Cycloheximide Treatment Causes a ZVAD-Sensitive Protease-Dependent Cleavage of Human Tau in Drosophila Cells
Abstract
Neurofibrillary tangles are the main pathological feature of Alzheimer’s disease. Insoluble tau protein is the major component of neurofibrillary tangles. Defects in the tau protein degradation pathway in neurons can lead to the accumulation of tau and its subsequent aggregation. Currently, contradictory results on the tau degradation pathway have been reported by different groups. This discrepancy is most likely due to different cell lines and methods used in those studies. In this study, we found that cycloheximide treatment induced mild activation of a ZVAD-sensitive protease in Drosophila Kc cells, resulting in cleavage of tau at its C-terminus; this cleavage could generate misleading tau protein degradation pattern results depending on the antibodies used in the assay. Because cycloheximide is a broadly used chemical reagent for the study of protein degradation, the unexpected artificial effect we observed here indicates that cycloheximide is not suitable for the study of tau degradation. Other methods, such as inducible expression systems and pulse-chase assays, may be more appropriate for studying tau degradation under physiological conditions.
INTRODUCTION
Insoluble tau is the key component of neurofibrillary tangles, which are characteristic of Alzheimer’s disease (AD). In AD patients, the extent of tau pathology correlates with cognitive decline [1], and reducing tau levels has been shown to attenuate neuronal dysfunction in mouse models of AD [2, 3]. There is evidence that proteasomal and autophagic activities are decreased in AD-sensitive brain regions compared to unaffected regions (reviewed in [4]). These functional deficiencies are thought to contribute to the accumulation of tau, resulting in its aggregation. Thus, determining the main pathway of tau degradation may allow for the development of effective tau-based therapeutic strategies.
Numerous studies have used various pharmacological and genetic interference approaches to analyze the tau degradation pathway. However, whether the ubiquitin-proteasome system or autophagy-lysosome system is the primary system for tau degradation is still unclear [5, 6]. The commonly accepted explanation for the contradictory results in this area is that the degradation of tau is context-dependent: tau might be degraded through different pathways in different cell types or under different conditions (in vivoversus in vitro). Furthermore, the methods used to study tau degradation were different between laboratories. Some laboratories used a pulse-chase assay to label the newly synthesized tau protein and study its half-life [7], while others used cycloheximide (CHX) to block tau synthesis [8–10]; inducible expression systems were also wildly used [11–13]. These differences in methodology could also explain the variability of data with regards to the mechanism of tau degradation.
Drosophila studies in the past decade have improved our understanding of the molecular mechanism of tau neurotoxicity. A previous study by our group revealed that the C-terminus of tau is important for tau protein stability and toxicity in Drosophila [14]. In the present study, we analyzed tau degradation in Drosophila Kc cells. Our results showed that CHX is able to activate caspase activity in Kc cells; this causes cleavage of the tau protein at its C-terminus, while the rest of the protein remains intact. If only an anti-tau C-terminal antibody is used in such an experiment, the recorded tau degradation rate would be much higher than the true tau degradation rate. In summary, our study revealed an important aspect of CHX that should be taken into account when interpreting data from studies on the degradation of proteins known to be caspase or other ZVAD-sensitive protease substrates.
MATERIALS AND METHODS
Antibodies and reagents
The primary antibodies used in this study were as follows: DM1A (Sigma), 9E10 (Sigma), and Tau46 (Abcam). DM1A is immunospecific for α-tubulin, which was used as the loading control in this study. The antibody 9E10 recognizes the c-Myc tag sequence, which is expressed at the amino terminus of the Tau protein. The C-terminal Tau antibody Tau46 specifically recognizes residues 404–441 and can detect full-length Tau. Cycloheximide was purchased from MP Biomedicals. Protease Inhibitor Set, Z-VAD-fmk, MG132 and epoxomicin were purchased from Calbiochem. A Caspase 3 Assay Kit (Fluorimetric), chloroquine, and 3-methyladenine (3-MA) were purchased from Sigma. An In Situ Cell Death Detection Kit was purchased from Roche AppliedScience.
Cell culture and transfections
Drosophila Kc167 (Kc) cells (gift from Ming Fang’s laboratory) were cultured in Schneider’s Insect Medium (Invitrogen) containing 5% (vol/vol) fetal bovine serum (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin. Kc cells were maintained at 25°C. Cell lines were transfected using X-tremeGENE DNA Transfection Reagents (Roche Applied Science), following the manufacturer’s instructions.
Cell lysis and immunoblotting
Cell extracts were prepared in RIPA buffer (50 mM Tris-HCl, pH 7.4, with 150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) supplemented with complete EDTA-free protease inhibitor cocktail tablets (Roche Applied Science). Protein concentration was determined using the bicinchoninic acid assay (Pierce), and samples were prepared for immunoblotting by dilution in 5 × loading buffer (0.25 mM Tris-HCl, pH 6.8, 15% SDS, 50% glycerol, 25% β-mercaptoethanol and 0.01% bromphenol blue).
Caspase-3 activity determination
Kc cells were incubated with or without cycloheximide (100 μg/ml) for the indicated times, lysed in lysis buffer (Sigma) for 20 min on ice, and then centrifuged at 14,000 × g for 10 min. The caspase-3 specific reaction mixture was prepared following the manufacturer’s instructions (Sigma). Kc cell lysate (20 μg of total protein) was incubated with caspase-3 specific reaction mixture (200 μl) at 37°C for 1 h. Recombinant caspase 3 was used as the positive control. Fluorescence was determined with a fluorescence plate reader (TECAN).
In situ cell death detection
In situ cell death was detected by labeling of DNA strand breaks (TUNEL technology) according to the manufacturer’s instructions. Kc cells were incubated with terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) reaction mixture for 60 min at 37°C in the dark. The samples were washed twice with PBS and once with water and then mounted. Images were acquired using a Zeiss AX10 Fluorescence microscope.
Data analysis
To determine whether the difference in the induction of apoptosis by CHX was statistically significant between treatments, the percentages of apoptotic cells were measured in five randomly selected microscopic fields using Metamorph imaging software. Statistical analyses were performed using one-way analysis of variance, and Student’s t test results are expressed as the means ± S.E. A p-value <0.05 was considered statistically significant (*).
RESULTS
Significant difference in htau degradation rate with the application of different antibodies for the detection of htau in Kc cells
Drosophila has been proven to be an important animal model for the study of neurodegenerative diseases. To investigate the degradation of full-length htau in Drosophila Kc cells, CHX was applied to inhibit protein biosynthesis. We obtained inconsistent results when different anti-tau antibodies were used. Using an antibody (Tau46) against the C-terminal region, 428–441 aa, of the longest isoform of human tau [15], we found that htau was degraded quickly in Kc cells. Only a weak tau signal could be detected after 12 h of CHX treatment, and the tau signal almost completely disappeared after 24 h of CHX treatment. In a parallel experiment, western blotting using the 9E10 antibody, which recognizes the Myc tag added to the N-terminus of htau, showed that only a small portion of tau was degraded at 24 h (Fig. 1B, C). We also observed the same degradation pattern when we used another isoform of human tau (Tau (0N4 R))(Fig. 1D, E).
After further exploration, we found that the tau protein showed a multiple-band pattern when it was detected with 9E10 antibody when the samples were separated in a low-concentration SDS-PAGE gel (lower panel in Fig. 1B). There are at least two explanations for this multi-band pattern: a change in the phosphorylation state of certain residues in htau during degradation and proteolysis caused by protease activities.
Cycloheximide treatment causes truncation of human tau (htau) at C-terminus
Next, Kc cell lysates obtained after 0 and 24 h of incubation with CHX were treated with nonspecific alkaline phosphatase to remove phosphate groups on the amino acid residues, and htau proteins from both samples were found to migrate faster than the htau proteins in those samples that had not been treated with akaline phosphatase. However, the 24-h sample migrated slightly, but significantly, faster than the 0-h sample (Fig. 2A), which implies that tau underwent proteolysis at its C-terminus. Taken together, these results show that htau is hydrolyzed at its C-terminus when Kc cells are treated with CHX.
Cycloheximide induces a low-level of activation of caspase in Kc cells
To determine which particular protease is responsible for the cleavage of tau at its C-terminus, a set of protease inhibitors were employed. The results showed that only the caspase inhibitor ZVAD prevented the formation of multi-band pattern after CHX incubation (Fig. 2B). In the alkaline phosphatase experiment, the presence of ZVAD slowed down the migration of human tau in the 24-h CHX-treated sample, with the same rate as the 0-h sample (Fig. 2 C). These results show that caspase is responsible for the cleavage of htau at its C-terminnus.
CHX has been reported to activate caspase-3 and induce apoptosis in multiple cell lines [16–19]. Our current study showed that treatment with CHX can induce a low level of caspase-3 activation in Kc cells within 24 h (Fig. 3A). Overexpression of tau has also been reported to activate caspases in certain cells [20]. However, our study did not observe activation of caspases by full-length tau (Fig. 3B). The results of our TUNEL assay showed that apoptosis was only significantly induced when Kc cells were incubated with CHX for more than 36 h (Fig. 4). These data imply that short-term CHX treatment of Kc cells can activate caspase mildly, which then cleaves htau at its C-terminus without obviously inducing apoptosis.
The Asp421 residue of hTau is the primary caspase cleavage site in Kc cells
Caspase-3 has been reported to preferentially cleave tau at Asp421 (D421), and phosphorylation of tau at Ser422 (S422) can prevent this cleavage [21]; therefore, we examined whether this is also true in Kc cells. After 24 h of CHX treatment, the unphosphorylated S422A mutant htau showed a multi-band pattern on SDS-PAGE gel similar to that of wildtype tau, but the pseudo-phosphorylated S422E htau mutant did not (Fig. 5A, B). Using the anti-tau C-terminal antibody Tau46, we found that the cleavage at C-terminus of S422E htau was significantly slower than S422A htau (Fig. 5 C, D). Our results indicate that D421 is the primary cleavage site at the C-terminus of tau and that phosphorylation at S422 can prevent this cleavage.
DISCUSSION
There are multiple methods to study protein degradation in cultured cells. In the present study, CHX was chosen because of its ability to inhibit protein synthesis. The results showed that CHX treatment activates caspase, which results in the cleavage of tau at its C-terminus. When antibodies against the C-terminus of tau were used for western blot, the results showed that the majority of tau proteins were degraded within 12 h of incubation with CHX; however, this finding is misleading because the use of N-terminal antibodies revealed that only the C-terminus of tau was missing after 12 h of incubation with CHX. Thus, it is important to use antibodies against different regions of the substrate protein in studies of protein degradation using CHX.
Our study revealed an important aspect of the drug CHX that should be taken into account when interpreting data from studies on the degradation of tau and other similar proteins that are potential substrates of caspase.
In addition to the observation of caspase activation by CHX in our study, previous studies have found that protein synthesis inhibitors can affect other stages of gene expression and protein turnover (transcription, post-transcriptional mRNA modification, translation, PTM, and so forth), and the detailed biological processes affected by protein synthesis inhibitors are described in the review by Alvarez-Castelao et al. [22]. Our findings and those of previous studies indicate that the CHX treatment approach of protein degradation is intricate. It is necessary to utilize other methods, such as inducible expression systems or pulse-chase experiments, to verify the results of protein degradation assays using CHX.
Tet-on and Tet-off systems can be used to achieve transcriptional regulation of a gene by using the trigger, tetracycline, to turn the gene’s expression on or off for a certain period of time. However, for a particular gene, the relatively longer half-life of the mRNA compared to that of the corresponding protein may affect the results obtained using a Tet-on/off system. Thus, it is necessary to check the decay rate of mRNA before making a conclusion about the protein degradation rate.
Another method to examine the degradation pathway of a protein is pulse-chase experiments. Traditionally pulse-chase experiments monitor newly synthesized protein level via a pulse (a brief interval treatment) of radioactive amino acids (35S-Met/Cys, 14 C amino acids, or others) after starvation to deplete the endogenous amino acid pool. Then the level of the protein of interest is determined by immunoprecipitation using a specific antibody and/or measurement of the radioactive signal (for detailed procedure, refer to [23]). However, the starvation treatment can activate the ubiquitin-proteasome pathway and, subsequently, the autophagic pathway, resulting in an increase in the degradation rate of the pre-existing proteins in order to guarantee the amino acids supply, while nascent polypeptides are largely protected from proteolysis [24]. Hence the appropriate amino acid starvation duration and pulse labeling time should be determined using serial trials. The above analysis indicates the complexity and limitation of each approach for the study of protein clearance pathways. One singular standard method will never exist, but accurate conclusions can be drawn if several methods used in combination indicate the same mechanism of protein degradation.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (31170737, 31370768), National Major Scientific and Technological Special Project for “Significant New Drugs Development” (2013ZX09103001-019), Beijing New Medical Discipline Based Group (100270569), Open Research Fund of the State Key Laboratory of Cognitive Neuroscience and Learning and the Fundamental Research Funds for the Central Universities.
Authors’ disclosures available online (http://j-alz.com/manuscript-disclosures/15-0423r2).
REFERENCES
1 | Guillozet AL , Weintraub S , Mash DC , Mesulam MM ((2003) ) Neurofibrillary tangles, amyloid, and memory in aging and mild cognitive impairment. Arch Neurol 60: , 729–736. |
2 | Ittner LM , Ke YD , Delerue F , Bi M , Gladbach A , van Eersel J , Wolfing H , Chieng BC , Christie MJ , Napier IA , Eckert A , Staufenbiel M , Hardeman E , Gotz J ((2010) ) Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell 142: , 387–397. |
3 | Roberson ED , Halabisky B , Yoo JW , Yao J , Chin J , Yan F , Wu T , Hamto P , Devidze N , Yu GQ , Palop JJ , Noebels JL , Mucke L ((2011) ) Amyloid-beta/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer’s disease. J Neurosci 31: , 700–711. |
4 | Chesser AS , Pritchard SM , Johnson GV ((2013) ) Tau clearance mechanisms and their possible role in the pathogenesis of Alzheimer disease. Front Neurol 4: , 122 |
5 | Wang Y , Mandelkow E ((2012) ) Degradation of tau protein by autophagy and proteasomal pathways. Biochem Soc Trans 40: , 644–652. |
6 | Lee MJ , Lee JH , Rubinsztein DC ((2013) ) Tau degradation: The ubiquitin-proteasome system versus the autophagy-lysosome system. Prog Neurobiol 105: , 49–59. |
7 | Drubin D , Kobayashi S , Kellogg D , Kirschner M ((1988) ) Regulation of microtubule protein levels during cellular morphogenesis in nerve growth factor-treated PC12 cells. J Cell Biol 106: , 1583–1591. |
8 | David DC , Layfield R , Serpell L , Narain Y , Goedert M , Spillantini MG ((2002) ) Proteasomal degradation of tau protein. J Neurochem 83: , 176–185. |
9 | Luo HB , Xia YY , Shu XJ , Liu ZC , Feng Y , Liu XH , Yu G , Yin G , Xiong YS , Zeng K , Jiang J , Ye K , Wang XC , Wang JZ ((2014) ) SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination. Proc Natl Acad Sci U S A 111: , 16586–16591. |
10 | Rodriguez-Martin T , Cuchillo-Ibanez I , Noble W , Nyenya F , Anderton BH , Hanger DP ((2013) ) Tau phosphorylation affects its axonal transport and degradation. Neurobiol Aging 34: , 2146–2157. |
11 | Hamano T , Gendron TF , Causevic E , Yen SH , Lin WL , Isidoro C , Deture M , Ko LW ((2008) ) Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur J Neurosci 27: , 1119–1130. |
12 | Hamano T , Gendron TF , Ko LW , Yen SH ((2009) ) Concentration-dependent effects of proteasomal inhibition on tau processing in a cellular model of tauopathy. Int J Clin Exp Pathol 2: , 561–573. |
13 | Gendron TF , McCartney S , Causevic E , Ko LW , Yen SH ((2008) ) Ethanol enhances tau accumulation in neuroblastoma cells that inducibly express tau. Neurosci Lett 443: , 67–71. |
14 | Geng J , Xia L , Li W , Dou F ((2015) ) The C-terminus of tau protein plays an important role in its stability and toxicity. J Mol Neurosci 55: , 251–259. |
15 | Carmel G , Mager EM , Binder LI , Kuret J ((1996) ) The structural basis of monoclonal antibody Alz50’s selectivity for Alzheimer’s disease pathology. J Biol Chem 271: , 32789–32795. |
16 | Blom WM , de Bont HJ , Meijerman I , Mulder GJ , Nagelkerke JF ((1999) ) Prevention of cycloheximide-induced apoptosis in hepatocytes by adenosine and by caspase inhibitors. Biochem Pharmacol 58: , 1891–1898. |
17 | Pengju Z , Weiwen C , Aiying W , Zhaobo C , Nana N , Zhaoqin H , Qingwei L , Anli J ((2010) ) NKX3.1 potentiates TNF-alpha/CHX-induced apoptosis of prostate cancer cells through increasing caspase-3 expression and its activity. Biochem Biophys Res Commun 398: , 457–461. |
18 | Chen YC , Shen SC , Lee WR , Lin HY , Ko CH , Shih CM , Yang LL ((2002) ) Wogonin and fisetin induction of apoptosis through activation of caspase 3 cascade and alternative expression of p21 protein in hepatocellular carcinoma cells SK-HEP-1. Arch Toxicol 76: , 351–359. |
19 | Kim HS , Jeong SY , Lee JH , Kim BE , Kim JW , Jeong SW , Kim IK ((2000) ) Induction of apoptosis in human leukemia cells by 3-deazaadenosine is mediated by caspase-3-like activity. Exp Mol Med 32: , 197–203. |
20 | Yoshizaki C , Tsukane M , Yamauchi T ((2004) ) Overexpression of tau leads to the stimulation of neurite outgrowth, the activation of caspase 3 activity, and accumulation and phosphorylation of tau in neuroblastoma cells on cAMP treatment. Neurosci Res 49: , 363–371. |
21 | Guillozet-Bongaarts AL , Cahill ME , Cryns VL , Reynolds MR , Berry RW , Binder LI ((2006) ) Pseudophosphorylation of tau at serine 422 inhibits caspase cleavage: in vitro Evidence and implications for tangle formation in vivo . J Neurochem 97: , 1005–1014. |
22 | Alvarez-Castelao B , Ruiz-Rivas C , Castaño JG ((2012) ) A critical appraisal of quantitative studies of protein degradation in the framework of cellular proteostasis. Biochem Res Int 2012: , 823597 |
23 | Mellins E ((2013) ) Pulse–chase analysis for studies of MHC class II biosynthesis, maturation, and peptide loading. Methods Mol Biol 960: , 411–432. |
Figures and Tables
Fig.1
Tau can be hydrolyzed at its C-terminus in the presence of Cycloheximide. A) Schematic representation of the antibodies used in this study. 9E10 was used to recognize myc tag which was fused into N-terminus of Tau, the epitope recognized by Tau46 is located within the range of 404–441aa in the 2N4R isoform of human tau. B) Cycloheximide chase analysis of human tau (2N4R) in Drosophila Kc cells using antibody Tau46 or 9E10. C) Quantification of the relative levels of tau after the treatment described in (B). The amount of tau was normalized to the tubulin levels. The results are the means ± S.E. from three independent experiments. D) Cycloheximide chase analysis of human tau (0N4R) in Drosophila Kc cells using antibody 9E10 and Tau46. E) Quantification of the relative levels of tau after the treatment described in (D). The amount of tau was normalized to the tubulin levels. The results are the means ± S.E. from three independent experiments.
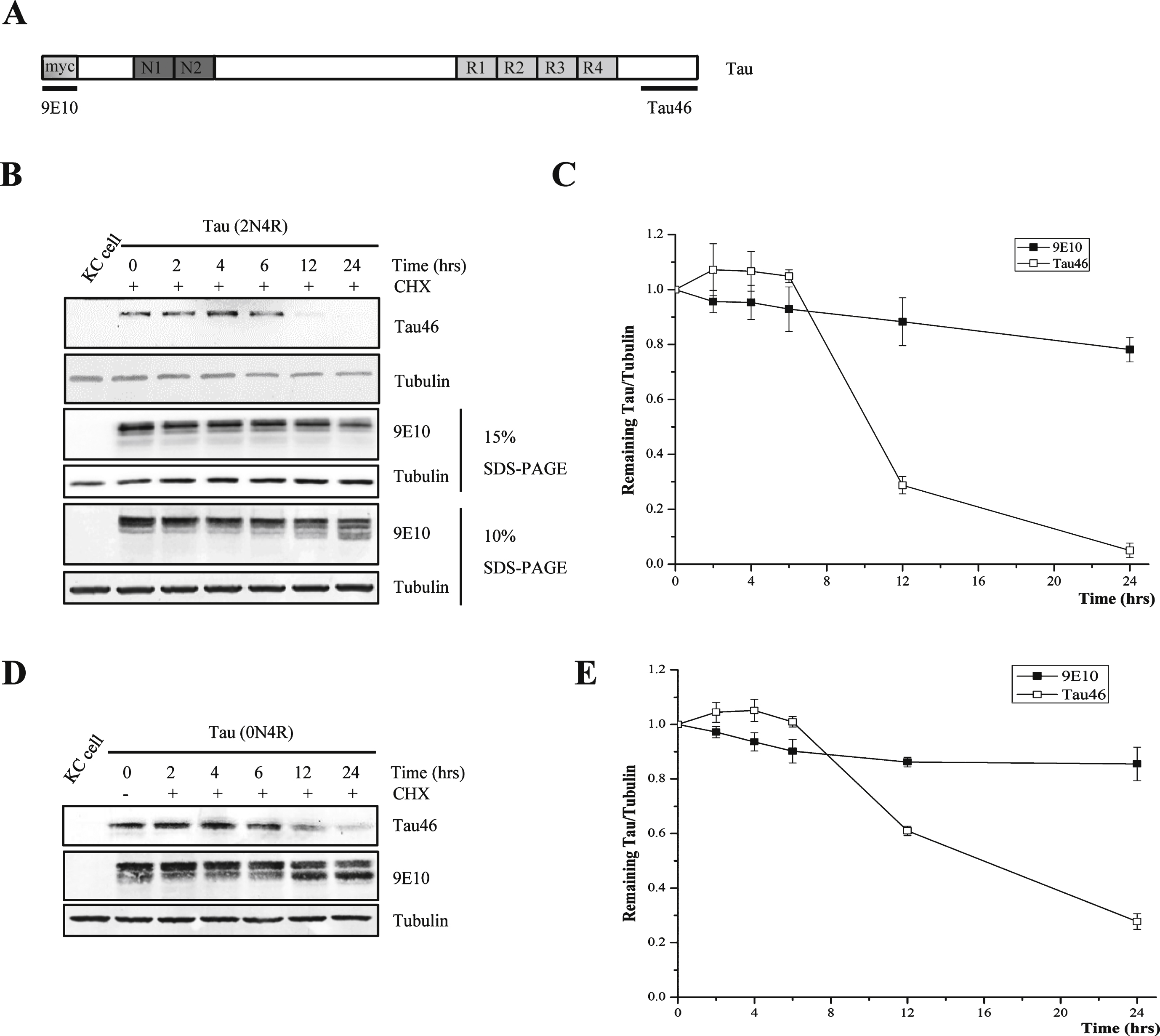
Fig.2
Tau can be truncated by protease during degradation and the truncation can be inhibited by caspase inhibitor. A) Western blot analysis of full-length human tau treated with alkaline phosphatase. At 48-h post transfection, Kc cells were treated with or without 100 μg/ml cycloheximide for 24 h, and the cell lysates were treated with or without alkaline phosphatase at 37°C for 1 h. B) Western blot analysis of full-length human tau in Drosophila Kc cells treated with different protease inhibitors in the presence of cycloheximide (100 μg/ml). At 48-h post transfection, Kc cells were treated with 100 μg/ml cycloheximide and protease inhibitors for 24 h. C) Alkaline phosphatase treatment of full-length human tau from cycloheximide-treated Drosophila Kc cells in the presence or absence of caspase inhibitor Z-VAD. At 48-h post transfection, Kc cells were treated with 100 μg/ml cycloheximide and Z-VAD for 24 h, then the cell lysates were treated with or without alkaline phosphatase at 37°C for 1 h.
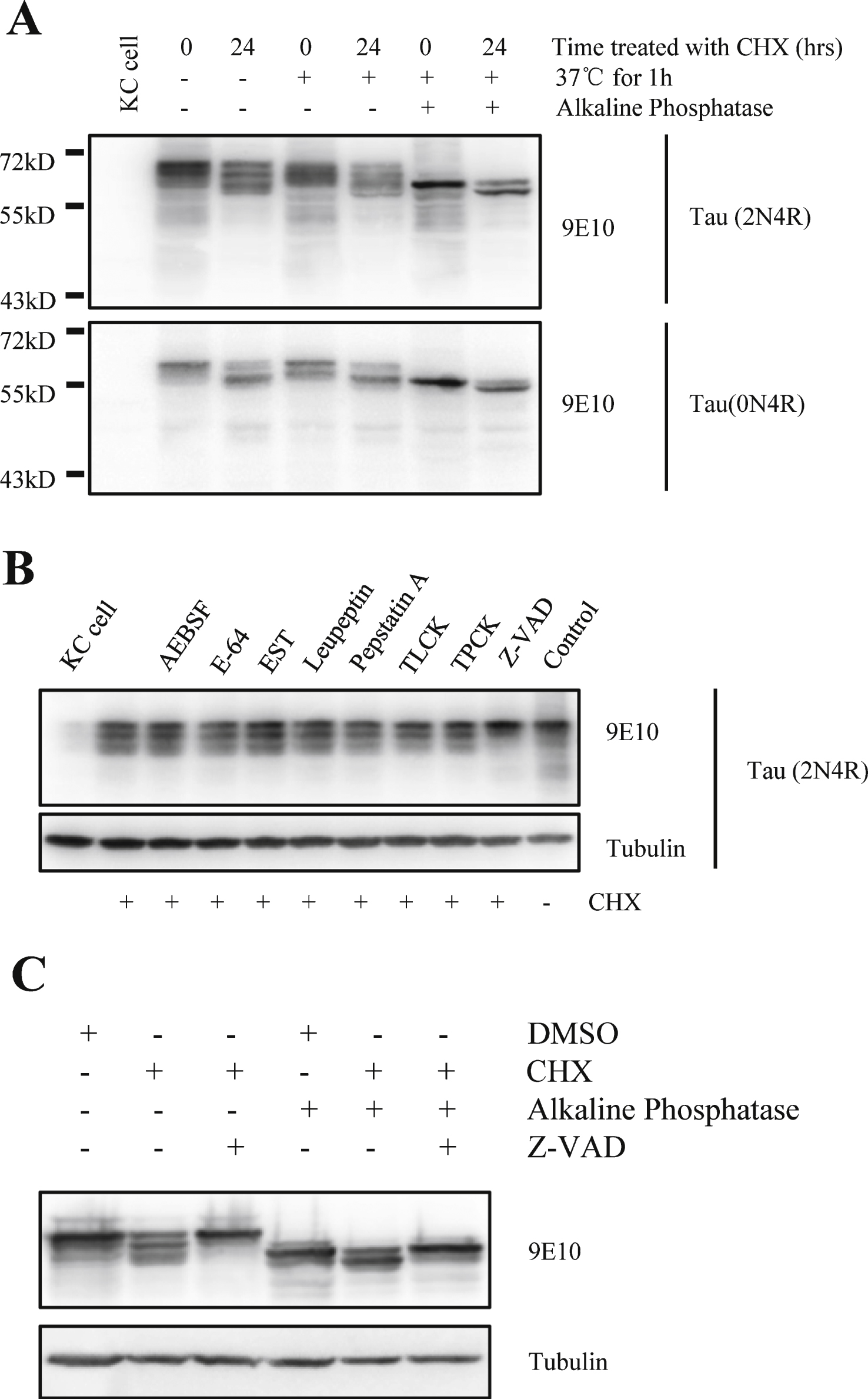
Fig.3
Cycloheximide induces activation of caspase in Kc cells. A) Caspase activity were assayed when Kc cells were treated with cycloheximide (100 μg/ml) for the indicated times, recombinant caspase-3 was used as the positive control (n = 3, *p < 0.05). B) Caspase activity were assayed in Kc cells overexpressing full-length htau (2N4R) for 24 h, recombinant caspase-3 was used as the positive control (n = 3).
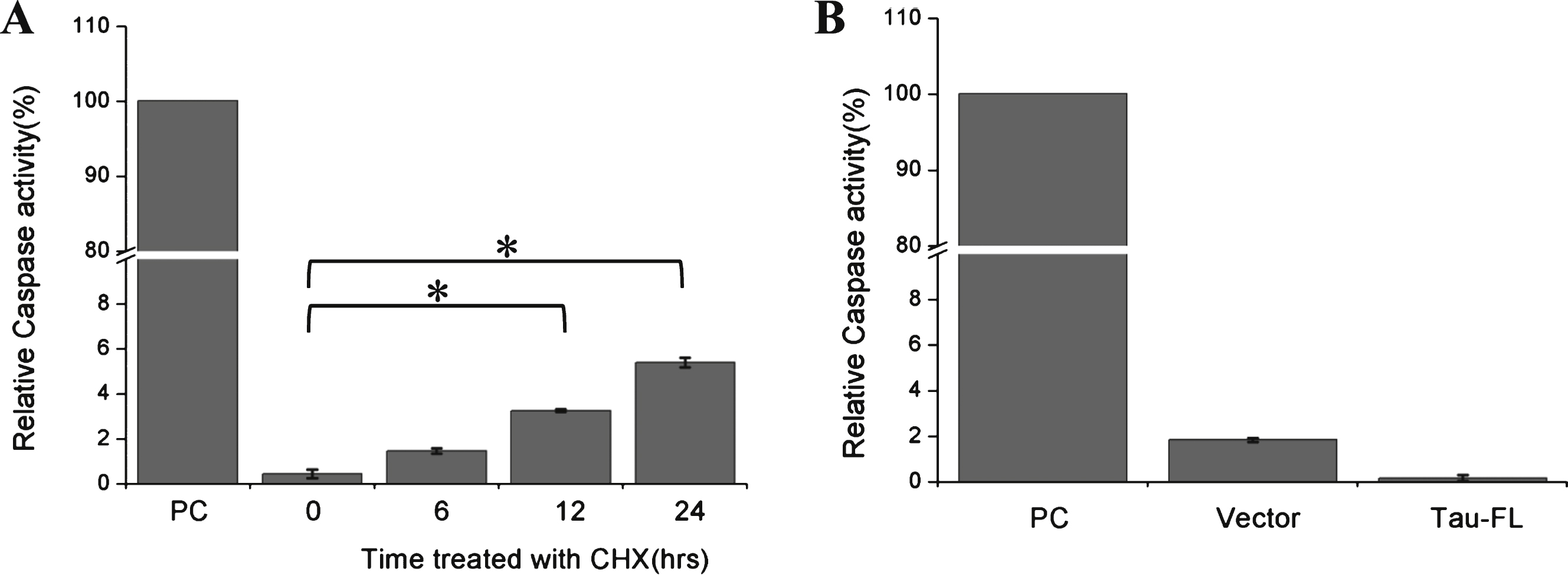
Fig.4
Cycloheximide induces the apoptosis of Kc cells. A) The apoptosis of Kc cells under CHX treatment. Kc cells were treated with cycloheximide (100 μg/ml) for the indicated times, TUNEL assay was used to detect apoptosis of Kc cells. PC, positive control, add 100 μL DNaseI incubated for 10 min before fixation of the Kc cells; NC, negative control, Kc cells were treated with DMSO for 48 h. B) The statistical histogram of TUNEL signal in Kc cells with different cycloheximide treatment duration (n = 3, **p < 0.001).
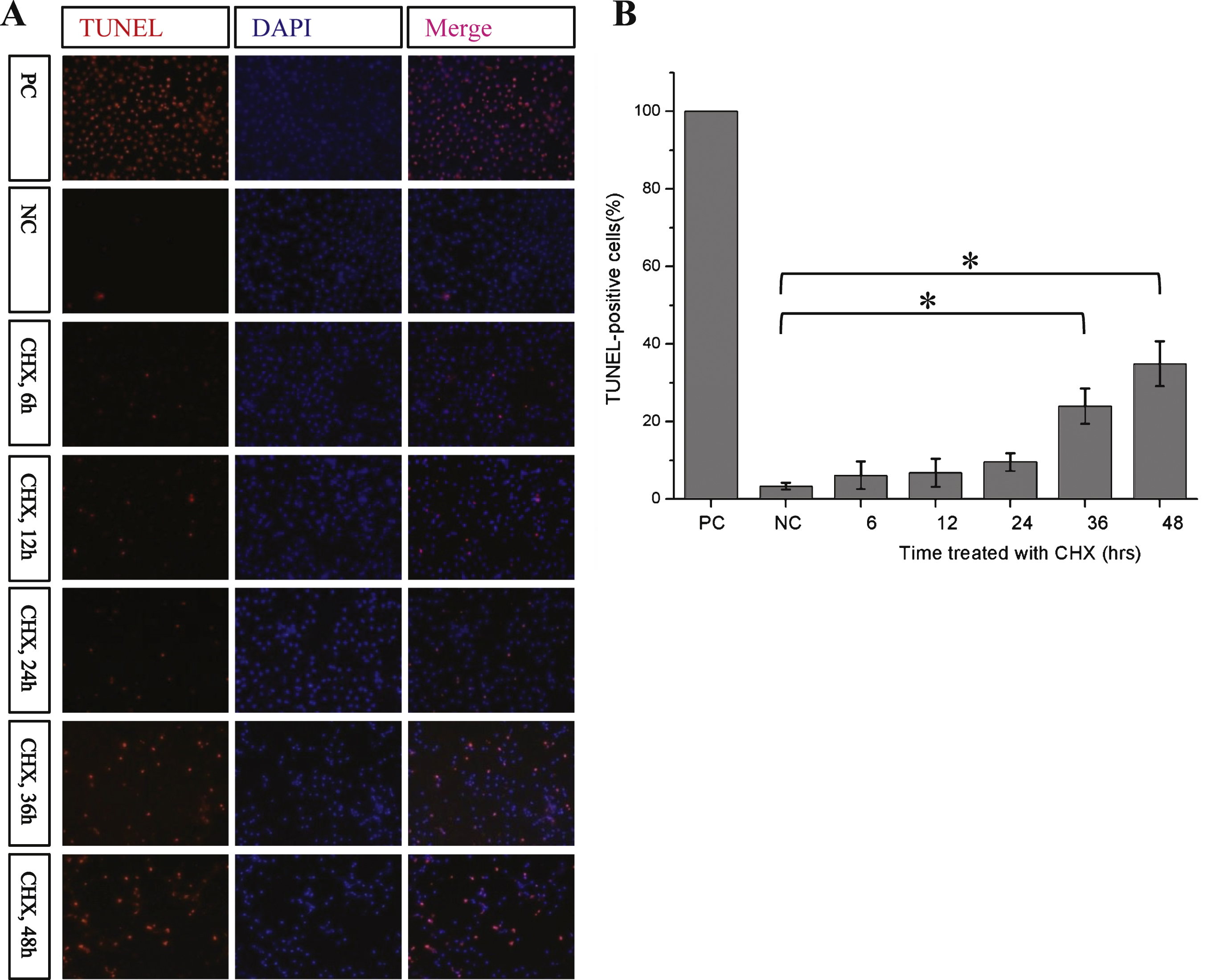
Fig.5
Tau can be cleaved primarily at Asp421with the treatment of Cycloheximide. A) Cycloheximide chase analysis of mutant human tau [tau(S422A) and tau(S422E)] in Drosophila Kc cells using 9E10 antibody. B) Quantification of the relative levels of tau after the treatment described in (A). The amount of tau was normalized to the tubulin levels. The results are the means ± S.E. from three independent experiments. C) Cycloheximide chase analysis of mutant human tau [tau(S422A) and tau(S422E)] in Drosophila Kc cells using Tau46 antibody. Tubulin was used as the loading control in all western blot analyses. D) Quantification of the relative levels of tau after the treatment described in (C). The amount of tau was normalized to the tubulin levels. The results are the means ± S.E. from three independent experiments and data points indicated with asterisks are significant. *p < 0.05.
![Tau can be cleaved primarily at Asp421with the treatment of Cycloheximide. A) Cycloheximide chase analysis of mutant human tau [tau(S422A) and tau(S422E)] in Drosophila Kc cells using 9E10 antibody. B) Quantification of the relative levels of tau after the treatment described in (A). The amount of tau was normalized to the tubulin levels. The results are the means ± S.E. from three independent experiments. C) Cycloheximide chase analysis of mutant human tau [tau(S422A) and tau(S422E)] in Drosophila Kc cells using Tau46 antibody. Tubulin was used as the loading control in all western blot analyses. D) Quantification of the relative levels of tau after the treatment described in (C). The amount of tau was normalized to the tubulin levels. The results are the means ± S.E. from three independent experiments and data points indicated with asterisks are significant. *p < 0.05.](https://content.iospress.com:443/media/jad/2016/49-4/jad-49-4-jad150423/jad-49-jad150423-g005.jpg)




