FAIR4PGHD: A framework for FAIR implementation over PGHD
Abstract
Patient Generated Health Data (PGHD) are being considered for integration with health facilities, however little is known about how such data can be made machine-actionable in a way that meets FAIR guidelines. This article proposes a 5-stage framework that can be used to achieve this.
1.Introduction
Electronic health records are a type of scientific data, and like most scientific data, implementing the guiding principles of FAIR (Findable, Accessible, Interoperable, Reusable) [18] for data management and stewardship becomes necessary because it enhances the public and appropriate use of this data (Fig. 1). Studies on these exist [14,15] and have also brought forth opportunities to explore other dimensions and needs of this data type. So, FAIRifying patient health records is investigated [15,16].
However, this does not usually include patient generated health data (PGHD). The implication is that key context – like onset of chronic diseases, disease progression, ailment deterioration or recovery or improvement or pandemic-onset are not immediately seen until a clinic visit or when patients get (terribly) sick. Patient generated health data (PGHD) is health data that is collected by a patient (or their authorized representative or family) outside the clinic setting and can be used by them or clinicians for their health management [12].
PGHD are collected using many mediums including but not limited to mobile health application (mHealth) apps and wearables. Research has been done to integrate PGHD with electronic health records [7,8]. Additionally, information models that simplify and broaden the scope of data exchange are studied [1,10]. Evidence also points to the use of current health IT standards for documenting and sharing PGHD with clinicians [2]. The creation of wearable metadata has also been examined in certain research [5], although this is not general and does not take into account the variety of PGHD sources.
GO FAIR identify seven (7) steps involved in FAIRification as follows: (i) retrieving raw (non-FAIR) data, (ii) analyzing the retrieved data, (iii) semantic model definition, (iv) making data linkable, (v) assigning (custom) license, (vi) defining the metadata for the dataset, and (vii) deploying or publishing the FAIR data resource [3]. However, because health data requires some key consideration viz ethical, legal and privacy concerns [13], provides an enhanced FAIRification guideline for health data research (Fig. 2).
Expanding on this, van Reisen, et. Al [15], demonstrate a design for electronic health record in facilities, which largely also adhere to FAIR guidelines but also complies with health data sensitivities and regulations (NGDPR and GDPR) [9,16]. For facility-based health records, diverse deployment techniques and technologies have been created to comply with FAIR. Two (2) approaches were demonstrated using CEDAR technology (for individual datasets) and bespoke Excel2RDF format – for bulk datasets [15]. The Center for Expanded Data Annotation and Retrieval (CEDAR) technology was established in 2014 to create a computational ecosystem for the development, evaluation, use, and refinement of biomedical metadata [4]. It is important to state that FAIR concept is not the same with FHIR – which stands for Fast Health Interoperability Resources (FHIR), one of the recent standards to enhance interoperability in health systems employing lightweight web services [11]. However, in applying FAIR concept on health data, FHIR standards can be employed (see Fig. 3).
Fig. 3.
PGHD-FAIR framework using localised CEDAR.
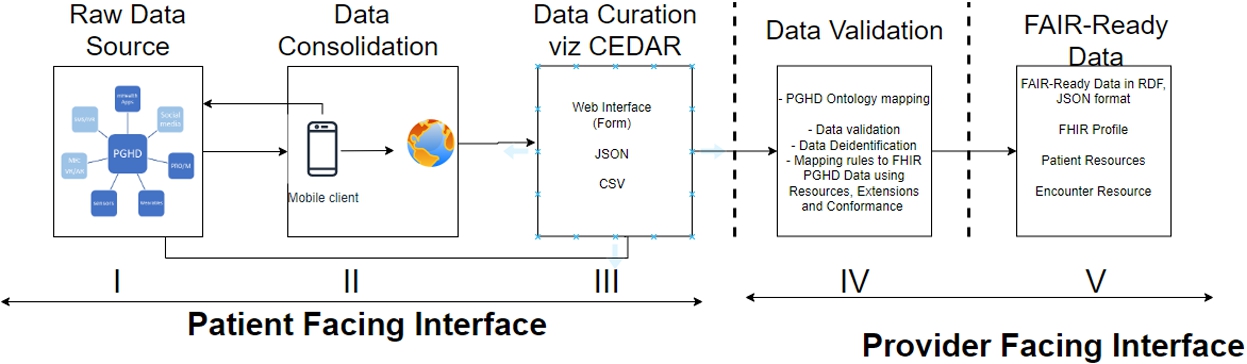
2.Methodology
Understanding PGHD peculiarities [6], adapting the framework of FAIR for health data [13] and learning from experiences of the use of localized CEDAR and Excel2RDF bulk data upload in VODAN [17], we provide a framework for making PGHD FAIRified (Fig. 3). The architecture employs a privacy by design approach, such that all PGHD data communicated to the service provider is one way i.e. not accessible outside the health facility. The details of each stage of the framework is given below.
2.1.Data source
Data Source refers to the multiple, sometimes combined sources of PGHD generated by patients or their representatives that are reported to the health facility, either as part of a health program or a patient portal for personal use. For instance, data from wearables and self-reported data on nutrition to account for data that a diabetic patient may report towards their diabetes management program.
2.2.Data consolidation
Some raw PGHD (for example, data from wearables) are sometimes staged and consolidated before being communicated over mobile applications to a web server before they are accessible via an API from a third party system. As shown in the architecture, this step does not apply to all PGHD sources, as some sources can directly be sent for data curation without being consolidated, this depends on the source and the needs of clinicians.
2.3.Data curation
At this stage, all PGHD sources will be curated using appropriate localised CEDAR templates (see Fig. 4). Depending on the type of PGHD, it is curated using a selected template. Templates are predesigned to be linkable to ontologies (using the VALUES property of CEDAR).
Fig. 4.
CEDAR implementation of PGHD-FAIRification framework.
2.4.Data validation
On submission or receipt of PGHD, with the right CEDAR templates, data from fields that are based on the requirement of the PGHD source(s) are further validated against known constraints, value ranges and conformance to privacy requirements such as data de-identification or pseudonymisation. Simultaneously, depending on the type of field, select fields can be linked/mapped to relevant ontologies and standards like SNOMED-CT, VODANA-Ontology and/or communication protocols like FHIR, such that they can be made to be interoperable and reusable. However, patient identifiable fields like patient name, patient number and address are not allowed to be exposed outside clinical settings hence not allowed to be linkable.
2.5.FAIR-ready data
With validated data, after the required mapping, data is made into a machine-actionable form in RDF and JSON-LD format – and can only be accessible based on the regulations in place for the PGHD data.
3.Discussion
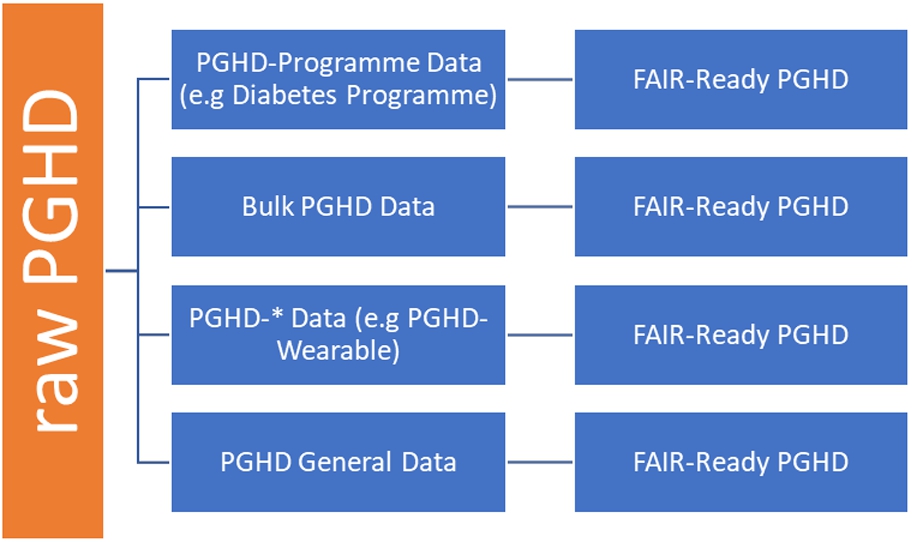
PGHD can come from multiple sources and formats, and as part of a health program (e.g Diabetes Management Plan – where CGM or blood sugar data, nutrition data and fitness data are generated by patient themselves but eventually transmitted for use by clinicians), or as a bulk data from one PGHD source, say in CSV format (e.g in the case of wearables (Fitbit) that allow export of data as CSV by users) or as a single raw data directly from one source (through APIs). This framework illustrates and suggests that, in each of the aforementioned cases, with the use of appropriate technologies, we can achieve FAIR-ready data that are verifiable by clinicians and fit for practical purposes.
As a practical example, we can create a CEDAR template on Blood Pressure Management Programme for Hypertensive Patients to cater for PGHD from multiple sources (Fig. 4). This fits our use-case of the framework where the PGHD-Programme data scenario is used. In this case, a template that fits the programme is created as PGHD-BP Management and added to existing templates say OPD, ANC, COVID-19 templates that are developed under VODAN-CEDAR technology, to receive PGHD for this program. PGHD related to the program are then received and made FAIR-ready (Fig. 4).
4.Conclusion
While working with PGHD, having the right template that makes data FAIR-ready is important. CEDAR tool enables stakeholders to comply with FAIR metadata standards. The FAIRification process for PGHDs differs from scientific data and a workplan is proposed that is specific to the nature of PGHDs. The value of a FAIRification process of PGHDs is in the possibility of integrating different sources of PGHD to expand the knowledge and understanding that can be derived. The model allows a diversity of PGHD sources to be linked for clinical use, and this framework portrays how we can integrate these sources using CEDAR or any other annotating tool. We provide four (4) use scenarios to illustrate common use of PGHD in practice – which can appropriately be further modified to meet users’ needs. With this, PGHD can be made available during encounters (real-time data) or through historical data (bulk data). With such frameworks used and templates in place, clinicians can easily reuse the template, modifying it to fit their PGHD data source(s) or programmes, create workflows from it, collect data, create metadata, and validate results in a FAIR-manner.
Acknowledgements
The authors have no acknowledgments to report.
Funding
This work was conducted with the financial support of the Science Foundation Ireland Centre for Research Training in Digitally Enhanced Reality (d-real) under Grant No. 18/CRT/6224 and at the ADAPT SFI Research Centre for AI-Driven Digital Content Technology under Grant No. 13/RC/2106_P2. For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Conflict of interest
The authors have no conflict of interest to report.
References
[1] | A. Alamri, Ontology middleware for integration of IoT healthcare information systems in EHR systems, Computers 7: (4) ((2018) ), 51. doi:10.3390/computers7040051. |
[2] | S. El-Sappagh, F. Ali, A. Hendawi, J.H. Jang and K.S. Kwak, A mobile health monitoring-and-treatment system based on integration of the SSN sensor ontology and the HL7 FHIR standard, BMC Medical Informatics and Decision Making 19: (1) ((2019) ), 1–36. doi:10.1186/s12911-018-0723-6. |
[3] | FAIRification process. Available at: https://www.go-fair.-org/fair-principles/fairification-process/. Accessed October 10, 2022. |
[4] | R.S. Gonçalves, M.J. O’Connor, M. Martínez-Romero, A.L. Egyedi, D. Willrett, J. Graybeal and M.A. Musen, The CEDAR workbench: An ontology-assisted environment for authoring metadata that describe scientific experiments, in: International Semantic Web Conference, 2017 October 21, Springer, Cham, (2017) , pp. 103–110. |
[5] | Y. Jayawardana, G. Jayawardena, A.T. Duchowski and S. Jayarathna, Metadata-driven eye tracking for real-time applications, in: Proceedings of the 21st ACM Symposium on Document Engineering, 2021 August 16, (2021) , pp. 1–4. |
[6] | A.A. Kawu, L. Hederman, D. O’Sullivan and J. Doyle, Patient generated health data and electronic health record integration, governance and socio-technical issues: A narrative review, Informatics in Medicine. Unlocked. 2022 Dec 21:101153. |
[7] | N. Lv, L. Xiao, M.L. Simmons, L.G. Rosas, A. Chan and M. Entwistle, Personalized hypertension management using patient-generated health data integrated with electronic health records (EMPOWER-H): Six-month pre-post study, Journal of Medical Internet Research 19: (9) ((2017) ), e7831. doi:10.2196/jmir.7831. |
[8] | S. Marceglia, P. D’Antrassi, M. Prenassi, L. Rossi and S. Barbieri, Point of care research: Integrating patient-generated data into electronic health records for clinical trials, in: AMIA Annual Symposium Proceedings, Vol. 2017: , American Medical Informatics Association, (2017) , p. 1262. |
[9] | NIGERIA DATA PROTECTION REGULATION 2019. Available at: https://ndpb.gov.ng/Files/NigeriaDataProtectionRegulation.pdf. Accessed October 10, 2022. |
[10] | P. Plastiras and D. O’Sullivan, Exchanging personal health data with electronic health records: A standardized information model for patient generated health data and observations of daily living, International Journal of Medical Informatics 120: ((2018) ), 116–125. doi:10.1016/j.ijmedinf.2018.10.006. |
[11] | R.K. Saripalle, Fast Health Interoperability Resources (FHIR): Current status in the healthcare system, International Journal of E – Health and Medical Communications (IJEHMC) 10: (1) ((2019) ), 76–93. |
[12] | M. Shapiro, D. Johnston, J. Wald and D. Mon, Patient-generated health data, April. 2012 Apr, RTI International. |
[13] | A.A. Sinaci, F.J. Núñez-Benjumea, M. Gencturk, M.L. Jauer, T. Deserno, C. Chronaki, G. Cangioli, C. Cavero-Barca, J.M. Rodríguez-Pérez, M.M. Pérez-Pérez and G.B. Erturkmen, From raw data to FAIR data: The FAIRification workflow for health research, Methods of Information in Medicine 59: (S 01) ((2020) ), e21–e32. |
[14] | SPHN Data Ecosystem for FAIR Data. Available at: https://www.youtube.com/watch?v=pqV0qp4oisM. Accessed October 10, 2022. |
[15] | M. van Reisen, F. Oladipo, M. Stokmans, M. Mpezamihgo, S. Folorunso, E. Schultes, M. Basajja, A. Aktau, S.Y. Amare, G.T. Taye and P.H. Purnama Jati, Design of a FAIR digital data health infrastructure in Africa for COVID-19 reporting and research, Advanced Genetics. 2021 Jun; 2(2):e10050. |
[16] | M. Van Reisen, F.O. Oladipo, M. Mpezamihigo, R. Plug, M. Basajja, A. Aktau, P.H. Jati, R. Nalugala, S. Folorunso, S.Y. Amare and I. Abdulahi, Incomplete Covid-19 data: The curation of medical health data by the virus outbreak data network – Africa, Data Intelligence 4: (4) ((2022) ), 673–697. doi:10.1162/dint_e_00166. |
[17] | VODAN, Available at https://www.vodan-totafrica.info/. Accessed October 10, 2022. |
[18] | M.D. Wilkinson, M. Dumontier, I.J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.W. Boiten, L.B. da Silva Santos, P.E. Bourne and J. Bouwman, The FAIR guiding principles for scientific data management and stewardship, Scientific Data 3: (1) ((2016) ), 1–9. |

![The FAIR guiding principles for scientific data management and stewardship [18].](https://content.iospress.com:443/media/fc/2023/1-1/fc-1-1-fc230500/fc-1-fc230500-g001.jpg)
![An architecture implementing the FAIRification workflow for health data [13].](https://content.iospress.com:443/media/fc/2023/1-1/fc-1-1-fc230500/fc-1-fc230500-g002.jpg)



