Genome-Wide Mendelian Randomization Identifies Ferroptosis-Related Drug Targets for Alzheimer’s Disease
Abstract
Background:
Alzheimer’s disease (AD) currently lacks effective disease-modifying treatments. Recent research suggests that ferroptosis could be a potential therapeutic target. Mendelian randomization (MR) is a widely used method for identifying novel therapeutic targets.
Objective:
Employ genetic information to evaluate the causal impact of ferroptosis-related genes on the risk of AD.
Methods:
564 ferroptosis-related genes were obtained from FerrDb. We derived genetic instrumental variables for these genes using four brain quantitative trait loci (QTL) and two blood QTL datasets. Summary-data-based Mendelian randomization (SMR) and two-sample MR methods were applied to estimate the causal effects of ferroptosis-related genes on AD. Using extern transcriptomic datasets and triple-transgenic mouse model of AD (3xTg-AD) to further validate the gene targets identified by the MR analysis.
Results:
We identified 17 potential AD risk gene targets from GTEx, 13 from PsychENCODE, and 22 from BrainMeta (SMR p < 0.05 and HEIDI test p > 0.05). Six overlapping ferroptosis-related genes associated with AD were identified, which could serve as potential therapeutic targets (PEX10, CDC25A, EGFR, DLD, LIG3, and TRIB3). Additionally, we further pinpointed risk genes or proteins at the blood tissue and pQTL levels. Notably, EGFR demonstrated significant dysregulation in the extern transcriptomic datasets and 3xTg-AD models.
Conclusions:
This study provides genetic evidence supporting the potential therapeutic benefits of targeting the six druggable genes for AD treatment, especially for EGFR (validated by transcriptome and 3xTg-AD), which could be useful for prioritizing AD drug development in the field of ferroptosis.
INTRODUCTION
Over the past three decades, there has been a noticeable increase in the global prevalence and societal and economic impact of Alzheimer’s disease (AD), underscoring the imperative for extensive research into its pathogenesis and potential treatments.1 The pathogenesis of AD is complex, involving multiple factors such as amyloid-β (Aβ) toxicity, tau protein, genetic mutations, synaptic damage, and neuroinflammation.2,3 Ferroptosis, an iron-dependent form of regulated cell death, has offered fresh insights into comprehending the pathogenesis of AD.4
Recent studies suggest that ferroptosis might be involved in the pathological process of AD.5,6 There are several similarities between the characteristics of AD pathogenesis and those of ferroptosis, such as iron accumulation and lipid peroxides.7,8 As a result, ferroptosis is gradually being acknowledged as a unique cell death mechanism that contributes to the pathogenesis of AD. Interestingly, we also observed a close correlation between neuronal death and ferroptosis in AD patients. Elevated levels of iron and markers of lipid peroxidation have been detected in the brains of AD patients.4 Dysregulation of iron metabolism, specifically the accumulation of iron within neurons, has been implicated in the initiation of ferroptosis. This aberrant iron accumulation leads to increased reactive oxygen species (ROS) production, which in turn triggers lipid peroxidation and the subsequent death of neurons through ferroptosis.4,9
Substantial efforts have been invested in the search for treatments that can modify AD, but significant progress remains elusive due to the unknown pathophysiology of the disease.10 Given the presence of iron deposits in the brains of AD patients, ferroptosis inhibitors offer promising prospects for AD treatment.11 Excess iron can worsen oxidative damage, and targeting ferroptosis may help alleviate cognitive deficits and oxidative stress in AD.12,13 However, the research on ferroptosis inhibitors and AD is still in its early stages. Large-scale human genetic studies provide a unique avenue for innovative drug development for complex diseases. This is because drug targets backed by genetic evidence have a higher likelihood of success in drug discovery pipelines.14
MR analysis has been widely utilized to adapt licensed drugs and uncover novel therapeutic targets by amalgamating summary data from disease genome-wide association study (GWAS) and quantitative trait loci (QTL) studies.15 The levels of gene expression can be viewed as a form of long-term exposure, and QTLs situated in the genomic region of “druggable” genes are always regarded as proxies.16 Additionally, previous studies have used MR methods to assess the causal effects of disease exposure or biomarkers on various neurodegenerative diseases, further validating the reliability of our research approach and enriching our background knowledge for this study.17–19
This MR study employing brain-specific and blood-specific expression QTLs (eQTLs) and protein QTLs (pQTLs) of ferroptosis-related genes as instrumental variables, with ferroptosis-related genes as the exposure factor and AD GWAS summary-level data as the outcome factor, enables us to identify potential drug targets associated with ferroptosis in AD. This strategy employs genetic information to evaluate the causal impact of ferroptosis-related genes on the risk of AD and can offer valuable insights for the development of targeted therapies for AD.
MATERIAL AND METHODS
Ferroptosis-related genes list
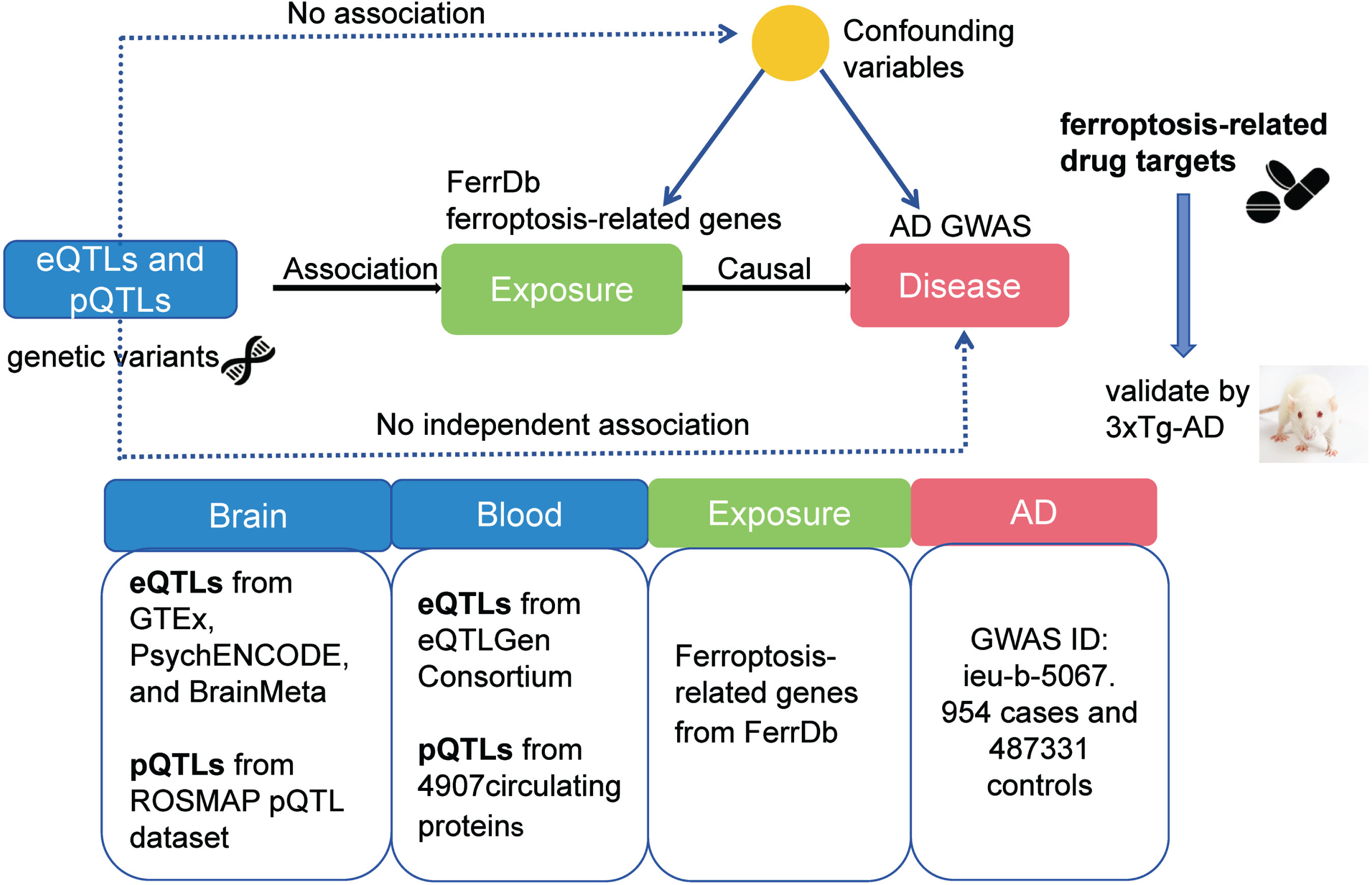
The principle and flow diagram of MR study are depicted in Fig. 1. FerrDb, a central database dedicated to the study of ferroptosis regulators and their disease associations, is accessible via http://www.zhounan.org/ferrdb/. Genes related to ferroptosis were extracted from this database for subsequent analysis.
Fig. 1
The principle and flowchart of MR analysis.
QTL datasets used for genetic instrumental variables selection
We leveraged four QTL datasets of brain to derive genetic instrumental variables for ferroptosis-related genes. Moreover, we utilized eQTL and pQTL data from available blood tissue to exhaustively search for ferroptosis-related targets related to AD. The GTEx eQTL datasets were acquired from the SMR website (https://yanglab.westlake.edu.cn/software/smr/#eQTLsummarydata).
The PsychENCODE eQTL datasets were generated using brain tissues (specifically, the prefrontal cortex) from 1,378 human individuals.20 We accessed the LD reference panel of the European (EUR) population from the 1000 Genomes project provided by OpenGWAS API.21 The BrainMeta v2 cis-eQTL summary data, downloaded from the SMR website, underwent eQTL mapping using RNA-seq data from 2,865 brain cortex samples, collected from 2,443 unrelated individuals of European ancestry with genome-wide single nucleotide polymorphism (SNP) data. The pQTL data of brain originated from a recent study, which can be downloaded from Synapse (https://doi.org/10.7303/syn23627957).22
The summary-level eQTL data of blood tissues were procured from the eQTLGen Consortium, comprising a total of 31,684 individuals (https://www.eqtlgen.org/). The pQTL summary study is grounded in a large-scale GWAS on the blood proteome (https://www.decode.com/summarydata/). We extracted summary-level statistics of genetic associations with levels of 4,907 circulating proteins from a large-scale pQTL study conducted in 35,559 Icelanders.
GWAS summary statistics of AD
The summary statistics for AD from GWAS were retrieved from the MRC IEU Open GWAS Project (https://gwas.mrcieu.ac.uk/). The GWAS results, identified by the GWAS ID ieu-b-5067, served as the outcome data in the MR analysis. This analysis incorporated data from 954 cases and 487,331 controls.
Mendelian randomization
SMR method
In our study, we utilized the SMR method to explore the relationship between gene expression levels and outcomes of interest, leveraging summary-level data from both GWAS and eQTL studies, specifically focusing on eQTLs as instrumental variables.23 To isolate brain-specific eQTLs, we analyzed all GTEx results and pinpointed cis-SNPs linked to gene expression. Following this, we delved deeper to ascertain if genes whose expression was influenced by these SNPs were implicated in ferroptosis-related gene targets. We then narrowed down to genetic variants associated with the expression of ferroptosis-related genes, setting a stringent threshold of eQTL significance at p < 5×10–08 for further analysis.
To ensure the validity of our findings, we performed the HEIDI test within the SMR software to discern whether the observed associations between gene expression and outcomes could be attributed to linkage scenarios.23 A p-value less than 0.05 from the HEIDI test suggests a likelihood of linkage influencing the observed association.24 Our selection criteria focused on genes exhibiting an SMR p-value below 0.05, coupled with a HEIDI p-value greater than 0.05, highlighting them as promising candidates linked to AD.
One SNP could be associated with the expression of multiple genes, a phenomenon known as horizontal pleiotropy. To assess the likelihood of horizontal pleiotropy, we identified nearby genes (within a 1 Mb window) whose expression was significantly associated with the genetic instrumental variant. We then performed SMR analysis to ascertain if the expression of these genes was related to AD outcomes.
Two-sample MR method
“TwoSampleMR” R package was used to conduct the two-sample MR analysis.25 In this study, we employed cis-pQTL data as genetic instruments (exposure) and GWASs of AD as the outcome trait data. The two-sample MR method evaluates the relationship between protein expression and AD. The Wald ratio was applied when only one pQTL was available for a given protein. In instances where two or more genetic instruments were available, we implemented the inverse variance weighted MR (MR-IVW) method followed by a heterogeneity analysis.
PPI analysis and expression of the identified gene targets in different cell types of the human brain
The online tool, Search Tools for the Retrieval of Interacting Genes (STRING), was used to scrutinize protein interactions. We filtered the Protein-Protein Interaction (PPI) pairs based on a confidence score exceeding 0.40 and visualized the PPI network using Cytoscape V3.8.2 software.26 We leveraged the Cortical Development Expression Viewer (CoDEx) data portal to investigate the expression patterns of genes significant in the SMR analysis at the single-cell level.27
Triple-transgenic mouse model of AD
3xTg-AD is an important preclinical animal model used for studying the pathogenesis and potential treatments for AD. This mouse model was created by incorporating three transgenes (APP, PS1, and Tau mutation) into the mouse genome to mimic key features of AD pathology observed in human patients.28 By combining these three mutations in a single mouse model, the 3xTg-AD model recapitulates various aspects of AD pathology, including Aβ plaque deposition, neurofibrillary tangle formation, synaptic dysfunction, and cognitive decline.
Males of the 3xTg-AD strain, aged between 6-7 weeks, were obtained from the Changzhou Cavens Laboratory Animal Co., Ltd. (Wuhan, China; license number: SCXK (Su) 2016-0010). These mice were maintained in a pathogen-free laboratory at Wuhan Myhalic Biotechnology Co., Ltd. (Wuhan, China), under precise conditions: a temperature range of 20–26°C, 50% ±10% humidity, and a 12-h day-night light cycle. Standard rodent chow and unrestricted access to water were provided until they reached 9 months of age. Males of the C57BL/6J strain, aged 7 months, were purchased from Wuhan Youdu Biotechnology Co., Ltd. (Wuhan, China; license number: SCXK (E) 2021-0025). These mice were housed under identical conditions to those of the 3xTg-AD mice, including comparable dietary regimens up to 9 months of age. All animal research was conducted with the approval of the Animal Ethics Committee of Wuhan Myhalic Biotechnology Co., Ltd. (ethical approval number: HLK-20230518-001). The experimental procedures complied with the ARRIVE guidelines and the American Veterinary Medical Association Guidelines for the Euthanasia of Animals (2020). No interventional experimental procedures were employed on the mice. Following anesthesia with isoflurane, the mice were decapitated, and their entire brains were rapidly harvested for western blot analyses.
Validation of the gene targets by transcriptome and western blotting
Validation of external transcriptomic datasets
To provide additional validation for the gene targets identified via SMR analysis, we employed transcriptomic datasets pertaining to AD from the Gene Expression Omnibus (GEO) database. Specifically, dataset GSE122063, sourced from the GPL16699 platform, encompassed transcriptomic data from 56 individuals diagnosed with AD and 44 control samples.29 Gene expression profiling was conducted on the frontal and temporal lobes of AD patients and non-demented controls (referred to as Controls) sourced from the University of Michigan Brain Bank.
Western blotting
We used a lysis buffer for the acquisition of overall protein from the complete brain of 3xTg-AD. After assessing with a BCA kit, we segregated the protein samples on a gel comprising of sodium dodecyl sulfate-polyacrylamide and shifted them onto membranes composed of polyvinylidene difluoride (PVDF). Following an hour-long incubation with 5% skim milk, both primary antibodies and GAPDH were employed overnight at a temperature of 4°C. Subsequently, a secondary antibody connected with horseradish peroxidase was administered to the membrane and allowed to incubate for an hour at ambient temperature. To render the proteins visible, we employed a chemiluminescence kit.
The following primary antibodies were used: CDC25A (55031-1-AP, proteintech, Wuhan, China), LIG3 (26583-1-AP, proteintech, Wuhan, China), TRIB3 (13300-1-AP, proteintech, Wuhan, China), DLD (16431-1-AP, proteintech, Wuhan, China), EGFR (A11351, Abclonal, Wuhan, China), PEX10 (A16949, Abclonal, Wuhan, China), and β-Tublin (10094-1-AP, proteintech, Wuhan, China).
RESULTS
Ferroptosis-related genes list
A total of 369 genes were identified as drivers, 348 as suppressors, 11 as markers, and 116 were left unclassified. Some genes may duplicate across different categories. Consequently, we have gathered 564 genes related to ferroptosis for further analysis (Supplementary Table 1).
SMR to prioritize candidate gene targets in brain tissue
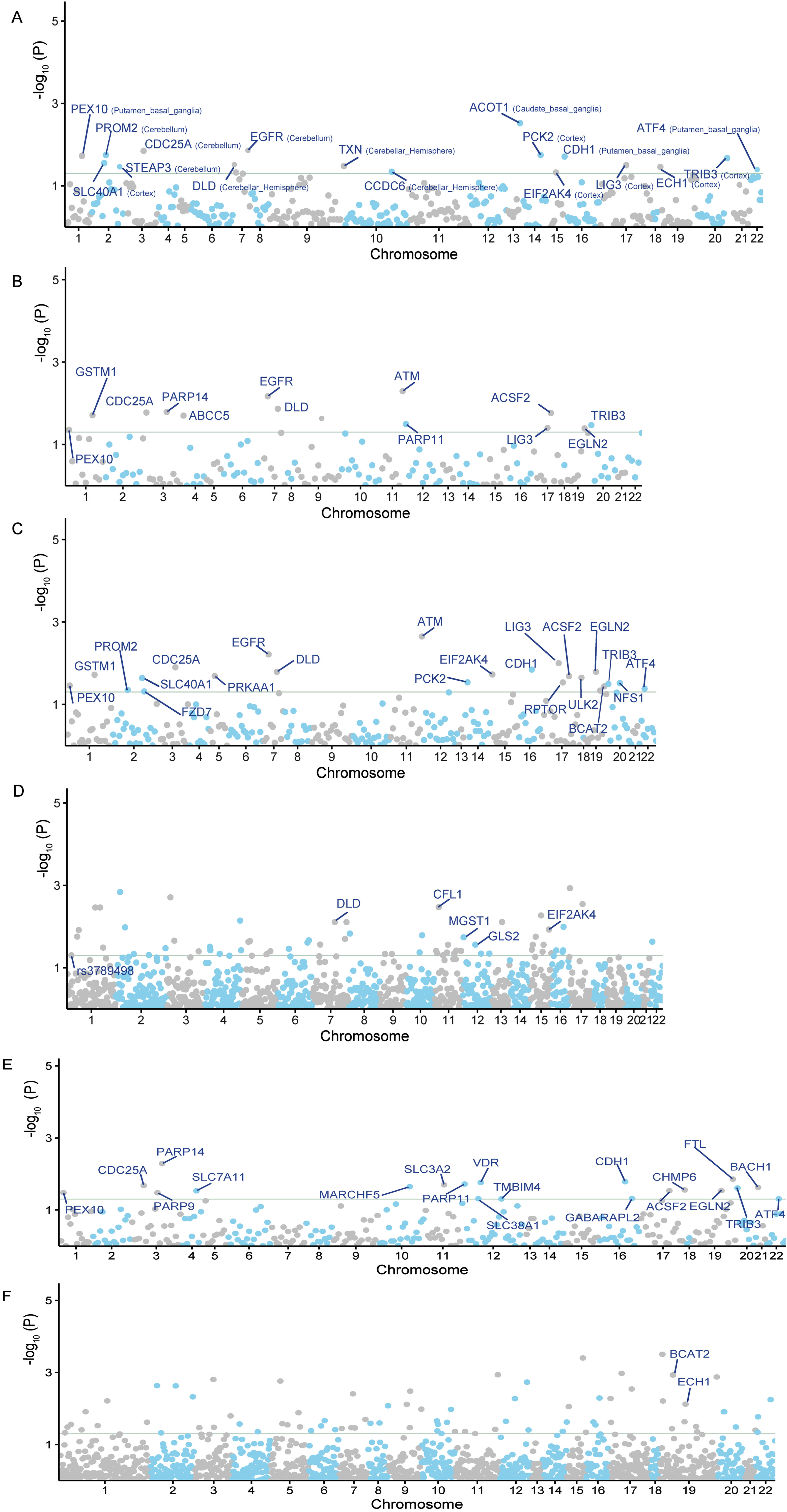
SMR analysis can be used to ascertain causal relationships and investigate whether alterations in gene or protein expression contribute to disease onset. Consequently, the significant genes or proteins identified by SMR can serve as potential therapeutic targets. By using cis-eQTL SNPs from GTEx (spanning 13 brain regions) as genetic instruments, we pinpointed 17 potential gene targets for AD (SMR p < 0.05 and HEIDI test p > 0.05) (Fig. 2A). We also found 13 significant correlations from PsychENCODE, and 22 from BrainMeta (Fig. 2B, C). Additional details can be found in the Supplementary Table 1.
Fig. 2
The Manhattan plots of MR analysis results using QTLs and AD GWAS summary statistics. The gray horizontal line is the Bonferroni corrected significant level. A) The MR result using GTEx brain eQTL as instruments; B) The MR result using PsychENCODE eQTL as instruments; C) The MR result using BrainMeta eQTL as instruments; D) The MR result using ROSMAP pQTL as instruments; E) The MR result using cis-eQTL SNPs from eQTLGen Consortium as genetic instruments; F) The MR result using cis-pQTL SNPs from circulating 4,907 proteins levels as genetic instruments.
When we combined the three gene lists, we were able to identify 30 potential genes. When we found the common genes among the three lists, we discovered six overlapping candidate genes, including PEX10, CDC25A, EGFR, DLD, LIG3, and TRIB3 (SMR p < 0.05 and HEIDI test p > 0.05) (Fig. 3A, Tables 1 and 2). These genes, including PEX10, EGFR, DLD, and LIG3, function as inducers of ferroptosis, promoting its occurrence. Conversely, CDC25A and TRIB3 serve as an inhibitor of ferroptosis, preventing its onset. The detailed SMR locus plot for the six genes can be found in Supplementary Figure 1.
Fig. 3
A) SMR method identified six overlapping ferroptosis-related gene targets associated with AD when using GTEx, PsychENCODE, and BrainMeta brain eQTL as genetic instruments, including PEX10, CDC25A, EGFR, DLD, LIG3, and TRIB3. B) Venn diagram showed the intersection of AD risk genes identified by GTEx, PsychENCODE, and BrainMeta brain eQTL and ROSMAP brain pQTL as genetic instruments. C)Venn diagram showed the intersection of AD risk genes identified by GTEx, PsychENCODE, and BrainMeta brain eQTL and eQTLGen blood eQTL as genetic instruments. D) Venn diagram showed the intersection of AD risk genes identified by eQTLGen blood eQTL and blood pQTL as genetic instruments.
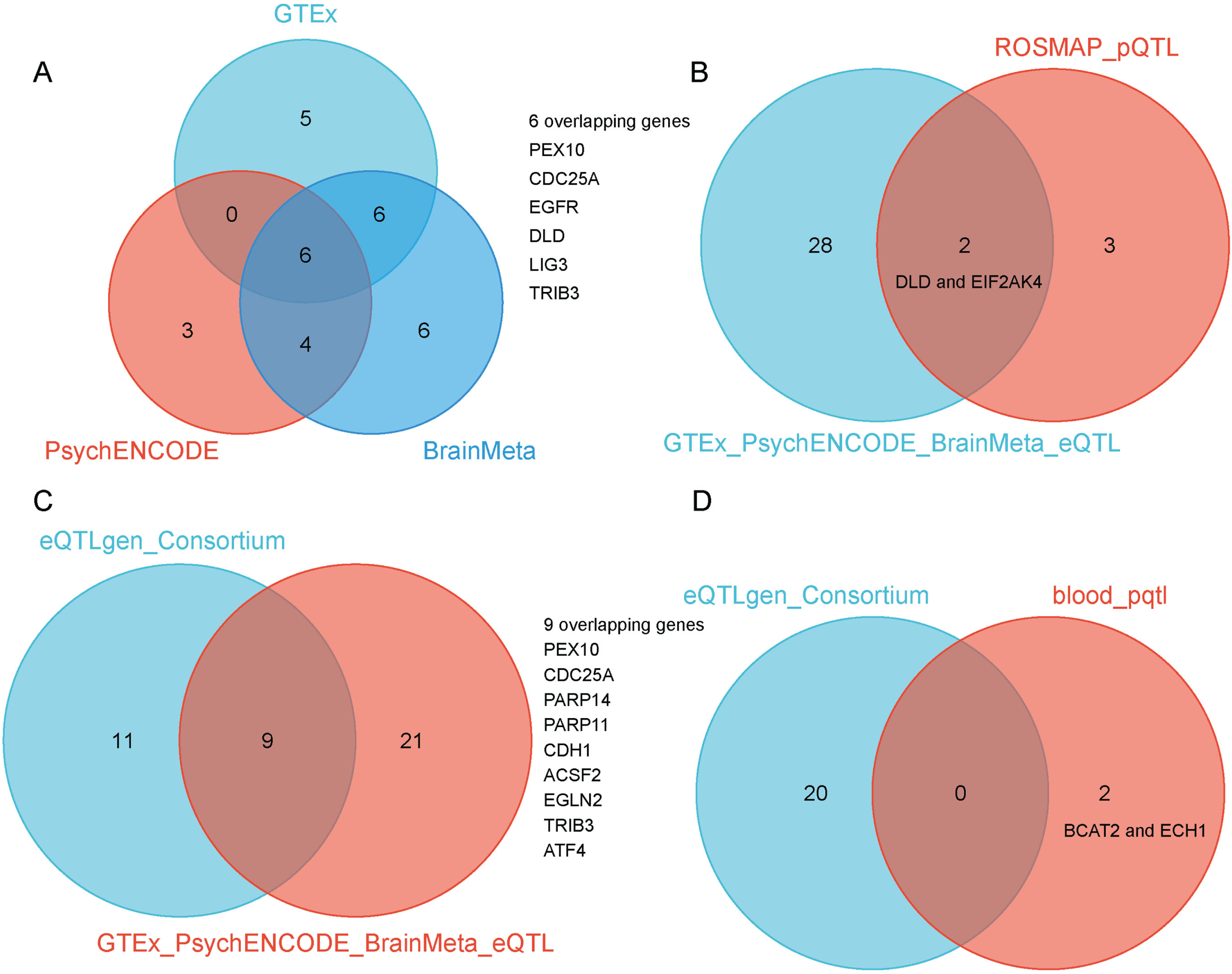
Table 1
SMR results from GTEx brain eQTLs
| GTEx _brain | Gene | topSNP | b_SMR | se_SMR | p_SMR | p_HEIDI |
| Amygdala | TRIB3 | rs62191434 | –0.0004 | 0.0002 | 0.0401 | 0.9869 |
| Anterior_cingulate | DLD | rs10271464 | –0.0007 | 0.0003 | 0.0306 | 0.7389 |
| TRIB3 | rs62191434 | –0.0004 | 0.0002 | 0.0441 | 0.9696 | |
| Caudate | DLD | rs4518 | –0.0006 | 0.0003 | 0.0474 | 0.7126 |
| TRIB3 | rs62191434 | –0.0003 | 0.0001 | 0.0349 | 0.9785 | |
| Cerebellar | CDC25A | rs13097450 | 0.0003 | 0.0001 | 0.0141 | 0.9997 |
| DLD | rs2237686 | –0.0011 | 0.0005 | 0.0224 | 0.7021 | |
| Cerebellum | CDC25A | rs3731550 | 0.0003 | 0.0001 | 0.0182 | 0.9988 |
| EGFR | rs76928645 | 0.0014 | 0.0005 | 0.0109 | 0.8670 | |
| TRIB3 | rs62191434 | –0.0005 | 0.0002 | 0.0431 | 0.9742 | |
| Cortex | DLD | rs4518 | –0.0006 | 0.0003 | 0.0485 | 0.9015 |
| LIG3 | rs1634800 | –0.0003 | 0.0002 | 0.0316 | 0.9639 | |
| TRIB3 | rs11698987 | –0.0003 | 0.0001 | 0.0212 | 1.0000 | |
| Frontal_Cortex | DLD | rs4266570 | –0.0007 | 0.0003 | 0.0328 | 0.9751 |
| TRIB3 | rs62191440 | –0.0004 | 0.0002 | 0.0415 | 0.8672 | |
| Hippocampus | TRIB3 | rs6051554 | –0.0005 | 0.0002 | 0.0363 | 0.9685 |
| Hypothalamus | LIG3 | rs1088450 | –0.0004 | 0.0002 | 0.0306 | 0.6199 |
| TRIB3 | rs62191440 | –0.0006 | 0.0003 | 0.0431 | 0.8981 | |
| Nucleus_accumbens | LIG3 | rs1634800 | –0.0004 | 0.0002 | 0.0332 | 0.7828 |
| TRIB3 | rs62191440 | –0.0004 | 0.0002 | 0.0361 | 0.9917 | |
| Putamen | PEX10 | rs10910063 | –0.0009 | 0.0004 | 0.0187 | 0.5846 |
| LIG3 | rs3135967 | –0.0003 | 0.0002 | 0.0348 | 0.8546 | |
| TRIB3 | rs62191440 | –0.0003 | 0.0002 | 0.0369 | 0.9939 | |
| Substantia | TRIB3 | rs6051544 | –0.0003 | 0.0002 | 0.0374 | 0.9950 |
Table 2
SMR results from PsychENCODE and BrainMeta brain eQTLs
| Gene | topSNP | b_SMR | se_SMR | p_SMR | p_HEIDI | |
| PsychENCODE | PEX10 | rs12085089 | –0.0021 | 0.0011 | 0.0441 | 0.2550 |
| CDC25A | rs3731497 | 0.0013 | 0.0005 | 0.0168 | 0.8560 | |
| EGFR | rs74504435 | 0.0012 | 0.0004 | 0.0068 | 0.1970 | |
| DLD | rs35765154 | –0.0018 | 0.0007 | 0.0136 | 0.7330 | |
| LIG3 | rs2339122 | –0.0019 | 0.0009 | 0.0397 | 0.5720 | |
| TRIB3 | rs62191434 | –0.0011 | 0.0005 | 0.0339 | 0.7850 | |
| BrainMeta | PEX10 | rs12092052 | –0.0007 | 0.0003 | 0.0348 | 0.1240 |
| CDC25A | rs17647717 | 0.0005 | 0.0002 | 0.0134 | 0.9618 | |
| EGFR | rs74504435 | 0.0004 | 0.0002 | 0.0061 | 0.2023 | |
| DLD | rs8440 | –0.0003 | 0.0001 | 0.0162 | 0.9991 | |
| LIG3 | rs3135967 | –0.0003 | 0.0001 | 0.0293 | 0.9971 | |
| TRIB3 | rs62191440 | –0.0004 | 0.0002 | 0.0319 | 0.8966 |
In addition to the gene expression-level candidate targets, we also conducted a two-sample MR analysis by incorporating the pQTL of all proteins from brain tissues. In total, we selected 65 LD-independent pQTL instruments from 65 proteins. We then intersected these 65 potential proteins with 564 ferroptosis-related targets, and finally obtained 5 candidate proteins associated with AD, including CFL1, MGST1, GLS2, DLD, and EIF2AK4 (p < 0.05) (Fig. 2D). When overlapping the results from eQTLs and pQTLs derived from brain tissue, we identified two common genes, DLD and EIF2AK4 (Fig. 3B).
We also conducted similar explorations in blood tissues. Using cis-eQTL SNPs from the eQTLGen Consortium as genetic instruments, we identified 20 potential AD targets (SMR p < 0.05 and HEIDI test p > 0.05) (Fig. 2E). We observed that the candidate genes identified from brain and blood tissue screenings have a high degree of overlap. The overlapping genes between the two tissues include PEX10, CDC25A, PARP14, PARP11, CDH1, ACSF2, EGLN2, TRIB3, and ATF4 (Fig. 3C). We also identified 2 potential ferroptosis-related proteins using cis-pQTL SNPs from circulating 4,907 proteins levels, including BCAT2 and ECH1 (p < 0.05) (Fig. 2F). When comparing the results of eQTLs and pQTLs obtained from blood tissue, we found no overlapping genes (Fig. 3D). More details can be found in Supplementary Table 1.
PPI analysis and expression of the identified genes in different cell types of the human brain
We carried out a PPI analysis of the 30 genes obtained from the brain eQTLs results, using the STRING database (https://string-db.org/). The PPI network exhibited significantly more connections than anticipated (p < 0.001). The top 10 genes (SLC7A11, SLC40A1, SLC3A2, RPTOR, GABARAPL2, EGFR, ATM, ATF4, TXN, and TRIB3) were identified as potential hub genes, based on their Degree with CytoHubba (Supplementary Figure 2). We examined the expression patterns of these prioritized genes using single cell RNA sequencing data. Among the genes associated with AD, PEX10, CDC25A, EGFR, DLD, LIG3, and TRIB3, were observed to be widely expressed across various brain cell types at relatively high levels (Supplementary Figure 3).
Validation of gene targets by transcriptome and western blotting
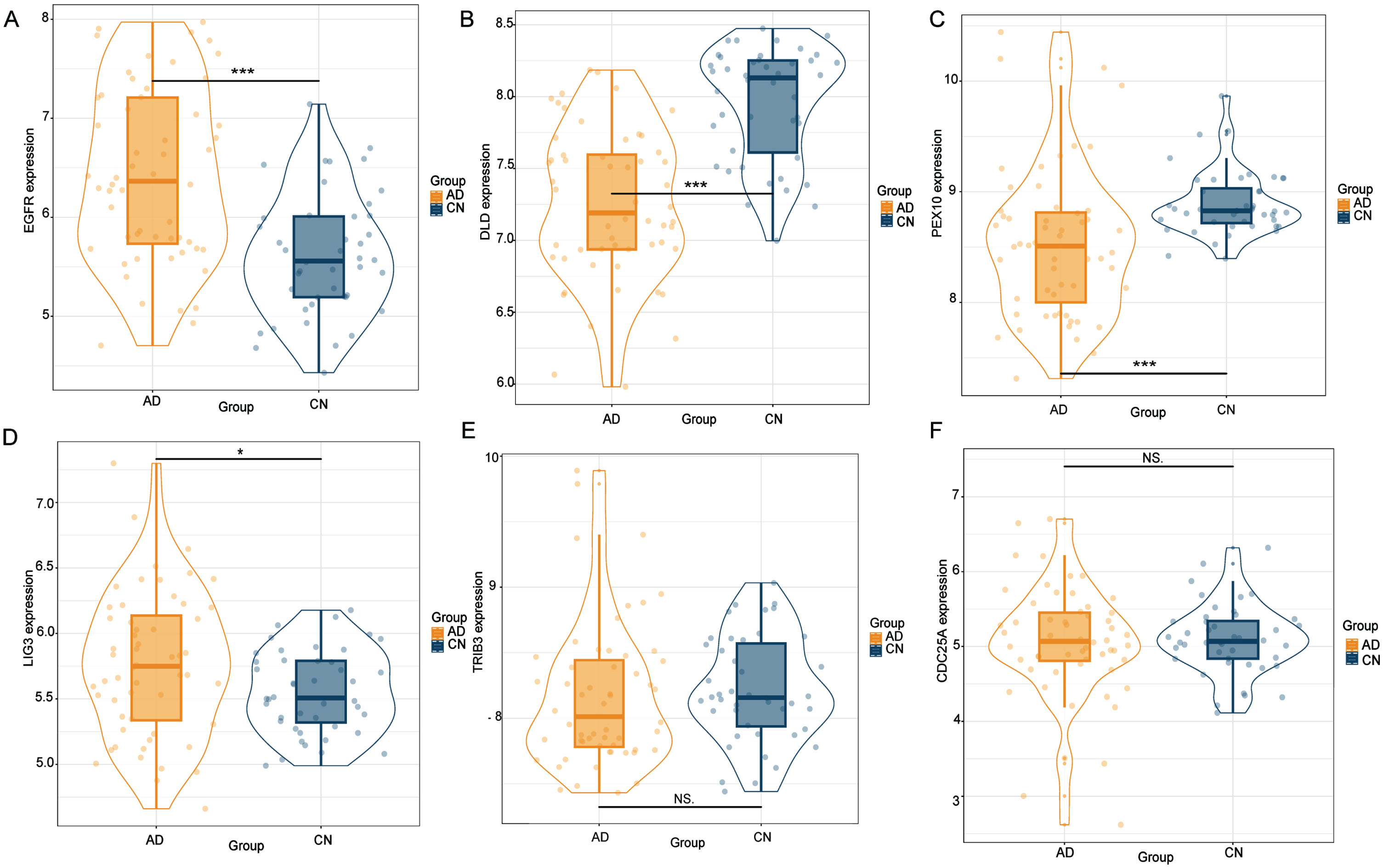
Using cis-eQTL SNPs from GTEx, PsychENCODE, and BrainMeta as genetic instruments, we identified six putative gene targets associated with AD. The expression levels of these genes were further validated by analyzing in external transcriptomic datasets. We found that EGFR, DLD, PEX10, and LIG3 were significantly expressed compared to the control group (p < 0.05), while TRIB3 and CDC25A did not appear to show statistical significance (Fig. 4). To enhance validation at the protein level, brain tissue was procured from 3xTg-AD mice, followed by western blotting experiment.
Fig. 4
The external transcriptome dataset corroborated the identification of six gene targets through SMR analysis. Notably, EGFR, DLD, PEX10, and LIG3 demonstrated significant expression differences relative to the control group (p < 0.05). Conversely, TRIB3 and CDC25A failed to reach statistical significance.
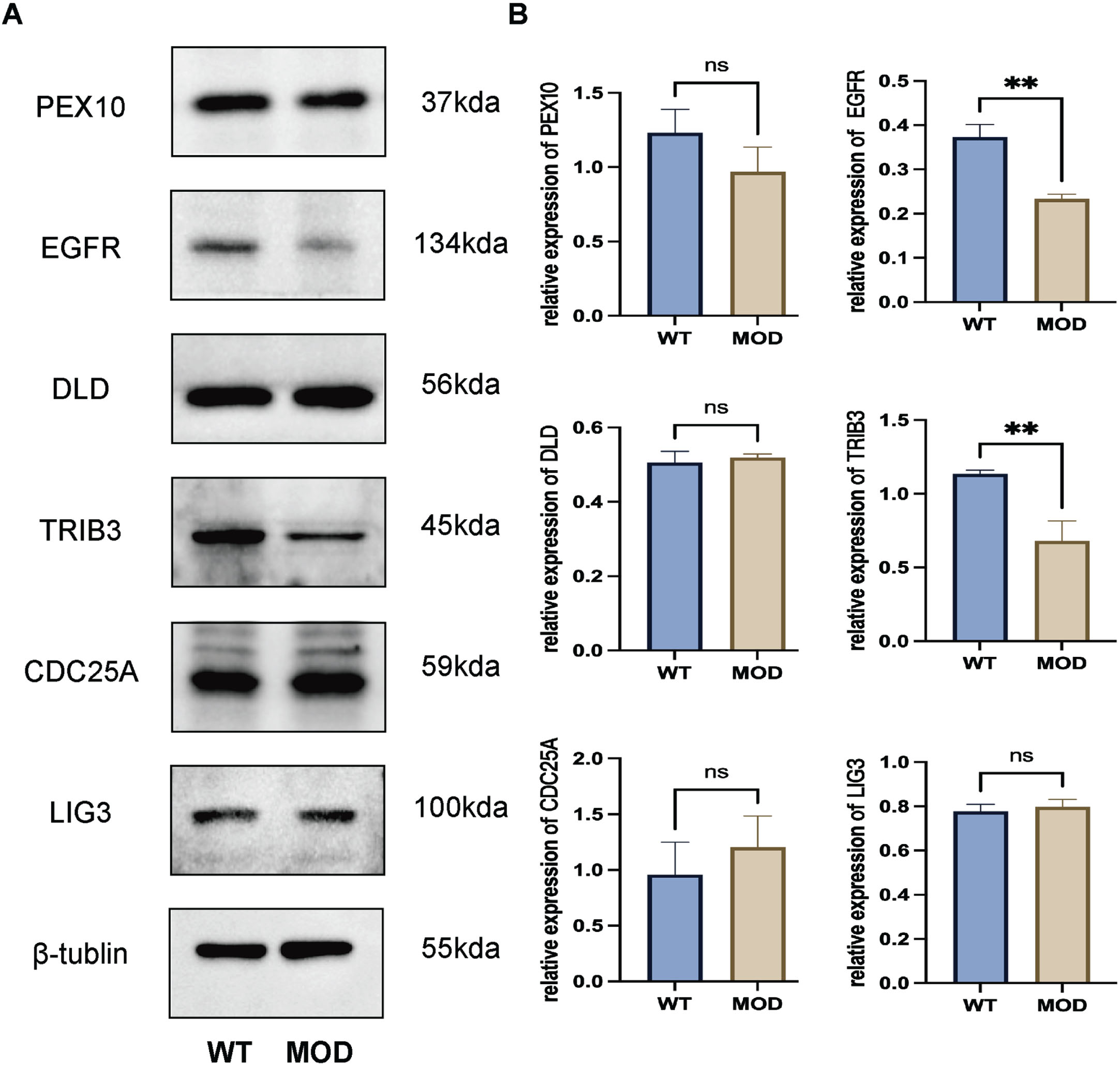
In the 3xTg-AD, western blotting demonstrated that both EGFR (p < 0.05) and TRIB3 (p < 0.05) were downregulated in the whole brain tissues compared with the control group (Fig. 5). Transcriptome and Western blotting analyses both revealed significant dysregulation of EGFR, underscoring its potential as a therapeutic target for AD.
Fig. 5
Validation of the gene targets by western blotting using 3xTg-AD mice. Both EGFR (p < 0.05) and TRIB3 (p < 0.05) were downregulated in the whole brain tissues compared with the control group. WT, wild type; MOD, model of 3xTg-AD mice.
DISCUSSION
Using the MR method, we have identified six ferroptosis-related gene targets associated with AD. Notably, EGFR showed significant dysregulation in external transcriptomic datasets and 3xTg-AD models, suggesting its potential role in regulating ferroptosis in AD. These findings suggest EGFR could serve as a promising target for therapeutic interventions aimed at modulating ferroptosis in AD treatment strategies.
The ferroptosis-related gene targets associated with AD
Existing research supports the involvement of the ferroptosis-related gene targets that we identified in the pathological process of AD. EGFR signaling has been linked to various cellular processes, such as neurodevelopment and neurodegenerative process.30 Recent research has demonstrated that EGFR signaling can influence ferroptosis by adjusting the expression of key genes involved in ferroptosis, such as GPX4 and SLC7A11.31 Some studies have indicated the potential involvement of TRIB3 in neurodegenerative disorders through the modulation of ferroptosis.32,33
The PEX10 encodes a peroxisomal membrane protein involved in peroxisome formation.34 Peroxisomal dysfunction is linked to neurodegenerative disorders such as AD, which shows impaired peroxisomal function and lipid metabolism.34,35 CDC25A regulates cell cycle and various cellular processes, which are interconnected with ferroptosis.36 LIG3, an enzyme crucial for DNA repair, has been linked to impaired DNA repair mechanisms in the process of ferroptosis in AD.37,38 Mitochondrial dysfunction has been associated with both ferroptosis and AD, indicating that DLD may indirectly contribute to the dysregulation observed in both conditions.39,40
The association of EGFR with ferroptosis
EGFR activation leads to downstream signaling through pathways such as the Ras-MAPK pathway, PI3K-AKT pathway, and STAT3 pathway. These pathways regulate various cellular processes, including proliferation, survival, and differentiation.41 However, they also influence the expression of genes involved in iron metabolism and oxidative stress, thereby indirectly affecting ferroptosis.42,43 EGFR signaling can modulate iron uptake and utilization within cells. For instance, EGFR activation can increase the expression of divalent metal transporter 1 (DMT1), which facilitates iron uptake.44 Additionally, EGFR signaling can affect the activity of enzymes involved in iron processing, such as ferritin and hephaestin, influencing intracellular iron levels.43
The accumulation of ROS due to increased iron availability can lead to lipid peroxidation, a hallmark of ferroptosis. EGFR signaling can enhance ROS production through various mechanisms, including upregulation of NADPH oxidases (NOXs).45,46 This increased oxidative stress can damage cellular membranes and organelles, promoting ferroptotic cell death.
Interestingly, therapeutic agents targeting EGFR, such as tyrosine kinase inhibitors (TKIs), can also modulate ferroptosis by altering iron metabolism and oxidative stress.47,48 For example, erlotinib, an EGFR TKI, has been shown to induce ferroptosis by disrupting iron uptake and enhancing lipid peroxidation. The interaction between EGFR and ferroptosis represents a promising area of research with potential clinical applications.49 Further elucidation of the underlying molecular mechanisms will facilitate the development of targeted therapeutic strategies aimed at leveraging ferroptosis for AD treatment.
EGFR serving as a therapeutic target for AD
In recent times, there’s been growing interest in repurposing existing anti-cancer drugs that target the EGFR for AD research, given the link between EGFR overactivity and several neurodegenerative conditions, including AD.50,51 Research has highlighted that blocking EGFR can reduce the activity of reactive astrocytes, lessen Aβ toxicity and inflammation, and encourage the regrowth of axons.52 Furthermore, other investigations have noted positive changes in behavior and cognitive functions, alongside decreased amyloid formation and enhanced autophagy, as a result of EGFR blockade in different animal models of AD.53
Mansour and his colleagues conducted a detailed review of past research, pinpointing overlooked neuroprotective routes and underscoring the importance of exploring EGFR inhibitors as possible treatments for AD.53 They found that Aβ peptides intensify oxidative stress mediated by NADPH oxidase (NOX).53,54 Activating EGFR leads to an increase in ROS via the PI3K pathway, which then sets off the Ras-Rac1 sequence, culminating in NOX activation.55 Interestingly, NOX appears to be closely associated with AD, suggesting it could serve as a valuable indicator for developing new medications.
Additionally, another EGFR-driven process that contributes to oxidative stress and axon loss involves the ZNRF1–AKT–GSK3β–CRMP2 pathway.56 ZNRF1, a ubiquitin ligase, is prevalent in neurons throughout the central nervous system (CNS). When neurons are damaged, ZNRF1 breaks down AKT, blocks GSK-3β, and disrupts microtubule stability, leading to axonal damage in the Wallerian model.56 More research is needed to understand the relationship between NOX-mediated oxidative stress, EGFR, and ZNRF1 in live models of AD.
Limitations
Our study acknowledges several limitations. Initially, despite a notable correlation between eQTLs in blood and brain, these observations are confined to genes expressed in both tissues and possessing eQTLs in them. While these studies can be beneficial for estimating the long-term effects of exposure beyond the scope of randomized controlled trials, they fail to directly reveal the impact of short-term drug treatments on disease risk. Secondly, the complete removal of confounding bias and/or horizontal pleiotropy remains a formidable challenge. Lastly, the discovery of additional candidate genes in the 3xTg-AD mice does not negate their association with AD. Additional laboratory and clinical data are necessary to substantiate these findings.
Conclusions
This study provides genetic evidence supporting the potential therapeutic benefits of targeting the six druggable genes for AD treatment, especially for EGFR (validated by transcriptome and 3xTg-AD), which could be useful for prioritizing AD drug development in the field of ferroptosis.
AUTHOR CONTRIBUTIONS
Ying Wang (Conceptualization; Data curation; Formal analysis; Methodology; Software; Visualization; Writing – original draft; Writing – review & editing); Xinhua Song (Methodology; Validation); Rui Wang (Conceptualization; Methodology); Xinzi Xu (Writing – review & editing); Yaming Du (Writing – review & editing); Guohua Chen (Funding acquisition; Project administration; Supervision); Junhua Mei (Funding acquisition; Project administration; Supervision).
ACKNOWLEDGMENTS
The authors are very grateful for the data support provided by the FerrDb, GTEx, PsychENCODE, BrainMeta, ROSMAP, eQTLGen Consortium, GEO, SMR and IEU Open GWAS Project.
FUNDING
The study was conducted without any financial support from external funding agencies.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
DATA AVAILABILITY
The datasets generated during and/or analyzed during the current study are available in the FerrDb repository (http://www.zhounan.org/ferrdb/), MRC IEU Open GWAS Project (https://gwas.mrcieu.ac.uk/), eQTLGen Consortium (https://www.eqtlgen.org/), and SMR website (https://yanglab.westlake.edu.cn/software/smr/#eQTLsummarydata).
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/ADR-240062.
REFERENCES
1. | Vejandla B , Savani S , Appalaneni R , et al. Alzheimer’s disease: the past, present, and future of a globally progressive disease. Cureus (2024) ; 16: : e51705. |
2. | Chen YG . Research progress in the pathogenesis of Alzheimer’s disease. Chin Med J (Engl) (2018) ; 131: : 1618–1624. |
3. | Frozza RL , Lourenco MV and De Felice FG. Challenges for Alzheimer’s disease therapy: insights from novel mechanisms beyond memory defects. Front Neurosci (2018) ; 12: : 37. |
4. | Jakaria M , Belaidi AA , Bush AI , et al. Ferroptosis as a mechanism of neurodegeneration in Alzheimer’s disease. J Neurochem (2021) ; 159: : 804–825. |
5. | Weiland A , Wang Y , Wu W , et al. Ferroptosis and its role in diverse brain diseases. Mol Neurobiol (2019) ; 56: : 4880–4893. |
6. | Lane DJR , Ayton S and Bush AI. Iron and Alzheimer’s disease: an update on emerging mechanisms. J Alzheimers Dis (2018) ; 64: (s1), S379–S395. |
7. | Ayton S , Wang Y , Diouf I , et al. Brain iron is associated with accelerated cognitive decline in people with Alzheimer pathology. Mol Psychiatry (2019) ; 25: : 2932–2941. |
8. | Hambright WS , Fonseca RS , Chen L , et al. Ablation of ferroptosis regulator glutathione peroxidase 4 in forebrain neurons promotes cognitive impairment and neurodegeneration. Redox Biol (2017) ; 12: : 8–17. |
9. | Zhao D , Yang K , Guo H , et al. Mechanisms of ferroptosis in Alzheimer’s disease and therapeutic effects of natural plant products: A review. Biomed Pharmacother (2023) ; 164: : 114312. |
10. | Su WM , Gu XJ , Dou M , et al. Systematic druggable genome-wide Mendelian randomisation identifies therapeutic targets for Alzheimer’s disease. J Neurol Neurosurg Psychiatry (2023) ; 94: : 954–961. |
11. | Vitalakumar D , Sharma A and Flora SJS. Ferroptosis: A potential therapeutic target for neurodegenerative diseases. J Biochem Mol Toxicol (2021) ; 35: : e22830. |
12. | Ma H , Dong Y , Chu Y , et al. The mechanisms of ferroptosis and its role in Alzheimer’s disease. Front Mol Biosci (2022) ; 9: : 965064. |
13. | Plascencia-Villa G and Perry G. Preventive and therapeutic strategies in Alzheimer’s disease: focus on oxidative stress, redox metals, and ferroptosis. Antioxid Redox Signal (2021) ; 34: : 591–610. |
14. | Ghoussaini M , Nelson MR and Dunham I. Future prospects for human genetics and genomics in drug discovery. Curr Opin Struct Biol (2023) ; 80: : 102568. |
15. | Gaziano L , Giambartolomei C , Pereira AC , et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med (2021) ; 27: : 668–676. |
16. | Schmidt AF , Finan C , Gordillo-Marañón M , et al. Genetic drug target validation using Mendelian randomisation. Nat Commun (2020) ; 11: : 3255. |
17. | Zeng Y , Cao S , Pang K , et al. Causal association between sepsis and neurodegenerative diseases: a bidirectional two-sample Mendelian randomization study. J Alzheimers Dis (2024) ; 97: : 229–237. |
18. | Zeng Y , Cao S and Yang H . No causal relationship between thyroid function and Parkinson’s disease: A bidirectional Mendelian randomization study. Neurol Sci (2024) ; 45: : 1481–1487. |
19. | Zeng Y , Guo R , Cao S , et al. CSF N-acylethanolamine acid amidase level and Parkinson’s disease risk: A Mendelian randomization study. Parkinsonism Relat Disord (2024) ; 123: : 106953. |
20. | Wang D , Liu S , Warrell J , et al. Comprehensive functional genomic resource and integrative model for the human brain. Science (2018) ; 362: : eaat8464. |
21. | Auton A , Brooks L D , Durbin RM , et al. A global reference for human genetic variation. Nature (2015) ; 526: : 68–74. |
22. | Wingo A P , Liu Y , Gerasimov ES , et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nate Genet (2021) ; 53: : 143–146. |
23. | Zhu Z , Zhang F , Hu H , et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet (2016) ; 48: : 481–487. |
24. | Chauquet S , Zhu Z , O’Donovan MC , et al. Association of antihypertensive drug target genes with psychiatric disorders: a Mendelian randomization study. JAMA Psychiatry (2021) ; 78: : 623–631. |
25. | Hemani G , Zheng J , Elsworth B , et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife (2018) ; 7: : e34408. |
26. | Shannon P , Markiel A , Ozier O , et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res (2003) ; 13: : 2498–504. |
27. | Polioudakis D , de la Torre-Ubieta L , Langerman J , et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron (2019) ; 103: : 785–801.e8. |
28. | Oddo S , Caccamo A , Kitazawa M , et al. Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer’s disease. Neurobiol Aging (2003) ; 24: : 1063–1070. |
29. | McKay EC , Beck JS , Khoo SK , et al. Peri-infarct upregulation of the oxytocin receptor in vascular dementia. J Neuropathol Exp Neurol (2019) ; 78: : 436–452. |
30. | Hao X , Wang H , Cui F , et al. Reduction of SLC7A11 and GPX4 Contributing to Ferroptosis in Sperm from Asthenozoospermia Individuals. Reprod Sci (2023) ; 30: : 247–257. |
31. | Zhang G , Fang Y , Li X , et al. Ferroptosis: A novel therapeutic strategy and mechanism of action in glioma. Front Oncol (2022) ; 12: : 947530. |
32. | Yan Hf , Zou T , Tuo Qz , et al. Ferroptosis: mechanisms and links with diseases. Sig Transduct Target Ther (2021) ; 6: : 49. |
33. | Stefanovska B , André F and Fromigué O . Tribbles pseudokinase 3 regulation and contribution to cancer. Cancers (2021) ; 13: : 1822. |
34. | Uzor NE , McCullough LD and Tsvetkov AS. Peroxisomal dysfunction in neurological diseases and brain aging. Front Cell Neurosci (2020) ; 14, 44. |
35. | Steinberg SJ , Snowden A , Braverman NE , et al. A PEX10 defect in a patient with no detectable defect in peroxisome assembly or metabolism in cultured fibroblasts. J Inherit Metab Dis (2009) ; 32: : 109–119. |
36. | Wang P , Zou F , Zhang X , et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res (2009) ; 69: : 8157–8165. |
37. | Sallmyr A , Rashid I , Bhandari SK , et al. Human DNA ligases in replication and repair. DNA Repair (2020) ; 93: : 102908. |
38. | Schumacher B , Pothof J , Vijg J , et al. The central role of DNA damage in the ageing process. Nature (2021) ; 592: : 695–703. |
39. | Tobore TO . On the central role of mitochondria dysfunction and oxidative stress in Alzheimer’s disease. Neurol Sci (2019) ; 40: : 1527–1540. |
40. | Bhatti JS , Bhatti GK and Reddy PH. Mitochondrial dysfunction and oxidative stress in metabolic disorders - A step towards mitochondria based therapeutic strategies. Biochim Biophys Acta Mol Basis Dis (2017) ; 1863: : 1066–1077. |
41. | Bahar ME , Kim HJ and Kim DR. Targeting the RAS/RAF/MAPK pathway for cancer therapy: from mechanism to clinical studies. Signal Transduct Target Ther (2023) ; 8: : 455. |
42. | Tang D , Chen X , Kang R , et al. Ferroptosis: molecular mechanisms and health implications. Cell Res (2021) ; 31: : 107–125. |
43. | Wang B , Zhang J , Song F , et al. EGFR regulates iron homeostasis to promote cancer growth through redistribution of transferrin receptor 1. Cancer Lett (2016) ; 381: : 331–340. |
44. | Brown RAM , Richardson KL , Kabir TD , et al. Altered iron metabolism and impact in cancer biology, metastasis, and immunology. Front Oncol (2020) ; 10: : 476. |
45. | Zeng W , Long X , Liu P S , et al. The interplay of oncogenic signaling, oxidative stress and ferroptosis in cancer. Int J Cancer (2023) ; 153: : 918–931. |
46. | Ye L , Wen X , Qin J , et al. Metabolism-regulated ferroptosis in cancer progression and therapy. Cell Death Dis (2024) ; 15: : 196. |
47. | Zhou Q , Meng Y , Li D , et al. Ferroptosis in cancer: From molecular mechanisms to therapeutic strategies. Signal Transduct Target Ther (2024) ; 9: : 55. |
48. | Zhao L , Zhou X , Xie F , et al. Ferroptosis in cancer and cancer immunotherapy. Cancer Commun (Lond) (2022) ; 42: : 88–116. |
49. | Koeberle SC , Kipp AP , Stuppner H , et al. Ferroptosis-modulating small molecules for targeting drug-resistant cancer: Challenges and opportunities in manipulating redox signaling. Med Res Rev (2023) ; 43: : 614–682. |
50. | Lorenzi M , Altmann A , Gutman B , et al. Susceptibility of brain atrophy to TRIB3 in Alzheimer’s disease, evidence from functional prioritization in imaging genetics. Proc Natl Acad Sci U S A (2018) ; 115: : 3162–3167. |
51. | Choi HJ , Jeong YJ , Kim J , et al. EGFR is a potential dual molecular target for cancer and Alzheimer’s disease. Front Pharmacol (2023) ; 14: : 1238639. |
52. | Chen YJ , Hsu CC , Shiao YJ , et al. Anti-inflammatory effect of afatinib (an EGFR-TKI) on OGD-induced neuroinflammation. Sci Rep (2019) ; 9: : 2516. |
53. | Mansour HM , Fawzy HM , El-Khatib AS , et al. Lapatinib ditosylate rescues memory impairment in D-galactose/ovariectomized rats: Potential repositioning of an anti-cancer drug for the treatment of Alzheimer’s disease. Exp Neurol (2021) ; 341: : 113697. |
54. | Tarafdar A and Pula G. The role of NADPH oxidases and oxidative stress in neurodegenerative disorders. Int J Mol Sci (2018) ; 19: : 3824. |
55. | Paletta-Silva R , Rocco-Machado N and Meyer-Fernandes JR. NADPH oxidase biology and the regulation of tyrosine kinase receptor signaling and cancer drug cytotoxicity. Int J Mol Sci (2013) ; 14: : 3683–3704. |
56. | Wakatsuki S , Furuno A , Ohshima M , et al. Oxidative stress-dependent phosphorylation activates ZNRF1 to induce neuronal/axonal degeneration. J Cell Biol (2015) ; 211: : 881–896. |




