RNA Analysis of Circulating Leukocytes in Patients with Alzheimer’s Disease
Abstract
Background:
One of the key symptoms of Alzheimer’s disease (AD) is the impairment of short-term memory. Hippocampal neurogenesis is essential for short-term memory and is known to decrease in patients with AD. Impaired short-term memory and impaired neurogenesis are observed in aged mice alongside changes in RNA expression of gap junction and metabolism-related genes in circulating leukocytes. Moreover, after penetrating the blood-brain barrier via the SDF1/CXCR4 axis, circulating leukocytes directly interact with hippocampal neuronal stem cells via gap junctions.
Objective:
Evaluation of RNA expression profiles in circulating leukocytes in patients with AD.
Methods:
Patients with AD (MMSE≧23, n = 10) and age-matched controls (MMSE≧28, n = 10) were enrolled into this study. RNA expression profiles of gap junction and metabolism-related genes in circulating leukocytes were compared between the groups (jRCT: 1050210166).
Results:
The ratios of gap junction and metabolism-related genes were significantly different between patients with AD and age-matched controls. However, due to large inter-individual variations, there were no statistically significant differences in the level of single RNA expression between these groups.
Conclusions:
Our findings suggest a potential connection between the presence of circulating leukocytes and the process of hippocampal neurogenesis in individuals with AD. Analyzing RNA in circulating leukocytes holds promise as a means to offer novel insights into the pathology of AD, distinct from conventional markers.
INTRODUCTION
Gap junctions are specialized intercellular connections [1]. They directly link the cytoplasm of two cells and allow the rapid movement of small water-soluble molecules, including most metabolites, according to their concentration gradient. Thus, gap junctions have a significant impact on the metabolic status of connected cells [2]. The therapeutic mechanisms of action of cell therapies, for instance hematopoietic or mesenchymal stem cells, can be mediated by gap junctions [3, 4]. Moreover, systemically administered hematopoietic stem cells and circulating blood cells directly interact with hippocampal neuronal stem cells via gap junction after penetrating the blood-brain barrier (manuscript under review). These findings are consistent with our previous report showing that hematopoietic stem cell transplantation increases post-stroke neurogenesis in mice [5].
One of the key symptoms of Alzheimer’s disease (AD) is the impairment of short-term memory. Hippocampal neurogenesis is essential for short-term memory but is known to sharply decrease in AD patients [6]. Moreover, aged mice exhibit short-term memory impairment, reduced neurogenesis, and significant changes in RNA expression of metabolism-related and gap junction genes in circulating leukocytes [7, 8]. According to the results obtained in aged mice, we have hypothesized that gap junction mediated cell-cell interaction between circulating white blood cells (WBC) and endothelial cells/neuronal stem cells has a significant role in neurogenesis at hippocampus and reduction of the cellular interaction induces the change of RNA expression in WBC [8]. Based on the possible similarity between AD patients and aged mice, we have investigated RNA expression profiles of circulating leukocytes in AD patients and compared them with age-matched controls.
MATERIALS AND METHODS
The clinical study was approved by the institutional review board of the Institute of Biomedical Research and Innovation at Kobe and Matsui Dietary & Dementia Clinic and was registered with the Japan registry of clinical trials (jRCT: 1050210166). Animal experiments were approved by the Animal Care and Use Committee of Foundation for Biomedical Research and Innovation and complies with the Guide for the Care and Use of Animals published by the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Clinical study enrollment
The outline of this exploratory study, including the number of enrolled patients and molecules for PCR analysis, was designed according to the findings obtained in aged mice with cognitive impairment and reduced neurogenesis at hippocampus [8]. Patients with probable AD and age-matched controls were enrolled in this study (n = 10, each group). The diagnosis of probable AD was made according to the National Institute on Aging-Alzheimer’s Association (NIA-AA) AD diagnostic criteria [9]. Only males were enrolled in this study in order to avoid possible variation in the experimental observations between sexes whilst being consistent with previous animal studies which also only involved male mice [8]. The minimum enrollment age was 65 years. The Mini-Mental State Examination (MMSE) scores of all AD patients was ≦23 whereas MMSE scores of age-matched controls were ≧28 [10]. The exclusion criteria for the clinical study are listed at Table 1. Written informed consent was obtained from all AD patients and/or their appropriate guardian, and healthy volunteers. The characteristic of the enrolled patients and volunteers are shown in Table 2.
Table 1
Exclusion criteria
| 1 | Diabetes |
| 2 | During cancer disease treatment, less than 5 years after the end of cancer treatment |
| 3 | Thyroid disease |
| 4 | Infectious diseases (AIDS, hepatitis B, hepatitis C) |
| 5 | Fever at the time of blood collection |
| 6 | Chronic subdural hematoma |
| 7 | Normal pressure hydrocephalus |
| 8 | During hemodialysis |
| 9 | Chronic Obstructive Pulmonary Disease |
| 10 | During home oxygen therapy |
| 11 | Smoking within the last 12 months |
| 12 | Taking drugs that have a clear effect on the immune system, such as anti-cancer drugs, steroids, and anti-rheumatic drugs |
| 13 | Patients with serious complications, or those who have a history of these and are judged by the investigator and the investigator to be ineligible for this study |
| 14 | Others who are judged by the principal investigator and the investigator to be ineligible for this study |
Table 2
Characteristic of enrolled patients and volunteers
| MMSE | Age | Body weight (Kg) | RBC (106/μl) | WBC (103/μl) | Gra (%) | Lym (%) | Mono (%) | Plt (103/μl) | FBS (g/dl) | HbA1c (%) | |
| Alzheimer | 16.5±5.3 | 81.3±6.8 | 62.6±11.8 | 4.0±0.5 | 5.8±1.6 | 55.5±10.4 | 33.1±9.4 | 7.6±1.7 | 193.8±50.5 | 94.4±7.8 | 5.6±0.3 |
| Control | 29.7±0.5 | 74.9±11.3 | 59.9±9.9 | 4.2±0.4 | 5.4±0.7 | 55.5±5.8 | 29.3±7.2 | 8.6±2.3 | 202.7±34.6 | 99.4±10.3 | 5.7±0.5 |
| p value | <0.001 | 0.20 | 0.52 | 0.42 | 0.54 | 0.69 | 0.43 | 0.14 | 0.60 | 0.22 | 0.91 |
Quantitative PCR (qPCR) analysis of circulating leukocytes
Fasting blood was obtained from the cubital vein and RNA expression in mononuclear cells and all leukocytes were evaluated in each group. In this study, two cell populations were evaluated, as the major cell populations in leukocytes that express gap junction are mononuclear cells. Mononuclear cells were isolated using a mononuclear cell preparation tube (BD Vacutainer #362760, BD Bioscience, NJ, USA). The target genes for RNA analysis were selected based on the results obtained in aged mice with cognitive impairment [8]. The absence of non-specific amplification with these primers was confirmed by the primer blast program provided by National Library of Medicine (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
The RNA of mononuclear cells and all leukocytes were stabilized with PAXgene Blood RNA Tube (#762165, BD Bioscience, NJ). Total RNA was isolated using a NucleoSpin RNA (Takara, Kyoto, Japan) according to the manufacturer’s instructions. cDNA was synthesized from 0.3μg total RNA using PrimeScript™ II 1st strand cDNA Synthesis Kit (TAKARA, Kyoto, Japan). Transcription of mRNA was analyzed using PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, CA, USA) and the Agilent AriaMx real time quantitative PCR System. 18 S ribosomal RNA was used for the reference gene. The list of target genes, primer sequences, and amplification protocols are shown in Table 3.
Table 3
Target genes, primer list and amplification protocol
| Human | |||||
| Gene | NCBI Accession No. | Sequences | bp | Tm | |
| hAMPKa | NM_001355034.2 | Forward | CGGCAAAGTGAAGGTTGGC | 96 | 60.01 |
| (AMP-activated protein kinase A) | Reverse | CCTACCACATCAAGGCTCCG | 60.18 | ||
| hCx37 | NM_002060.3 | Forward | CACCATGCCCCACCTACAAT | 134 | 60.03 |
| (Connexin 37) | Reverse | TGGGGGTTTTTGGCCATTCT | 60.10 | ||
| hCx43 | NM_000165.5 | Forward | AGGAGTTCAATCACTTGGCGT | 128 | 59.93 |
| (Connexin 43) | Reverse | CCCTCCAGCAGTTGAGTAGG | 59.46 | ||
| hGlut1 | NM_006516.3 | Forward | CCTGCAGTTTGGCTACAACAC | 103 | 60.00 |
| (Glucose transporter 1) | Reverse | CAGGATGCTCTCCCCATAGC | 59.68 | ||
| hGlut3 | NM_006931.3 | Forward | ATTACAGCGATGGGGACACA | 84 | 59.09 |
| (Glucose transporter 3) | Reverse | GCCAAATTGGAAAGAGCCGA | 59.11 | ||
| hHif1α | NM_001530.4 | Forward | CCAGACGATCATGCAGCTACT | 138 | 59.93 |
| (Hypoxia inducible factor 1α) | Reverse | TGATTGCCCCAGCAGTCTAC | 59.75 | ||
| hMCT4 | NM_001042423.3 | Forward | CGGAGCATCATCCAGGTCTAC | 71 | 60.00 |
| (Monocarboxylate transporter 4) | Reverse | GGCTGGAAGTTGAGTGCCAA | 60.82 | ||
| hPDK1 | NM_001278549.1 | Forward | GCAAAATCACCAGGACAGCC | 117 | 59.76 |
| (Pyruvate dehydrogenase kinase 1) | Reverse | TCTGTTGGCATGGTGTTCCA | 59.82 | ||
| hPHD3 | NM_022073.3 | Forward | GATCGTAGGAACCCACACGA | 161 | 59.19 |
| (Prolyl hydroxylase 3) | Reverse | TCAGAGCACGGTCAGTCTTC | 59.40 | ||
| h18s | NR_003286.4 | Forward | GGCCCTGTAATTGGAATGAGTC | 155 | 59.05 |
| (18 s ribosomal RNA) | Reverse | CCAAGATCCAACTACGAGCTT | 57.75 | ||
| Mouse | |||||
| Gene | NCBI Accession No. | Sequences | bp | Tm | |
| mAMPKa | NM_001013367.3 | Forward | ACCAGGTCATCAGTACACCA | 142 | 57.98 |
| Reverse | ACACCGGAAAGGATCTGCTG | 60.04 | |||
| mCx37 | NM_008120.3 | Forward | CAGCTGCGCGCTATTTAAGG | 134 | 60.04 |
| Reverse | CCATGTTTCCAGGGCCTCTC | 60.39 | |||
| mCx43 | NM_010288.3 | Forward | GAGTTCCACCACTTTGGCGT | 117 | 60.82 |
| Reverse | GTGGAGTAGGCTTGGACCTT | 59.02 | |||
| mGlut1 | NM_011400.3 | Forward | TGGCGGGAGACGCATAGTTA | 132 | 61.04 |
| Reverse | CTCCCACAGCCAACATGAGG | 60.68 | |||
| mPHD3 | NM_028133.2 | Forward | ATCCACATGAAGTCCAGCCC | 149 | 59.74 |
| Reverse | ACACCACAGTCAGTCTTTAGCA | 59.57 | |||
| m18s | NR_003278.3 | Forward | ACTCAACACGGGAAACCTCACC | 121 | 62.70 |
| Reverse | CCAGACAAATCGCTCCACCA | 60.32 | |||
| Amplification protocol | |||||
| Segment | Plateau | Temperature | Duration | Cycle | |
| Hot Start | 1 | 50 | 00:03:00 | 1 | |
| Hot Start 2 | 1 | 95 | 00:03:00 | 1 | |
| Amplification | 1 | 95 | 00:00:05 | 40 | |
| Amplification | 2 | 60 | 00:00:30 | 40 | |
| Melt | 1 | 95 | 00:00:30 | 1 | |
| Melt | 2 | 65 | 00:00:30 | 1 | |
| Melt | 3 | 95 | 00:00:30 | 1 |
qPCR analysis of cultured human umbilical vein endothelial cells and mice brain
Human umbilical vein endothelial cells (HUVEC, Kurabo, Osaka, Japan) were cultured in medium, serum and growth factors (HuMedia-EB2, Kurabo). HUVEC in passage 3 and 10 were used for qPCR analysis (n = 4, each). Male CB-17 mice aged 5 and 80 weeks old were used for brain tissue analysis (n = 5, each). Total RNA from HUVEC and mice hippocampus were isolated using RNeasy Plus Universal Mini Kit or Micro Kit (Qiagen, CA, USA) according to the manufacturer’s protocol, followed by qPCR analysis as described above. The list of target genes, primer sequences, and amplification protocols are shown in Table 3.
Data analysis
The individual comparisons were performed using Student’s t-test. Normal distribution of the data was confirmed using JMP 7 (JMP Statistical Discovery, NC, USA) with Shapiro-Wilk test. All data are shown as 95% CI.
RESULTS
RNA expression patterns in mononuclear cells of AD patients
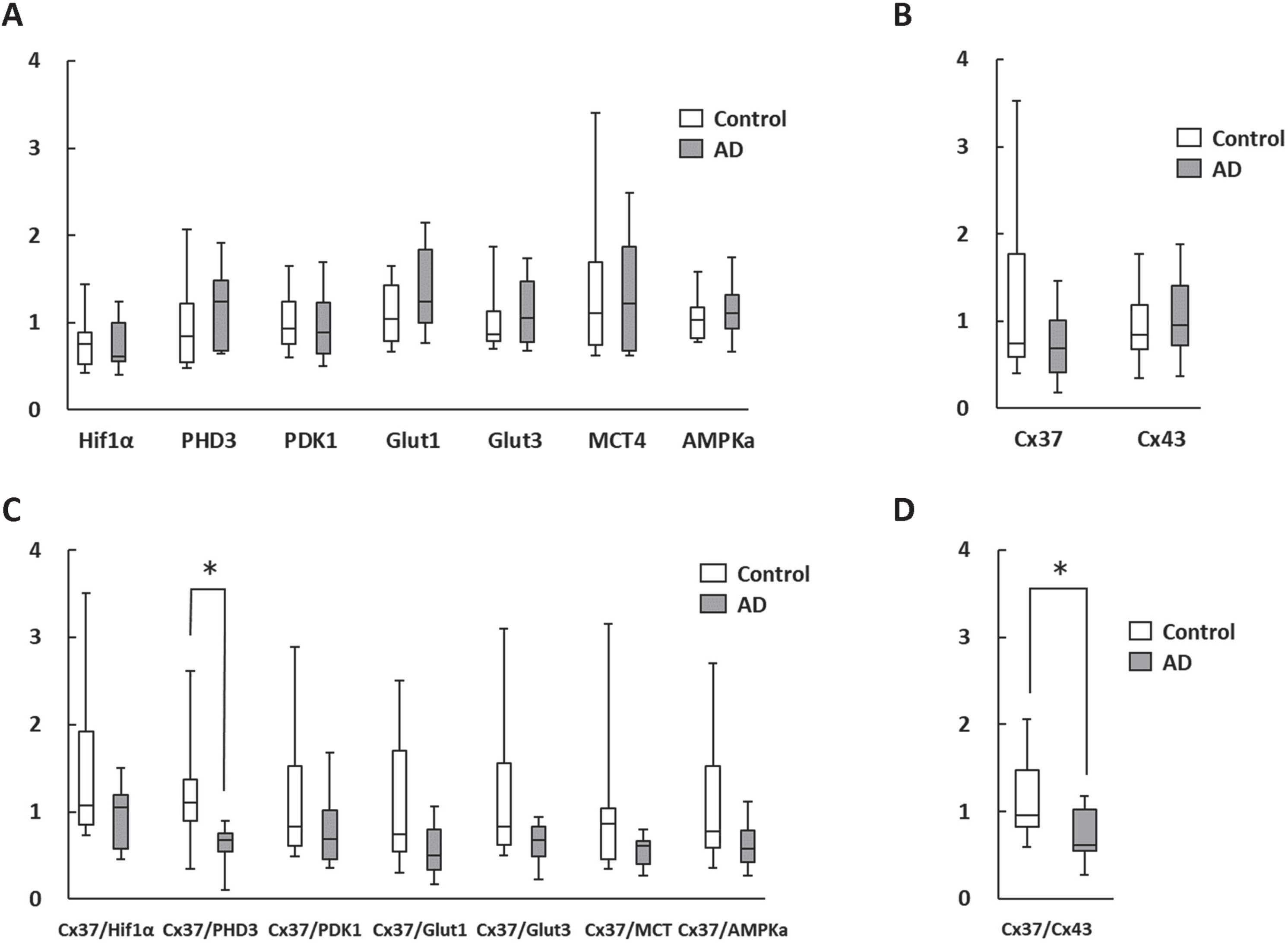
The qPCR results of the level of RNA expression in mononuclear cells in AD and control patients indicated no statistical difference between the groups, due to the significant variation between individuals (Fig. 1A, B). In order to normalize the results for inter-individual differences, the ratios of connexin gene and metabolism related genes, that were known to be decreased and increased in aged mice with cognitive impairment, respectively [8], were determined. It is noteworthy that in the case of Cx37/PHD3 ratio, statistical differences between the patient groups were observed (Fig. 1C). Furthermore, the Cx37/Cx43 ratio was also found to be statistical different between the groups (Fig. 1D).
Fig. 1
Change in RNA expression in circulating mononuclear cells. A, B) There were no statistically significant differences in metabolism-related (A) and gap junction (B) genes between AD patients and age-matched controls. C, D) Cx37/PHD3 (C) and Cx37/Cx43 ratios (D) were significantly lower in AD patients. *p < 0.05.
Cx37/metabolism related genes ratios are significantly altered in aged mice and in vitro cell passage
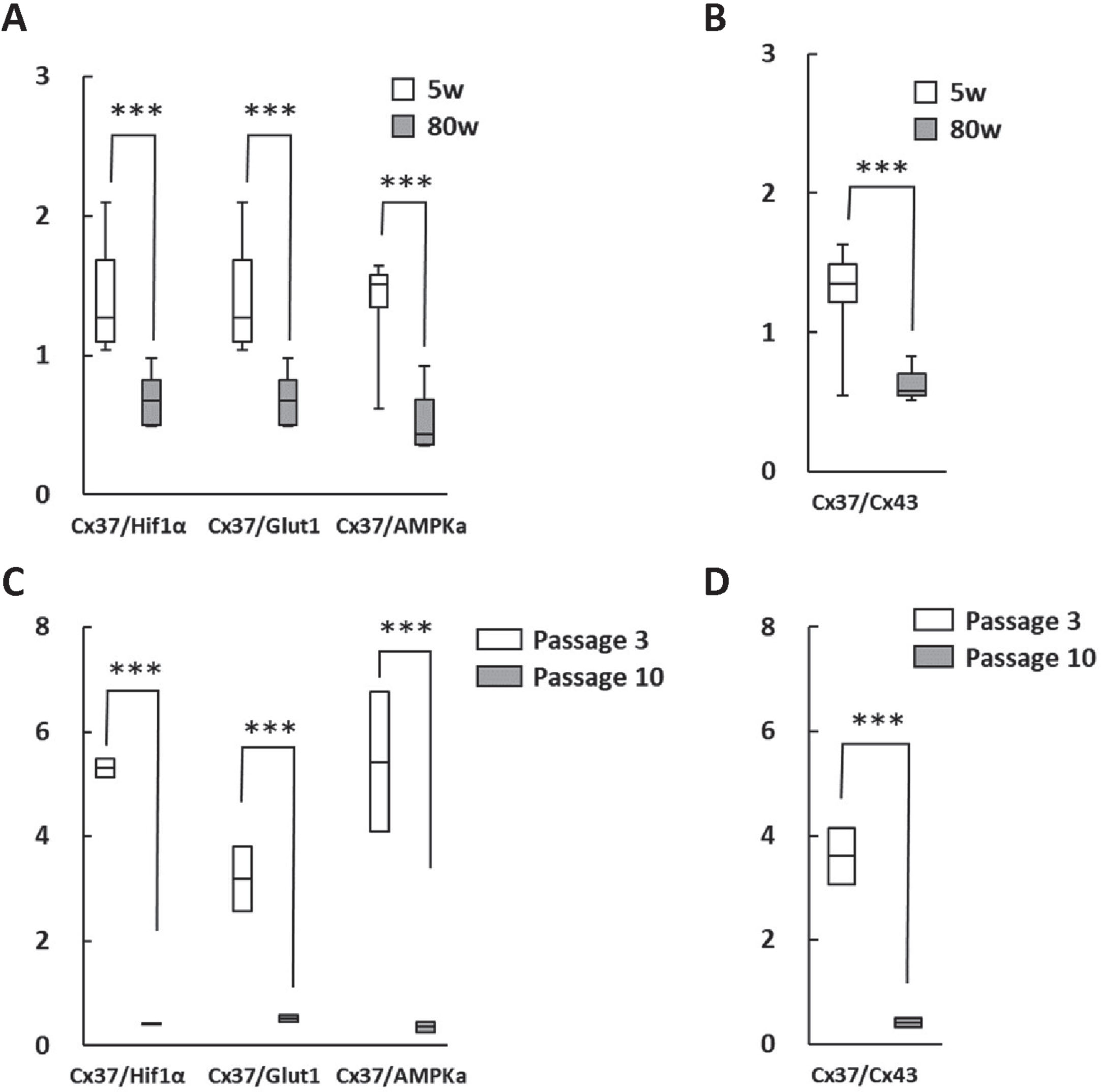
In order to verify the significance of the changes in RNA expression ratio during aging, RNA expression of PHD3, Glut1, AMP-activated protein kinase alpha (AMPKa), and Cx37 in brain hippocampus of young and aged mice with cognitive impairment were evaluated and found to be significantly different (Fig. 2A). It was striking that the Cx37/Cx43 ratio was also significantly altered in aging brains (Fig. 2B). Moreover, RNA expression ratios were investigated in HUVEC after short (passage 3) and long term culture (passage 10), revealing comparable differences (Fig. 2C, D).
Fig. 2
Change in RNA expression in the hippocampus of aged mice brain and cultured human endothelial cells. A, B) Cx37/PHD3, Cx37/Glut1 and Cx37/AMPk ratios (A) as well as the Cx37/Cx43 ratio (B) were significantly lower in aged mice (80 weeks). C, D) Moreover, Cx37/PHD3, Cx37/Glut1 and Cx37/AMPKa ratios (C) as well as the Cx37/Cx43 ratio (D) were significantly lower in long term-cultured HUVEC. ***p < 0.001.
Change in RNA expression pattern in white blood cells (WBC) of AD patients
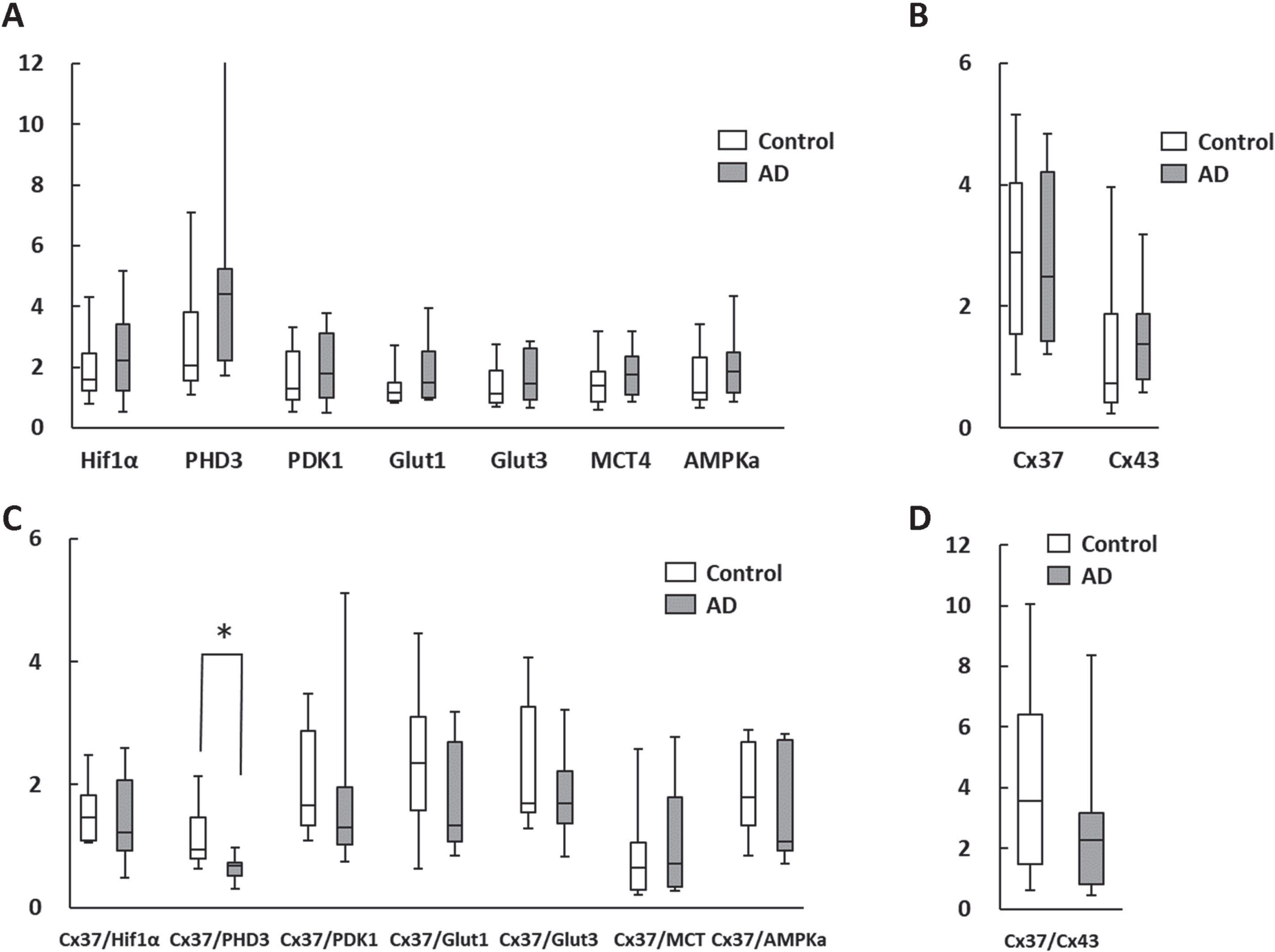
The qPCR analysis of RNA expression levels in AD patient leukocytes and age-matched controls are shown in Fig. 3. These are similar to the results obtained from the analysis of mononuclear cells, with the Cx37/PHD3 ratio being significantly lower in AD patients (Fig. 3C). However, Cx37/Cx43 ratios were indifferent between the groups (Fig. 3D). These findings indicate that analysis of isolated mononuclear cells and whole leukocytes would have a potential to provide information on AD pathology from a different perspective than existing blood markers, such as phosphorylated tau 181 [11] and ratio of various amyloid-β proteins [12].
Fig. 3
Change in RNA expression in circulating whole leukocytes. A, B) There were no statistical differences in metabolism-related (A) and the gap junction (B) genes between the AD patients and age-matched controls. C, D) The Cx37/PHD3 ratio (C), but not Cx37/Cx43 ratio (D) was significantly lower in AD patients. *p < 0.05.
DISCUSSION
We report herein that RNA expression of circulating leukocytes in AD patients contrasts with that of age-matched controls. These results indicate a possible link between circulating leukocytes and neurogenesis in AD patients and provide a novel concept that RNA analysis of circulating leukocytes would serve as a blood-based biomarker for AD.
Cellular interaction via the gap junction has a prominent role in development, cellular differentiation and regeneration processes [1, 3, 4]. Hematopoietic stem cell transplantation is known to initiate angiogenesis [13] and its mechanisms of action include the supply of metabolites to injured endothelium via gap junction resulting in hypoxia inducible factor-1α (Hif1α) activation [3]. This contrasts with mesenchymal stem cell transplantation which can be detrimental as it removes metabolites from activated endothelium and circulating mononuclear cell via gap junction [4]. Furthermore, we have recently found that circulating leukocytes directly interact with neuronal stem cells at the hippocampus via the gap junction, resulting in the inactivation of Hif1α in nestin-positive neuronal stem cells (manuscript under review). These findings adequately rationalize our current observations, in that the RNA expression of metabolism-related genes in AD patients significantly change concomitant with changes in expression of the gap protein genes. Our observations can explain the positive impact of circulating leukocytes in the maturation of cerebral endothelium and differentiation of neuronal stem cells into neuron at the hippocampus (Fig. 4). Thus far, our results indicate the significant role that the gap junction mediated cellular interaction has on optimization of the cellular metabolite status. A possible mechanism that links the metabolism of leukocyte and endothelium is shown in Fig. 5. The concentration of various glycolytic substrates and AMP in cytosol of leukocyte was higher than those of endothelium [14]. The transfer of glycolytic substrates and AMP from leukocyte to endothelium would reduce O2 consumption with inactivation of Hif1α and AMPKa mediated cascade in leukocyte. In contrast, the transfer to endothelium would activate Hif1α and AMPKa mediated cascade in endothelium. These possible mechanisms are consistent with our previous result that transfer of glycolytic substrates from hematopoietic stem cell to endothelium activates Hif1α in endothelium [3].
Fig. 4
Proposed hypothesis that links circulating leukocytes and AD. A) In healthy elderly, the physiological cellular interaction between circulating leukocytes and endothelium/neuronal stem cell via Cx37 contributes towards the maintenance of hippocampal neurogenesis. B) In patients with AD, the impairment of physiological cellular interaction between circulating leukocytes and endothelium/neuronal stem cell via Cx37 induces impaired neurogenesis with lower ratio of Cx37/PHD3 gene.
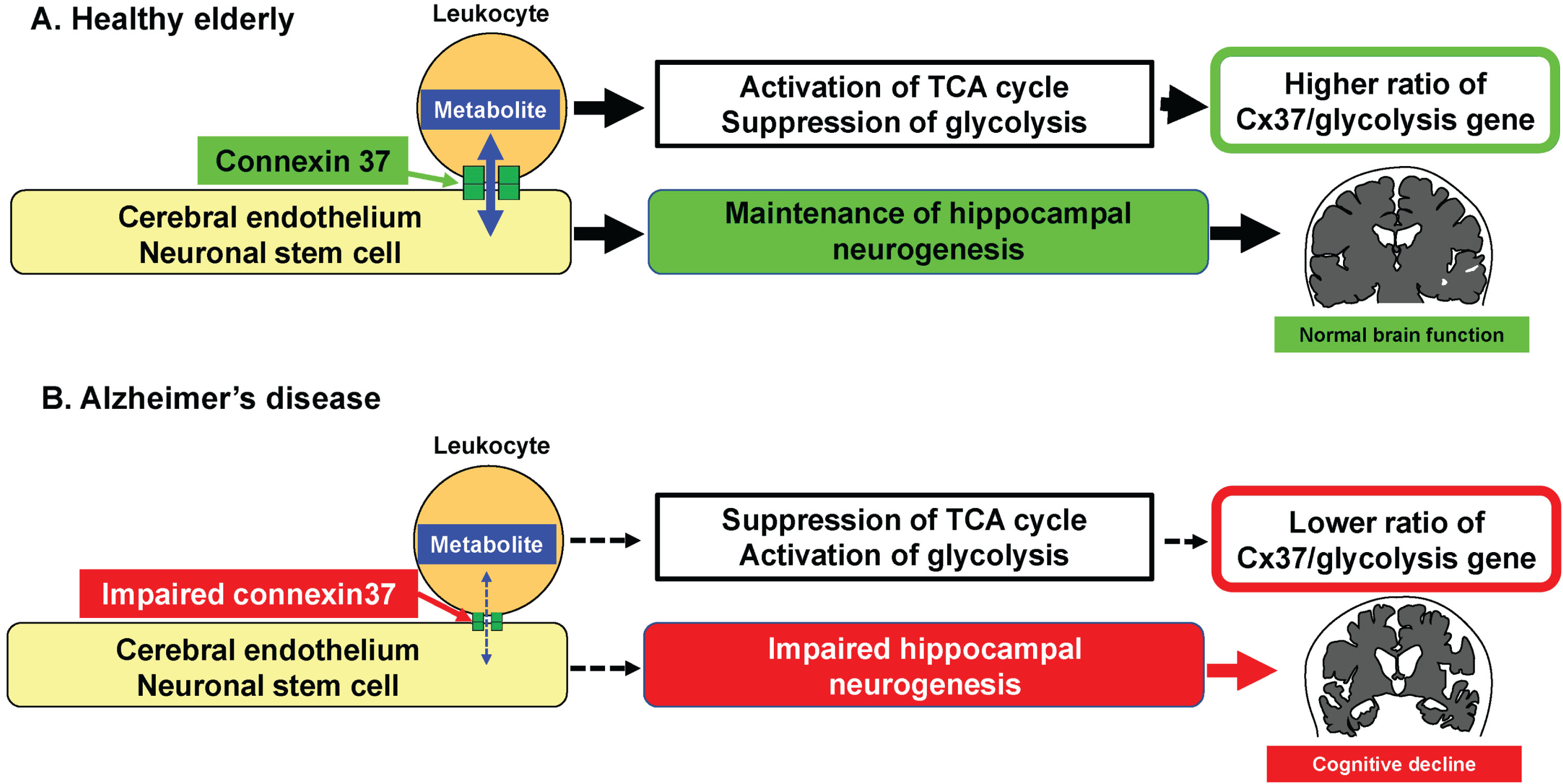
Fig. 5
Possible mechanism linking the metabolism of leukocyte and endothelium. Without (A) and with (B) formation of gap junction mediated cell-cell interaction. Transfer of glycolytic substrate from leukocyte to endothelium via gap junction reduces O2 consumption followed by inactivation of Hif1α mediated pathways. Transfer of AMP from leukocyte to endothelium inactivates AMPKa in leukocyte. Blue and grey boxes indicate activated and inactivated molecules/states, respectively. Blue line and grey dashed lines indicate active and inactive pathways, respectively. Red and yellow lines indicate glycolytic substrate and AMP, respectively.
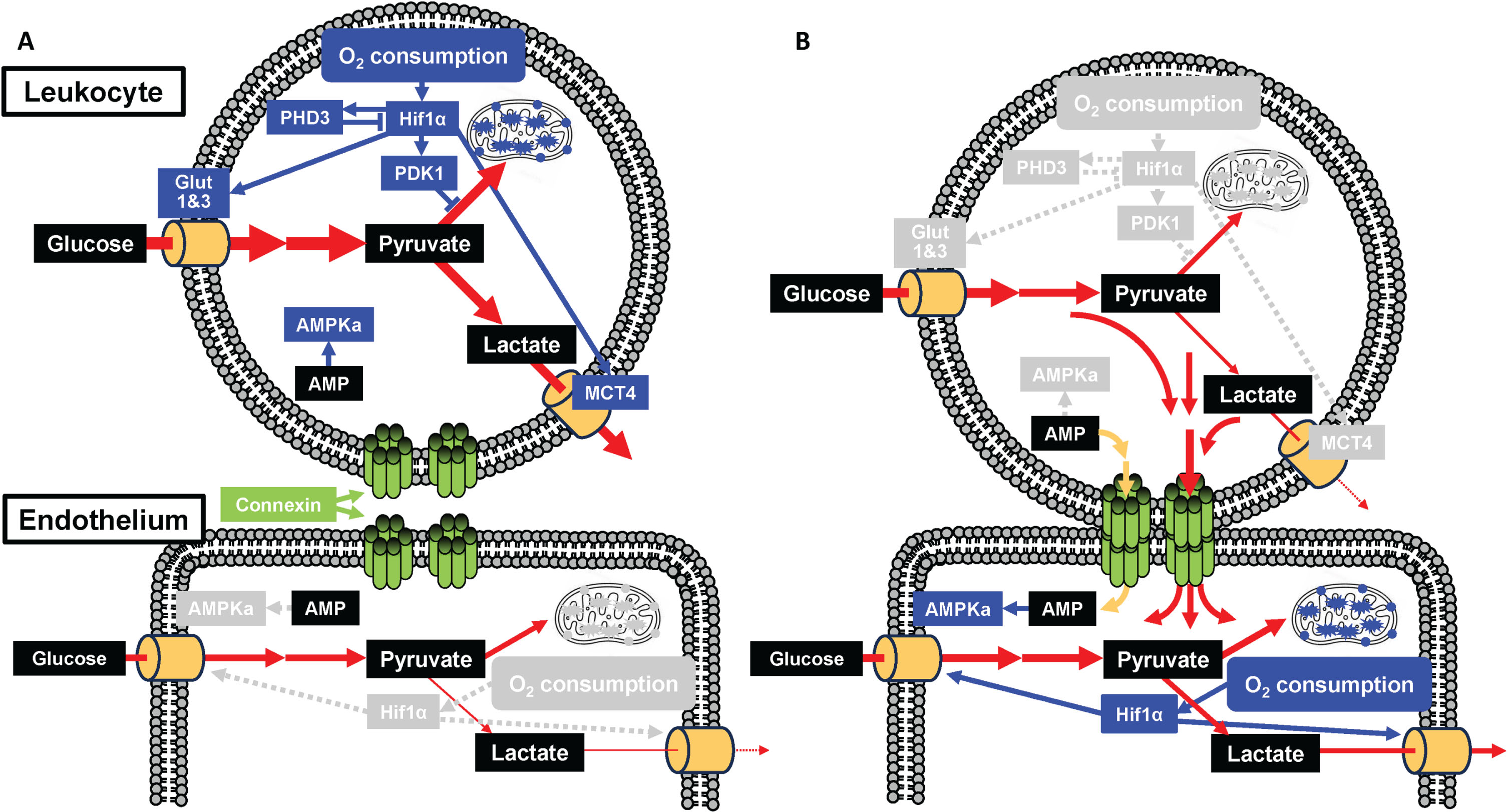
Our results suggest that the Cx37/PHD3 ratio may serve as a biomarker of AD, which has been validated directly in circulating leukocytes in AD patients and indirectly in aged mice brain and long term cultured HUVEC as model systems. Direct cellular interaction between hematopoietic stem cells and endothelial cells via gap junctions and concomitant activation of cellular metabolism has been shown to play a prominent role in tissue repair [3]. Potentially, the changes in RNA expression profiles in circulating leukocytes of AD patients are a general phenomenon that also occurs during age-related cognitive decline, as well as in other forms of dementia. This is also consistent with the proposition that AD in elderly is not only due to the accumulation of amyloid-β and/or tau protein, but also an age-related impairment characterized by vascular dysfunction, chronic inflammation, abnormal glucose metabolism and decrease of tissue stem cells [8, 15–17]. Although the fundamental cause of AD and senescence are still poorly understood [18, 19], the results from clinical trials of anti-amyloid-β therapies indicated that amyloid-β would not be the cause of AD, but a modifier of injured brain during aging [20–22]. These findings show the importance of using aged wild type mice and long-term cultured cells in AD research, instead of amyloid-β modulated mice and cells. Although the role of Cx37 and Cx43 are also poorly understood, Cx37 of leukocytes have been shown to have protective effect against atherosclerosis [23]. In contrast, Cx43 has been shown to enhance monocyte intimal migration, proliferation and apoptosis of endothelial cells [24]. Herein, reduction of Cx37 relative to Cx43 was found in AD which may contribute to AD pathology. In addition, RNA transcription of PHD3 is also activated following Hif1α activation and this is one of the major regulators of cellular metabolism [25]. Our results from the study reported herein also supports the association of the gap junction and Hif1α mediated cellular metabolism.
Chronic inflammation is one of the major factors that cause AD [26]. A significant number of reports have revealed these inflammatory characteristics in the leukocytes of AD patients, including clonal expansion of CD8-positive cells [27], expression of inositol polyphosphate-5-phosphatase [28], activation of double-stranded RNA-dependent protein kinase [29] and triggering receptor expression on myeloid cells [30–32]. In addition, transcriptome analysis of RNA expression in leukocytes have revealed a variety of molecular changes in AD patients, including double-stranded RNA-specific editase 1 which is responsible for pre-mRNA editing of the glutamate receptor [33], neutrophil-derived microvesicles that may impact blood-brain barrier integrity [34] and human leukocyte antigen-B (HLA-B) that can modulate natural killer cell activity [35]. These observations are supported by in vitro analysis of AD associated inflammation [36]. In contrast, our previous reports in aged animal model revealed the link between increased RNA expression of glycolysis related genes in peripheral blood, decreased expression of glycolysis related genes in brain, decreased neurogenesis at hippocampus and impaired short-term memory [7, 8]. As well as chronic inflammation, the change of energy metabolism with aging can be another factor that causes AD [37]. These findings provided the idea and rationale for evaluating metabolism-related genes in peripheral blood of AD patients. Unlike in animal samples, a comparison of simple molecules in human samples did not yield statistically difference between groups, probably because of larger variation between individual in human compared to the animal model. To reduce inter-individual variation, we employed the ratio of RNA expression and found similar results relative to the animal model. It is notable that the link between cellular metabolism of leukocyte and inflammation has also been shown in other diseases [38]. Further studies are required to reveal the link between metabolic state and inflammatory markers of leukocyte in order to identify the overall changes in AD patients.
In this study, we employed qPCR analysis for each gene based on the results obtained in aged mice with cognitive impairment [8]. Though comprehensive RNA analysis, such as RNA-seq, has an advantage in the number of analyzed genes, optimal RNA-seq quantification methods are ambiguous [39, 40], especially when detecting small differences as in the case of this study. In contrast, the techniques utilized in simple conventional qPCR methods are well established. Further studies of comprehensive RNA with sensitive protein analysis of WBC using established methods will be expected to provide information linking circulating WBC and the pathology of AD.
There are limitations of our study, for example, only males were enrolled to avoid possible variation between the sexes and the low patient numbers did not yield a significant difference in single RNA expression between the AD and age matched control groups. This will be addressed in future studies with an increased number of enrolled patients including females. A comparison with other blood biomarkers for AD were also not performed. The link between circulating WBC and amyloid-β levels in peripheral blood/cerebrospinal fluid as well as detailed evaluation of cognitive function and morphological analysis with brain MRI would provide new information relating to this. Finally, it should be noted that the results obtained in this exploratory research needs to be confirmed in more extensive confirmatory studies to avoid false-positive conclusions.
Despite these limitations, the main outcome of our work is that the RNA expression profile of circulating WBC with AD can be different from age matched controls according to established qPCR methods. These findings can now be further developed into a simple and novel diagnostic tool for AD. Recent clinical trials assessing the therapeutic impact of an anti-amyloid-β antibody [20, 21] showed that the anti-amyloid-β therapy has a mild potential to slow down the progression of AD but do not improve cognitive function. These clinical results indicate the need of novel perspectives in AD research, besides neuronal cell death.
In conclusion, our results reveal a possible link between circulating leukocytes and brain function in patients with AD. Our results also indicate that RNA analysis of circulating leukocytes would have a potential to provide information on AD pathology from a different perspective than existing blood markers.
ACKNOWLEDGMENTS
We thank Prof. Johannes Boltze at the University of Warwick for useful discussions.
FUNDING
The research was supported by funding from the Kaneka corporation.
CONFLICT OF INTEREST
A.T. and Y.M. receive research funding from the Kaneka corporation.
DATA AVAILABILITY
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
[1] | Okamoto T , Park EJ , Kawamoto E , Usuda H , Wada K , Taguchi A , Shimaoka M ((2021) ) Endothelial connexin-integrin crosstalk in vascular inflammation. Biochim Biophys Acta Mol Basis Dis 1867: , 166168. |
[2] | Kumar NM , Gilula NB ((1996) ) The gap junction communication channel. Cell 84: , 381–388. |
[3] | Kikuchi-Taura A , Okinaka Y , Takeuchi Y , Ogawa Y , Maeda M , Kataoka Y , Yasui T , Kimura T , Gul S , Claussen C , Boltze J , Taguchi A ((2020) ) Bone marrow mononuclear cells activate angiogenesis via gap junction-mediated cell-cell interaction. Stroke 51: , 1279–1289. |
[4] | Kikuchi-Taura A , Okinaka Y , Saino O , Takeuchi Y , Ogawa Y , Kimura T , Gul S , Claussen C , Boltze J , Taguchi A ((2021) ) Gap junction-mediated cell-cell interaction between transplanted mesenchymal stem cells and vascular endothelium in stroke. Stem Cells 39: , 904–912. |
[5] | Taguchi A , Soma T , Tanaka H , Kanda T , Nishimura H , Yoshikawa H , Tsukamoto Y , Iso H , Fujimori Y , Stern DM , Naritomi H , Matsuyama T ((2004) ) Administration of CD34+ cells after stroke enhances neurogenesis via angiogenesis in a mouse model. J Clin Invest 114: , 330–338. |
[6] | Moreno-Jimenez EP , Flor-Garcia M , Terreros-Roncal J , Rabano A , Cafini F , Pallas-Bazarra N , Avila J , Llorens-Martin M ((2019) ) Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat Med 25: , 554–560. |
[7] | Takeuchi Y , Okinaka Y , Ogawa Y , Kikuchi-Taura A , Kataoka Y , Gul S , Claussen C , Boltze J , Taguchi A ((2020) ) Intravenous bone marrow mononuclear cells transplantation in aged mice increases transcription of glucose transporter 1 and Na(+)/K(+)-ATPase at hippocampus followed by restored neurological functions. Front Aging Neurosci 12: , 170. |
[8] | Takeuchi Y , Saino O , Okinaka Y , Ogawa Y , Akamatsu R , Kikuchi-Taura A , Kataoka Y , Maeda M , Gul S , Claussen C , Boltze J , Taguchi A ((2022) ) Increased RNA transcription of energy source transporters in circulating white blood cells of aged mice. Front Aging Neurosci 14: , 759159. |
[9] | Jack CR Jr. , Bennett DA , Blennow K , Carrillo MC , Dunn B , Haeberlein SB , Holtzman DM , Jagust W , Jessen F , Karlawish J , Liu E , Molinuevo JL , Montine T , Phelps C , Rankin KP , Rowe CC , Scheltens P , Siemers E , Snyder HM , Sperling R ((2018) ) NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement 14: , 535–562. |
[10] | Tsoi KK , Chan JY , Hirai HW , Wong SY , Kwok TC ((2015) ) Cognitive tests to detect dementia: A systematic review and meta-analysis. JAMA Intern Med 175: , 1450–1458. |
[11] | Janelidze S , Mattsson N , Palmqvist S , Smith R , Beach TG , Serrano GE , Chai X , Proctor NK , Eichenlaub U , Zetterberg H , Blennow K , Reiman EM , Stomrud E , Dage JL , Hansson O ((2020) ) Plasma P-tau181 in Alzheimer’s disease: Relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer’s dementia. Nat Med 26: , 379–386. |
[12] | Nakamura A , Kaneko N , Villemagne VL , Kato T , Doecke J , Dore V , Fowler C , Li QX , Martins R , Rowe C , Tomita T , Matsuzaki K , Ishii K , Arahata Y , Iwamoto S , Ito K , Tanaka K , Masters CL , Yanagisawa K ((2018) ) High performance plasma amyloid-beta biomarkers for Alzheimer’s disease. Nature 554: , 249–254. |
[13] | Taguchi A , Ohtani M , Soma T , Watanabe M , Kinosita N ((2003) ) Therapeutic angiogenesis by autologous bone-marrow transplantation in a general hospital setting. Eur J Vasc Endovasc Surg 25: , 276–278. |
[14] | Ogawa Y , Akamatsu R , Fuchizaki A , Yasui K , Saino O , Tanaka M , Kikuchi-Taura A , Kimura T , Taguchi A ((2022) ) Gap junction-mediated transport of metabolites between stem cells and vascular endothelial cells. Cell Transplant 31: , 9636897221136151. |
[15] | Sweeney MD , Montagne A , Sagare AP , Nation DA , Schneider LS , Chui HC , Harrington MG , Pa J , Law M , Wang DJJ , Jacobs RE , Doubal FN , Ramirez J , Black SE , Nedergaard M , Benveniste H , Dichgans M , Iadecola C , Love S , Bath PM , Markus HS , Al-Shahi Salman R , Allan SM , Quinn TJ , Kalaria RN , Werring DJ , Carare RO , Touyz RM , Williams SCR , Moskowitz MA , Katusic ZS , Lutz SE , Lazarov O , Minshall RD , Rehman J , Davis TP , Wellington CL , Gonzalez HM , Yuan C , Lockhart SN , Hughes TM , Chen CLH , Sachdev P , O’Brien JT , Skoog I , Pantoni L , Gustafson DR , Biessels GJ , Wallin A , Smith EE , Mok V , Wong A , Passmore P , Barkof F , Muller M , Breteler MMB , Roman GC , Hamel E , Seshadri S , Gottesman RF , van Buchem MA , Arvanitakis Z , Schneider JA , Drewes LR , Hachinski V , Finch CE , Toga AW , Wardlaw JM , Zlokovic BV ((2019) ) Vascular dysfunction-The disregarded partner of Alzheimer’s disease. Alzheimers Dement 15: , 158–167. |
[16] | Calsolaro V , Edison P ((2016) ) Neuroinflammation in Alzheimer’s disease: Current evidence and future directions. Alzheimers Dement 12: , 719–732. |
[17] | Wang Q , Duan L , Li X , Wang Y , Guo W , Guan F , Ma S ((2022) ) Glucose metabolism, neural cell senescence and Alzheimer’s disease. Int J Mol Sci 23: , 4351. |
[18] | Breijyeh Z , Karaman R ((2020) ) Comprehensive review on Alzheimer’s disease: Causes and treatment. Molecules 25: , 5789. |
[19] | McHugh D , Gil J ((2018) ) Senescence and aging: Causes, consequences, and therapeutic avenues. J Cell Biol 217: , 65–77. |
[20] | Withington CG , Turner RS ((2022) ) Amyloid-related imaging abnormalities with anti-amyloid antibodies for the treatment of dementia due to Alzheimer’s disease. Front Neurol 13: , 862369. |
[21] | van Dyck CH , Swanson CJ , Aisen P , Bateman RJ , Chen C , Gee M , Kanekiyo M , Li D , Reyderman L , Cohen S , Froelich L , Katayama S , Sabbagh M , Vellas B , Watson D , Dhadda S , Irizarry M , Kramer LD , Iwatsubo T ((2023) ) Lecanemab in early Alzheimer’s disease. N Engl J Med 388: , 9–21. |
[22] | Taguchi A , Okinaka Y , Takeda A , Okamoto T , Boltze J , Claussen C , Gul S ((2023) ) Activation of neurogenesis in the hippocampus is a novel therapeutic target for Alzheimer’s disease. Neuroprotection 1: , 139–142. |
[23] | Wong CW , Christen T , Roth I , Chadjichristos CE , Derouette JP , Foglia BF , Chanson M , Goodenough DA , Kwak BR ((2006) ) Connexin37 protects against atherosclerosis by regulating monocyte adhesion. Nat Med 12: , 950–954. |
[24] | Wong CW , Burger F , Pelli G , Mach F , Kwak BR ((2003) ) Dual benefit of reduced Cx43 on atherosclerosis in LDL receptor-deficient mice. Cell Commun Adhes 10: , 395–400. |
[25] | Place TL , Domann FE ((2013) ) Prolyl-hydroxylase 3: Evolving roles for an ancient signaling protein. Hypoxia (Auckl) 2013: , 13–17. |
[26] | Forloni G , Balducci C ((2018) ) Alzheimer’s disease, oligomers, and inflammation. J Alzheimers Dis 62: , 1261–1276. |
[27] | Gate D , Saligrama N , Leventhal O , Yang AC , Unger MS , Middeldorp J , Chen K , Lehallier B , Channappa D , De Los Santos MB , McBride A , Pluvinage J , Elahi F , Tam GK , Kim Y , Greicius M , Wagner AD , Aigner L , Galasko DR , Davis MM , Wyss-Coray T ((2020) ) Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577: , 399–404. |
[28] | Yoshino Y , Yamazaki K , Ozaki Y , Sao T , Yoshida T , Mori T , Mori Y , Ochi S , Iga JI , Ueno SI ((2017) ) INPP5D mRNA expression and cognitive decline in Japanese Alzheimer’s disease subjects. J Alzheimers Dis 58: , 687–694. |
[29] | Couturier J , Page G , Morel M , Gontier C , Claude J , Pontcharraud R , Fauconneau B , Paccalin M ((2010) ) Inhibition of double-stranded RNA-dependent protein kinase strongly decreases cytokine production and release in peripheral blood mononuclear cells from patients with Alzheimer’s disease. J Alzheimers Dis 21: , 1217–1231. |
[30] | Sao T , Yoshino Y , Yamazaki K , Ozaki Y , Mori Y , Ochi S , Yoshida T , Mori T , Iga JI , Ueno SI ((2018) ) TREM1 mRNA expression in leukocytes and cognitive function in Japanese patients with Alzheimer’s disease. J Alzheimers Dis 64: , 1275–1284. |
[31] | Mori Y , Yoshino Y , Ochi S , Yamazaki K , Kawabe K , Abe M , Kitano T , Ozaki Y , Yoshida T , Numata S , Mori T , Iga J , Kuroda N , Ohmori T , Ueno S ((2015) ) TREM2 mRNA expression in leukocytes is increased in Alzheimer’s disease and schizophrenia. PLoS One 10: , e0136835. |
[32] | La Rosa F , Agostini S , Piancone F , Marventano I , Hernis A , Fenoglio C , Galimberti D , Scarpini E , Saresella M , Clerici M ((2023) ) TREM2 expression and amyloid-beta phagocytosis in Alzheimer’s disease. Int J Mol Sci 24: , 8626. |
[33] | Song Z , Ding Q , Yang Y ((2023) ) Orchestration of a blood-derived and ADARB1-centred network in Alzheimer’s and Parkinson’s disease. Cell Signal 110: , 110845. |
[34] | Vazquez-Villasenor I , Smith CI , Thang YJR , Heath PR , Wharton SB , Blackburn DJ , Ridger VC , Simpson JE ((2022) ) RNA-Seq profiling of neutrophil-derived microvesicles in Alzheimer’s disease patients identifies a mirna signature that may impact blood-brain barrier integrity. Int J Mol Sci 23: , 5913. |
[35] | Hooshmand K , Halliday GM , Pineda SS , Sutherland GT , Guennewig B ((2022) ) Overlap between central and peripheral transcriptomes in Parkinson’s disease but not Alzheimer’s disease. Int J Mol Sci 23: , 5200. |
[36] | Jorfi M , Park J , Hall CK , Lin CJ , Chen M , von Maydell D , Kruskop JM , Kang B , Choi Y , Prokopenko D , Irimia D , Kim DY , Tanzi RE ((2023) ) Infiltrating CD8(+) T cells exacerbate Alzheimer’s disease pathology in a 3D human neuroimmune axis model. Nat Neurosci 26: , 1489–1504. |
[37] | Yin F , Sancheti H , Patil I , Cadenas E ((2016) ) Energy metabolism and inflammation in brain aging and Alzheimer’s disease. Free Radic Biol Med 100: , 108–122. |
[38] | Zezina E , Sercan-Alp O , Herrmann M , Biesemann N ((2020) ) Glucose transporter 1 in rheumatoid arthritis and autoimmunity. Wiley Interdiscip Rev Syst Biol Med 12: , e1483. |
[39] | Shi H , Zhou Y , Jia E , Pan M , Bai Y , Ge Q ((2021) ) Bias in RNA-seq library preparation: Current challenges and solutions. Biomed Res Int 2021: , 6647597. |
[40] | Deshpande D , Chhugani K , Chang Y , Karlsberg A , Loeffler C , Zhang J , Muszynska A , Munteanu V , Yang H , Rotman J , Tao L , Balliu B , Tseng E , Eskin E , Zhao F , Mohammadi P , P PL , Mangul S ((2023) ) RNA-seq data science: From raw data to effective interpretation. Front Genet 14: , 997383. |




