoretische und Angewandte Genetik. (2016) ;129: (6):1191–201. |
[8] | Chambers AH , Pillet J , Plotto A , Bai J , Whitaker VM , Folta KM . Identification of a strawberry flavor gene candidate using an integrated genetic-genomic-analytical chemistry approach. BMC Genomics. (2014) ;15: :217. |
[9] | Hancock JF , Sooriyapathirana SS , Bassil NV , Stegmeir T , Cai L , Finn CE , et al. Public Availability of a Genotyped Segregating Population May Foster Marker Assisted Breeding (MAB) and Quantitative Trait Loci (QTL) Discovery: An Example Using Strawberry. Frontiers in Plant Science. (2016) ;7: :619. |
[10] | Lerceteau-Kohler E , Moing A , Guerin G , Renaud C , Petit A , Rothan C , et al. Genetic dissection of fruit quality traits in the octoploid cultivated strawberry highlights the role of homoeo-QTL in their control. TAG Theoretical and Applied Genetics Theoretische und Angewandte Genetik. (2012) ;124: (6):1059–77. |
[11] | Sun H , Wang HT , Kwon WS , Kim YJ , In JG , Yang DC . A simple and rapid technique for the authentication of the ginseng cultivar, Yunpoong, using an SNP marker in a large sample of ginseng leaves. Gene. (2011) ;487: (1):75–9. |
[12] | Porcar M , Ramos S , Latorre A . A simple DNA extraction method suitable for PCR detection of genetically modified maize. Journal of the Science of Food and Agriculture. (2007) ;87: (14):2728–31. |
[13] | Wang H , Qi M , Cutler AJ . A simple method of preparing plant samples for PCR. Nucleic Acids Research. (1993) ;21: (17):4153–4. |
[14] | Xin Z , Velten JP , Oliver MJ , Burke JJ . High-throughput DNA extraction method suitable for PCR. BioTechniques. (2003) ;34: (4):820–7. |
[15] | Collard B , Das A , Virk P , Mackill D . Evaluation of ‘quick and dirty’DNA extraction methods for marker-assisted selection in rice (Oryza sativa L). Plant Breeding. (2007) ;126: (1):47–50. |
[16] | Paris M , Carter M . Cereal DNA: A rapid high-throughput extraction method for marker assisted selection. Plant Molecular Biology Reporter. (2000) ;18: (4):357–60. |
[17] | Fang G , Hammar S , Grumet R . A quick and inexpensive method for removing polysaccharides from plant genomic DNA. BioTechniques. (1992) ;13: (1):52–4, 6. |
[18] | Nunes CF , Ferreira JL , Fernandes MCN , Breves SdS , Generoso AL , Soares BDF , et al. An improved method for genomic DNA extraction from strawberry leaves. Ciência Rural. (2011) ;41: (8):1383–9. |
[19] | Simko I . High-Resolution DNA Melting Analysis in Plant Research. Trends in Plant Science. (2016) . |
[20] | Zhang T , Yu LX , McCord P , Miller D , Bhamidimarri S , Johnson D , et al. Identification of molecular markers associated with Verticillium wilt resistance in alfalfa (Medicago sativa L.) using high-resolution melting. PloS One. (2014) ;9: (12):e115953. |
[21] | Han Y , Khu DM , Monteros MJ . High-resolution melting analysis for SNP genotyping and mapping in tetraploid alfalfa (Medicago sativa L.). Molecular breeding: New Strategies in Plant Improvement. (2012) ;29: (2):489–501. |
[22] | Sanchez-Sevilla JF , Cruz-Rus E , Valpuesta V , Botella MA , Amaya I . Deciphering gamma-decalactone biosynthesis in strawberry fruit using a combination of genetic mapping, RNA-Seq and eQTL analyses. BMC Genomics. (2014) ;15: :218. |
[23] | McKeown BJ . An acetylated (nuclease-free) bovine serum albumin in a PCR buffer inhibits amplification. BioTechniques. (1994) ;17: (2):246–8. |
[24] | Keb-Llanes M , González G , Chi-Manzanero B , Infante D . A rapid and simple method for small-scale DNA extraction in Agavaceae and other tropical plants. Plant Molecular Biology Reporter. (2002) ;20: (3):299–300. |
Figures and Tables
Fig.1
PCR amplification of FaFAD1 directly from strawberry leaf tissue. The eight cultivars and four advanced selections of strawberry (Fragaria×ananassa) were used in this study. Florida Elyana: FL Elyana, Sweet Sensation® ‘Florida 127’: FL 127, Florida Radiance: FL Radiance, Sweet Charlie, Winterstar™ ‘FL 05-107’: Winterstar, Strawberry Festival: Festival, Florida Ninety: FL Ninety, Winter Dawn, FL10-121, 11.28-34, 11.31-54 and FL10-46. Strawberry seedlings were grown in a greenhouse located at the UF/IFAS Gulf Coast Research and Education Center. CTAB-DNA samples were prepared as previously described [24]. Crude DNA extractions from leaf tissues followed a well-described NaOH-based method [14]. An amplified fragment of expected size (∼150 bp) was observed in eight accessions producing γ-decalactone for CTAB-DNA and for crude extractions after 5-fold dilution. Weak amplifications of FaFAD1 were found in all γ-decalactone producing accessions after 2-fold dilution. Acetylated or non-acetylated BSA was added to the PCR reactions using crude extracts. All experiments were repeated at least three times, and PCR amplicons were confirmed by gel electrophoresis at 230 V for 70 min using 1.5% agarose gels.
![PCR amplification of FaFAD1 directly from strawberry leaf tissue. The eight cultivars and four advanced selections of strawberry (Fragaria×ananassa) were used in this study. Florida Elyana: FL Elyana, Sweet Sensation® ‘Florida 127’: FL 127, Florida Radiance: FL Radiance, Sweet Charlie, Winterstar™ ‘FL 05-107’: Winterstar, Strawberry Festival: Festival, Florida Ninety: FL Ninety, Winter Dawn, FL10-121, 11.28-34, 11.31-54 and FL10-46. Strawberry seedlings were grown in a greenhouse located at the UF/IFAS Gulf Coast Research and Education Center. CTAB-DNA samples were prepared as previously described [24]. Crude DNA extractions from leaf tissues followed a well-described NaOH-based method [14]. An amplified fragment of expected size (∼150 bp) was observed in eight accessions producing γ-decalactone for CTAB-DNA and for crude extractions after 5-fold dilution. Weak amplifications of FaFAD1 were found in all γ-decalactone producing accessions after 2-fold dilution. Acetylated or non-acetylated BSA was added to the PCR reactions using crude extracts. All experiments were repeated at least three times, and PCR amplicons were confirmed by gel electrophoresis at 230 V for 70 min using 1.5% agarose gels.](https://content.iospress.com:443/media/jbr/2017/7-1/jbr-7-1-jbr145/jbr-7-jbr145-g001.jpg)
Fig.2
The detection of FaFAD1 using high-throughput HRM markers, UFGDHRM2, UFGDHRM4 and UFGDHRM5, in strawberry. Three primer sets were developed for HRM analysis. The HRM results from CTAB-DNA and NaOH-crude DNA extracts (5-fold dilution) were compared for three markers (A: UFGDHRM2, B: UFGDHRM4, and C: UFGDHRM5). BSA (0.1%) and PVP (1%) were included in all PCR reactions. The blue curve indicates presence of FaFAD1, while the red line indicates the absence (deletion) of FaFAD1. (D) The annotated FAD1 gene model from F. vesca was obtained from the Genome Database for Rosaceae (www.rosaceae.org). The primer location for three HRM markers was noted by black arrows.
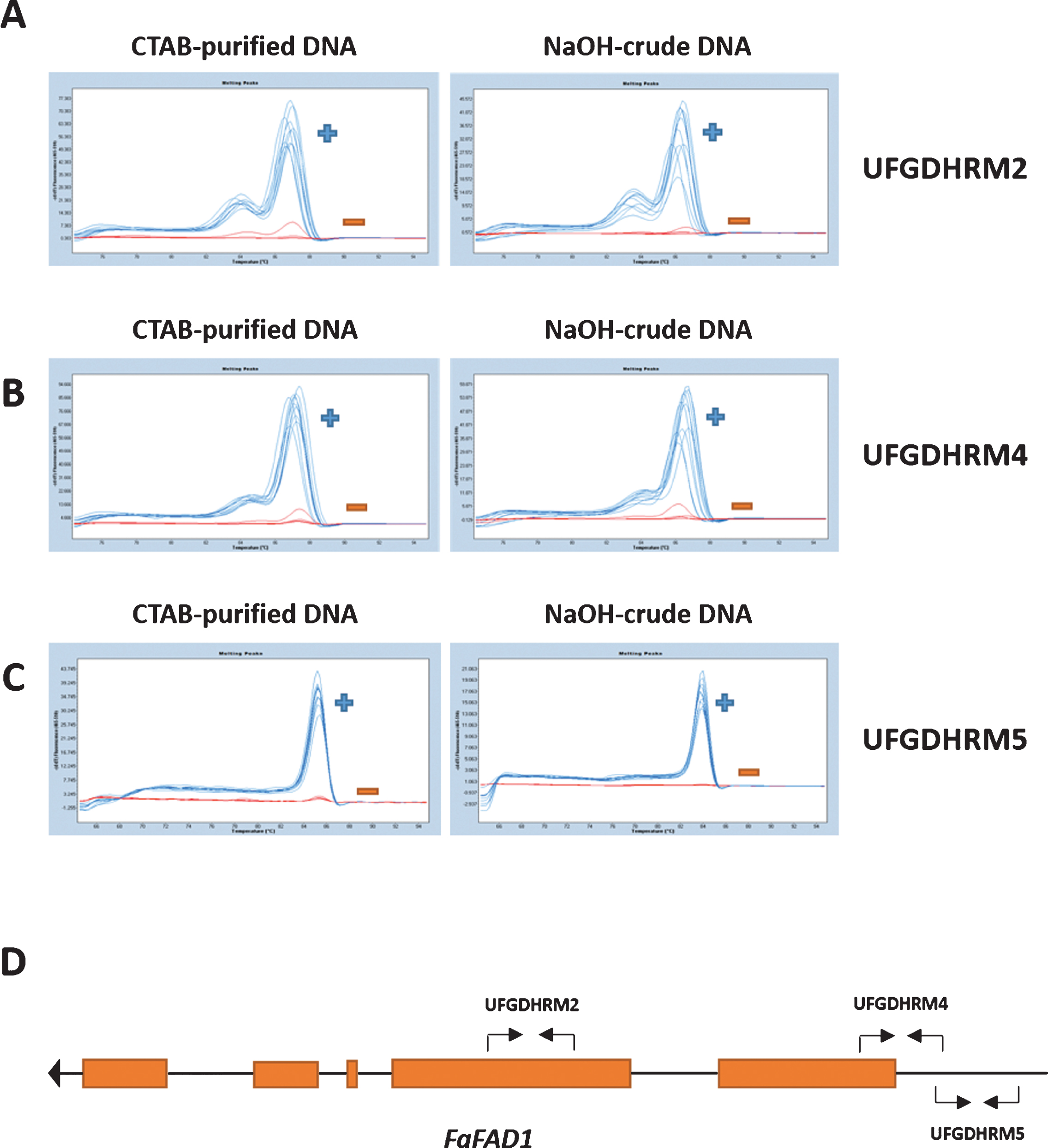
Fig.3
The detection of FaFAD1 directly from strawberry leaf tissue via an SSR marker. A previously reported SSR marker [17] was used to detect FaFAD1 in UF breeding cultivars and selections. The forward primer was M13 5’-labeled with 6-FAM, VIC, NED, forward: ACGACGTTGTAAAACGACGAAGAAGATGACACTAGGGACGAGGAA, reverse: GTTCTATGTGAGAACATGGGAAGAAACATGAC. The 205 bp fragment was only detected in cultivars or selections possessing FaFAD1. Although the fluorescence level in NaOH-crude DNA was relatively weaker than the CTAB-purified DNA, the presence or absence of FaFAD1 was successfully determined in all tested individuals.
![The detection of FaFAD1 directly from strawberry leaf tissue via an SSR marker. A previously reported SSR marker [17] was used to detect FaFAD1 in UF breeding cultivars and selections. The forward primer was M13 5’-labeled with 6-FAM, VIC, NED, forward: ACGACGTTGTAAAACGACGAAGAAGATGACACTAGGGACGAGGAA, reverse: GTTCTATGTGAGAACATGGGAAGAAACATGAC. The 205 bp fragment was only detected in cultivars or selections possessing FaFAD1. Although the fluorescence level in NaOH-crude DNA was relatively weaker than the CTAB-purified DNA, the presence or absence of FaFAD1 was successfully determined in all tested individuals.](https://content.iospress.com:443/media/jbr/2017/7-1/jbr-7-1-jbr145/jbr-7-jbr145-g003.jpg)
Fig.4
Summary schematic of a high-throughput genotyping system developed for strawberry and other Rosaceae crop breeding programs. Instead of using four leaf discs, we only collected one leaf disc (3 mm) in each well of a 96-well PCR plate and incubated 10 min with the extraction buffer in a PCR thermocycler. The crude DNA sample is diluted (2-fold for a single disc, or at least 5-fold when four leaf discs were used) with dilution buffer, and directly used for PCR amplification and HRM analysis using a LightCycler 480 system II. The total procedure, for either 96 or 384 format, can be completed in 2 to 3 hours. The procedure for the HRM scan of a 384-well plate (without PCR amplification) can be done in 20 min.