Scholarly triage: Advances in manuscript submission using text analytics techniques
Abstract
The drastic increase in the volume of submissions for inclusion in scholarly journals offers new challenges for scholarly publishers. These include sorting through thousands of new manuscripts to prioritize those most likely to be published first and detecting dubious research. Although recent advances in peer review and editorial management platforms have advanced processes to help alleviate the problem, new solutions to prioritize high-value papers – and to flag suspect ones – are emerging. Using text analytics methods grounded in natural language processing (NLP) and other techniques to augment the submission review process can include leveraging taxonomy-based indexing terms to match manuscripts to appropriate reviewers and editors, preventing fraud by detecting machine-generated entries, screening for irreproducible research practices, and predicting the likelihood of acceptance by examining non-content factors.
1.Introduction
According to the 2015 STM Report, scholarly publishing in the science, technology, and medicine (STM) fields generated about 2.5 million articles in approximately 28,100 peer-reviewed journals [6, p. 6]. These figures neither account for other content produced by scholarly publishers (conference proceedings, books, standards, educational content, and other materials) nor do they describe articles produced in non-STM fields such as the arts and humanities.
Nevertheless, these statistics illustrate the massive growth in the volume of published scholarly articles, which is expected to continue at a rate of 3%–3.5% per year [6, p. 6]. This massive quantity of content exerts pressure on an industry already beleaguered by transitions to digital delivery formats, declining revenue streams, and an endangered model challenged by the rise of Open Access (OA) journals (which are neither excluded from the figures cited above nor immune to the problems in the greater industry), websites providing free access to pirated content, and shrinking library subscriptions.
2.Emerging technologies
To address the ever-increasing volume of manuscript submissions, new applications are emerging to help publishers analyze and sort incoming papers. These include leveraging semantic tagging to assign manuscripts to peer reviewers and editors, detecting fraudulent machine-generated papers and irreproducible research, and using statistical methodologies to identify the papers most likely to be published as priorities for the review process.
2.1.Using indexing terms to facilitate peer review
The adoption of peer review management systems (e.g., Thomson Reuters’ Scholar One Manuscripts, eJournal Press’ ePress, Aries System’s Editorial Manager, etc.) to streamline the workload generated by, and reduce the cost of, reviewing large quantities of manuscripts is widespread. In addition, the increasing adoption of semantic enrichment (usually tagging journal articles using one or more taxonomies or other controlled vocabularies) provides an opportunity to augment the utility of peer review systems to include semantic tagging at the point of submission (as opposed to tagging finalized versions of content at the end of the article pipeline) and using the terms so applied to facilitate the assignment of peer reviewers (and, later, editors) with the appropriate expertise [6, p. 50]. Figure 1 shows one example of an online form with automated semantic tagging; this implementation features multiple ways for an editor to curate the automatically suggested indexing terms from a taxonomy.
This is especially useful for online submission tools, which are easily integrated with indexing engines. The resulting applications can include options for the submitting author or receiving publishing personnel to review the terms assigned and then curate them (adding missing terms, removing inappropriate terms) before they are ingested into the peer review management system.
Fig. 1.
An example of semantic enrichment at the point of submission integrated with a web-based manuscript submission system. This implementation allows the author to review and curate the terms assigned to the manuscript.
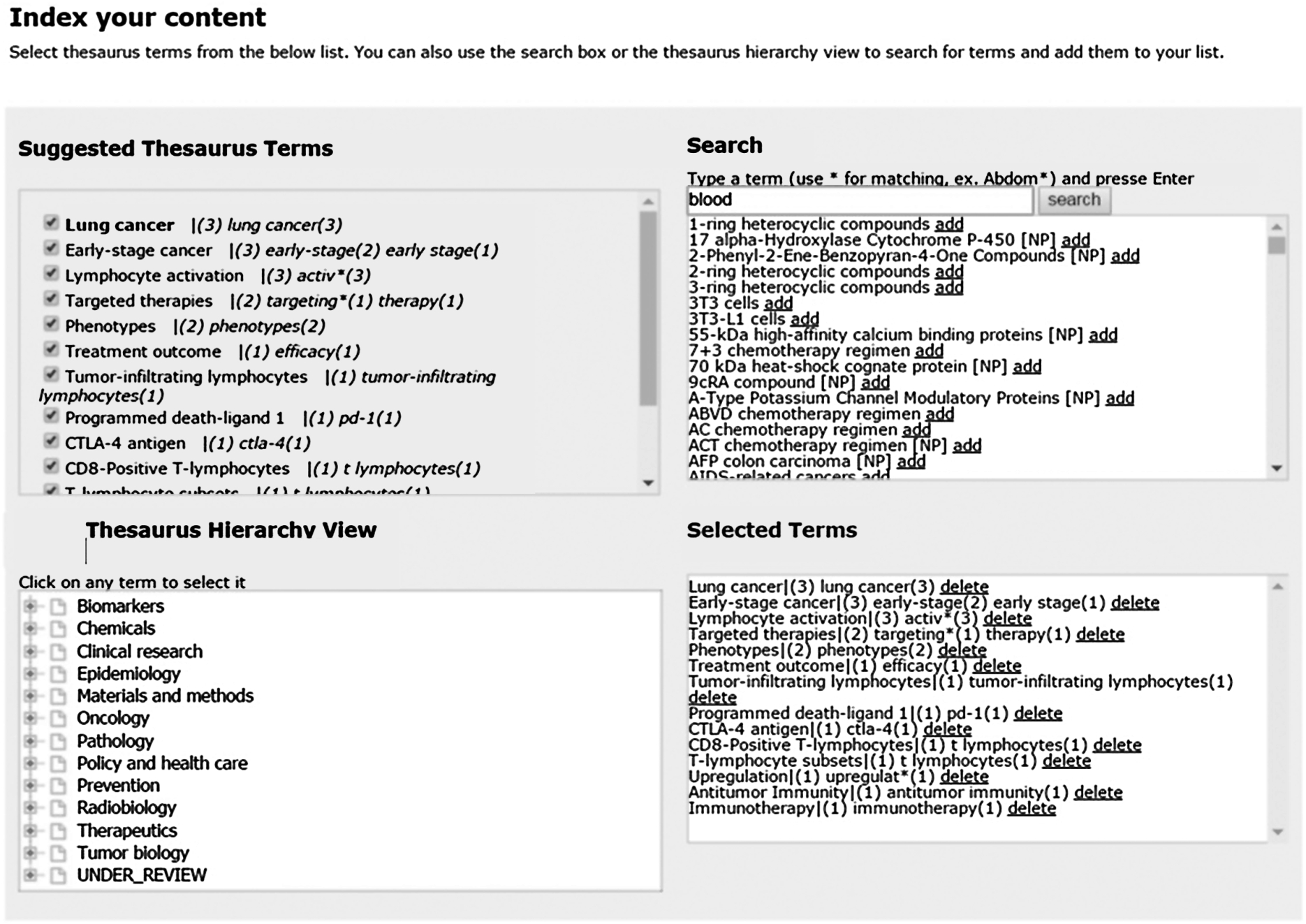
In order to match tagged incoming manuscripts to appropriate editors and/or reviewers, it is necessary to classify editors and reviewers using the same vocabulary. This can either be accomplished automatically, based on content written or reviewed in the past, or manually, by allowing editors and reviewers to self-select their areas of expertise.
Streamlining the peer review process addresses two central pain points for publishers: (1) the time and cost involved in the peer review process (on average, peer review takes between thirty days to six months or more); and (2) the average cost to a publisher to manage the peer review of a single paper (such costs average about $250) [5].
2.2.Detection of fraudulent machine-generated manuscripts
SCIgen is an online application that uses context-free grammar to generate “spoof” or nonsense papers based on a few user inputs, including references, examples, and an abstract. Developed at MIT in 2005, SCIgen’s stated purpose is for “amusement” or to “auto-generate submissions to conferences that you suspect might have very low submission standards.”11
However, SCIgen was quickly adopted and used to submit papers to journals, some of which have been accepted. Prominently, in 2014 Nature News reported that Springer and IEEE had discovered some one hundred and twenty SCIgen papers and removed them from circulation. This has, understandably, caused some consternation among publishers [4].
Fig. 2.
Sample SCIgen paper that was accepted as a “non-reviewed paper” at a conference. See full paper at: https://pdos.csail.mit.edu/archive/scigen/rooter.pdf.
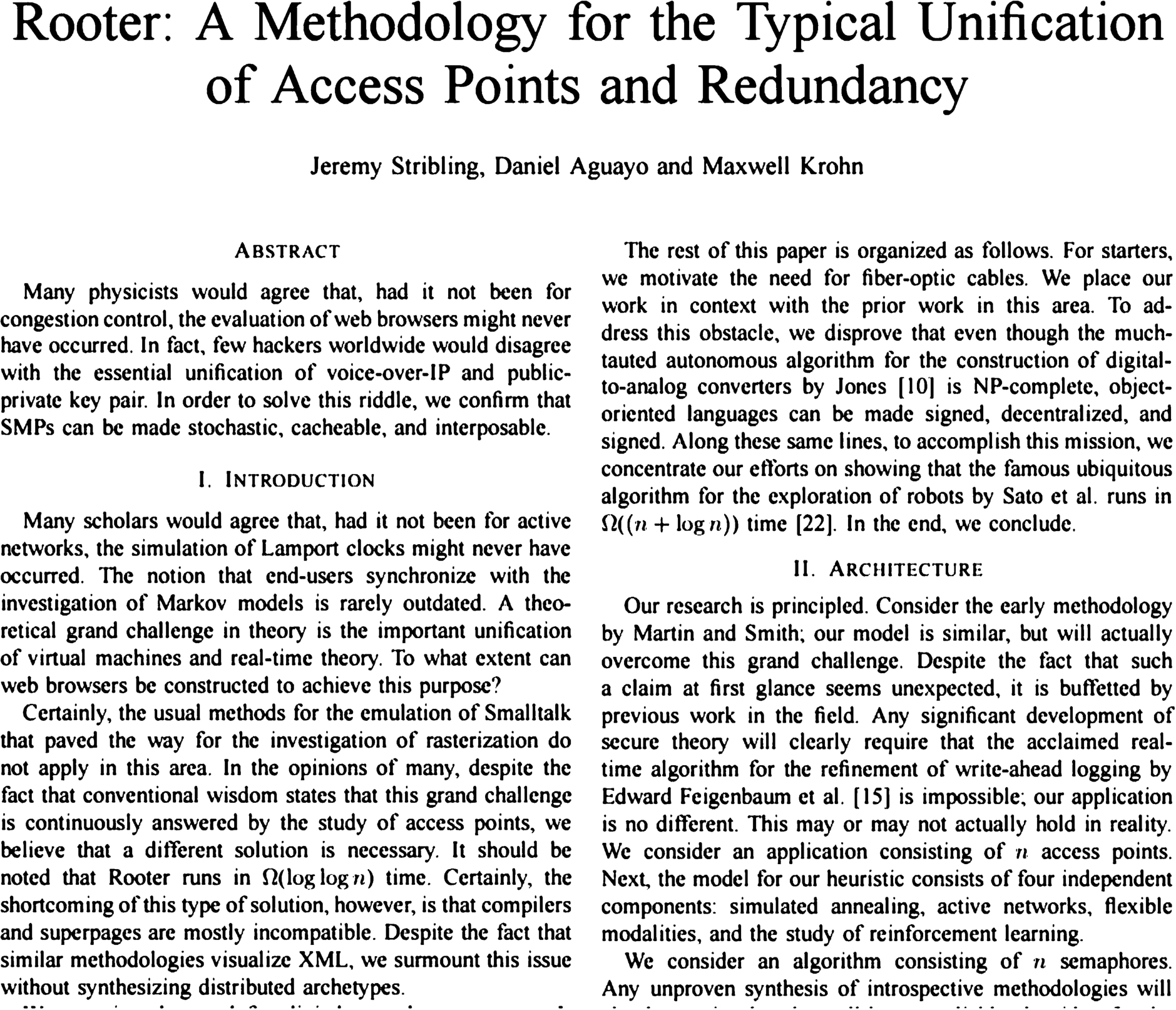
Accordingly, several applications to detect SCIgen papers at the point of submission have been developed and are available for integration into manuscript submission systems [4]. Some such systems feature reverse engineering of the SCIgen algorithms, which are made freely-available on the SCIgen site (see footnote 1). Others, such as the technique developed by Access Innovations, use Bayesian inferential techniques to model known SCIgen papers and compare them to a given manuscript to assign a probability of SCIgen origin. Figure 2 is the first page of a SCIgen-generated paper that was accepted as a “non-reviewed paper” at a conference.
2.3.Detecting irreproducible research
The problem of reproducibility, especially in biomedical research, is a topic of emerging interest in the scholarly community; this is evident by the number of sessions on the topic of reproducibility recent at scholarly publishing conferences (e.g., NFAIS 2017, STM 2017) as well as a number of scholarly papers (e.g., Freedman 2015) [2].
In the field of medical research in particular, the continued use of known problematic cell lines causes irreproducible results; this translates to a great deal of wasted time and money. These cell lines, used to carry out genetic tests and other experiments, are known to be misidentified or cross-contaminated with other cell lines. The International Cell Line Authentication Committee (ICLAC) publishes a list of known problematic cell lines (available at http://iclac.org/databases/cross-contaminations/), but few researchers check their materials [1].
The Global Biological Standards Institute (GBSI), a non-profit organization promoting good research practices, is partnering with ICLAC and Access Innovations to produce a tool for authors to check whether the cell lines used in their research are on the ICLAC list of known problematic cell lines; the same process will be available to publishers to scan incoming manuscripts and previously published papers.
2.4.Prioritizing likely-to-be-published papers
One solution to processing the large volume of submitted manuscripts involves identifying those papers most likely to be published and prioritizing them for review. This approach does not seek to dismiss any potentially-publishable papers out of hand; rather, the goal is to prioritize the highest-value papers as first to be processed (sent to editors and/or peer reviewers, etc.).
Without reading the content, factors (essentially the metadata) associated with a paper that can be analyzed include number of authors, country of origin (usually of the corresponding author), topic (based on some taxonomic indexing), length, affiliation, and other measurable data points.
Given the metadata for a large corpus of accepted and rejected papers (from a particular publisher or across a research area) and using statistical analysis, it is possible to produce analytics to predict the “fate” of a given manuscript; that is: the likelihood that it will be published, expressed (usually) as a percentage.
For example, in a 2008 study published in the Journal of Bone and Joint Surgery, the authors found that the country of origin and number of prior publications in other frequently-cited journals in orthopedics correlated with the likelihood of acceptance and publication [3].
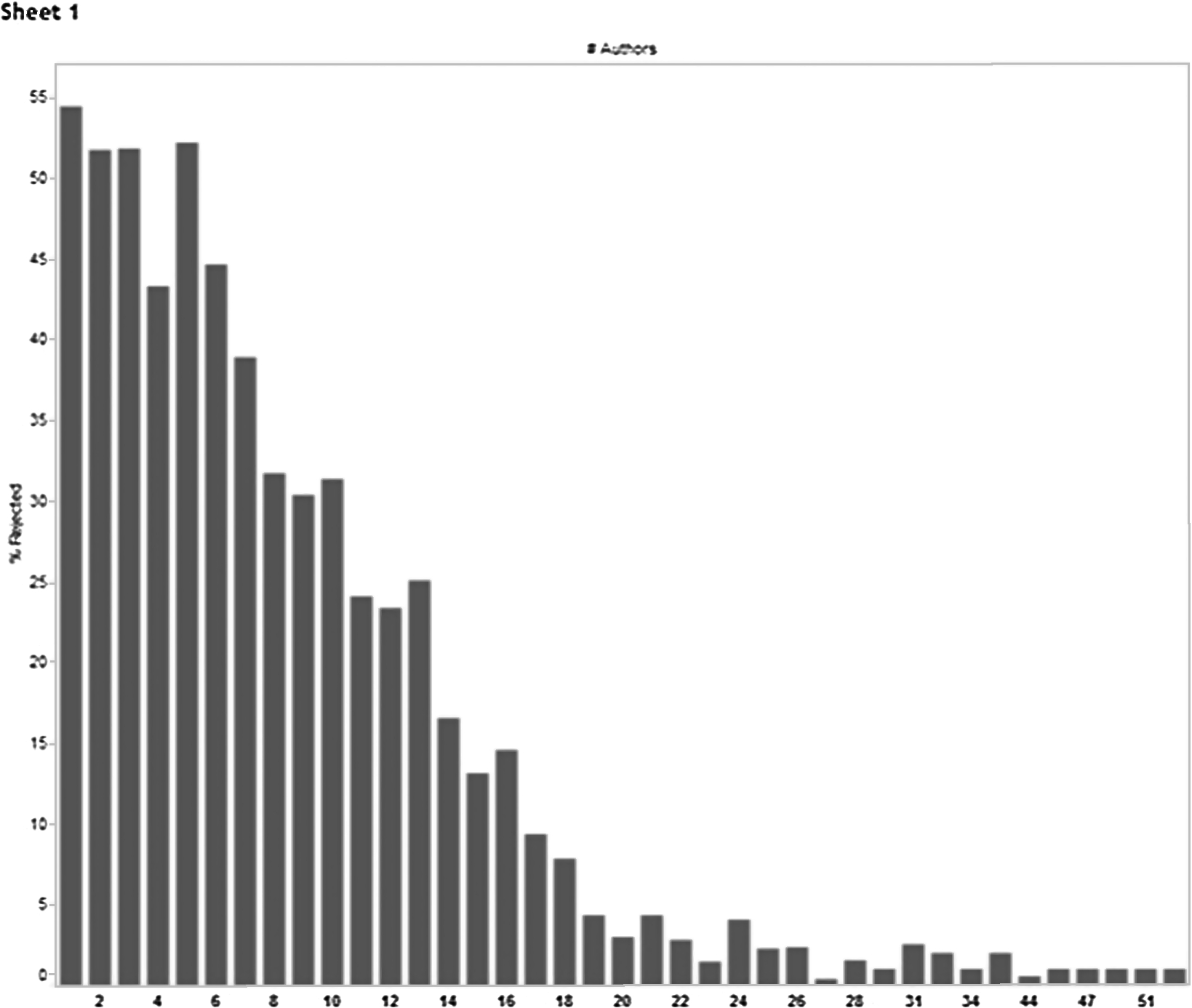
In a separate 2012 study with a large Open Access publisher, Access Innovations found that, for the corpus in question, the number of authors listed for an article was inversely correlated with the rejection rate. Figure 3 is a graph illustrating the inverse correlation between acceptance rate and number of authors from this study.
Similar discernable patterns based on data harvested from accepted and rejected papers – which, nominally, is based solely on the research itself – can be inferred from any corpus of sufficient size.
Fig. 3.
Inverse correlation of the number of authors with manuscript rejection rate from a 2012 study.
3.Conclusion
If, as projected, the growth in scholarly publishing continues apace, technologies to deal with the large volumes of content submitted to publishers will continue to emerge. As processing, categorizing, and relating very large content sets are familiar problems in the information industry, the newest advances in this space are driven by information professionals and professional services organizations – in concert with the scholarly publishers they serve – versed in the latest advances in language processing, document categorization and retrieval, and semantic technologies.
4.About the author
Bob Kasenchak is a taxonomist and Director of Business Development at Access Innovations, Inc. He has led taxonomy development and other projects for JSTOR, McGraw-Hill, Wolters Kluwer, ASCE, and AAAS, among others. Kasenchak attended St. John’s College, the New England Conservatory of Music and the University of Texas at Austin.
Notes
1 “SCIgen – An Automatic CS Paper Generator,” retrieved on April 30, 2017 from https://pdos.csail.mit.edu/archive/scigen/.
References
[1] | A. Capes-Davis, G. Theodosopoulos, I. Atkin, H.G. Drexler, A. Kohara, R.F. MacLeod, J.R. Masters, Y. Nakamura, Y.A. Reid, R.R. Reddel et al., Check your cultures! A list of cross-contaminated or misidentified cell lines, International Journal of Cancer 127: (1) ((2010) ), 1–8. doi:10.1002/ijc.25242. |
[2] | L.P. Freedman, I.M. Cockburn and T.S. Simcoe, The economics of reproducibility in preclinical research, PLOS Biology 13: (6) ((2015) ), e1002165. doi:10.1371/journal.pbio.1002165. |
[3] | K. Okike, M.S. Kocher, C.T. Mehlman, J.D. Heckman and M. Bhandari, Nonscientific factors associated with acceptance for publication in The Journal of Bone and Joint Surgery (American Volume), The Journal of Bone and Joint Surgery (American Volume) 90: (11) ((2008) ), 2432–2437. doi:10.2106/JBJS.G.01687. |
[4] | R. Van Noorden, Publishers withdraw more than 120 gibberish papers, Nature News, retrieved on April 30, 2017 from http://www.nature.com/news/publishers-withdraw-more-than-120-gibberish-papers-1.14763. |
[5] | J. Wallace, PEER project: Final report, PEER, 2012, retrieved on April 30, 2017 from www.peerproject.eu; see http://www.peerproject.eu/fileadmin/media/reports/20120618_PEER_Final_public_report_D9-13.pdf. |
[6] | M. Ware and M. Mabe, The STM Report, 4th edn, (2015) , retrieved on April 30, 2017 from http://www.stm-assoc.org/2015_02_20_STM_Report_2015.pdf. |



