Towards FAIR principles for research software
Abstract
The FAIR Guiding Principles, published in 2016, aim to improve the findability, accessibility, interoperability and reusability of digital research objects for both humans and machines. Until now the FAIR principles have been mostly applied to research data. The ideas behind these principles are, however, also directly relevant to research software. Hence there is a distinct need to explore how the FAIR principles can be applied to software. In this work, we aim to summarize the current status of the debate around FAIR and software, as basis for the development of community-agreed principles for FAIR research software in the future. We discuss what makes software different from data with regard to the application of the FAIR principles, and which desired characteristics of research software go beyond FAIR. Then we present an analysis of where the existing principles can directly be applied to software, where they need to be adapted or reinterpreted, and where the definition of additional principles is required. Here interoperability has proven to be the most challenging principle, calling for particular attention in future discussions. Finally, we outline next steps on the way towards definite FAIR principles for research software.
1.Introduction
The FAIR Guiding Principles [1] were published and promoted to improve the reuse of scholarly data by making it more findable, accessible, interoperable and reusable by humans and machines. Implementing FAIR helps researchers demonstrate the impact of their work by enabling the reuse and citation of the data they produce, and can promote collaboration among them. It also helps publishers and funders to define policies for data sharing, and to promote discoverability and reuse. In addition, it helps data stewards and managers to provide guidance on quality criteria for data deposits in digital repositories.
The intention of Wilkinson et al. [1] was that the principles not only apply to data, but also to other digital objects, e.g. algorithms, tools, and workflows, that led to that data, as all these elements must be available to ensure transparency, reproducibility and reusability [2]. At the policy level, software is indeed seen as part of FAIR, with the European Commission expert group on FAIR data stating that “Central to the realisation of FAIR are FAIR Digital Objects, which may represent data, software or other research resources.” [3]. Applying the FAIR principles in a useful way to research software will provide similar benefits of enabling transparency, reproducibility and reusability of research, making it easier for industry, science, education and society to have effective access to software-based knowledge. In particular, FAIR software should facilitate making FAIR data.
However, software is data and software is not data. Over the last three years, numerous discussions have taken place with the aim of understanding how the FAIR principles relate to software (see Table 2 on page 52 for an overview). It is clear that the four foundational principles in [1] are intended to apply to software, but can we apply them in a practical and useful way? The terminology and detail used in the 15 FAIR Guiding Principles is focused on their application to data – particularly in the life sciences – and can be confusing if applied to software without translation. The drivers, stakeholders and incentives, whilst overlapping, are not identical. In addition, the variety of software and its distribution channels poses a challenge when adapting the current FAIR principles.
In this work, we aim to summarize the current state of the debate around FAIR and software, as a basis for the development of specific principles for FAIR research software. First, we discuss what makes software different from data with regard to the application of the FAIR principles (Section 2), and argue why quality considerations about research software go beyond FAIR (Section 3). We then present an analysis of where the existing principles can be directly applied to software, where they need to be adapted or reinterpreted, and where the definition of additional principles is required (Section 4). The conclusions provide a summary and directives for future work on FAIR for research software.
2.Software is not data
Technically, software is a special kind of data. In computing, digital data (ultimately sequences of ones and zeros) are used to represent all information, including factual data as well as computer instructions. In the more abstract context of FAIR, software and data are regarded as different kinds of digital research objects next to each other. As such, they share particular characteristics that allow them to be treated alike for certain aspects of FAIR, such as the possibility of having a Digital Object Identifier (DOI) assigned, or having a license. However, as elaborated by Katz et al. [4], there are also several significant differences between data and software as digital research objects: Data are facts or observations that provide evidence. In contrast, software is the result of a creative process that provides a tool for doing something, for example with data. As such, software is executable, while data is not. Software is often built using other software. This is especially obvious for software that implements multi-step processes to coordinate multiple tasks and their data dependencies, which are usually referred to as workflows [5,6]. Generally, all software applications that are not written completely from scratch are of a composite nature that easily leads to complex dependencies. The lifetime of software is generally shorter than that of data, as versioning is applied more frequently and regularly leads to changes in behaviour and/or interfaces. Hence, dependencies as well as dependent software packages are subject to frequent changes.
Naturally, the work on FAIR principles for software is focused on research software. Research software is defined as “software that is used to generate, process or analyse results that you intend to appear in a publication (either in a journal, conference paper, monograph, book or thesis)” [7]. Importantly, for the purpose of having a reference definition, software that does not generate, process or analyse results – such as word processing software, or the use of a web search – is not considered research software. Research software is also a digital research object, and provenance around software usage plays a key role in the transparency, reproducibility and reusability of scientific activities, spanning from academic to industrial research. Research software includes but is not limited to source code, binaries and web services, and covers a broad spectrum from short scripts written ad hoc by researchers to produce results for a publication, to software rigorously developed for a mission-critical process [8]. Accordingly, research software can be distributed in many ways such as digital repositories e.g., Github, BitBucket, GitLab; or archives like the Software Heritage Project [9]; project websites, FTP folders, language specific archive networks e.g., the Comprehensive R Archive Network (CRAN) [10], the Comprehensive Perl Archive Network (CPAN) [11], the Python Package Index (PyPI) [12], Maven, the Node Package Manager (NPM), and others.
Traditionally, research software has been created and maintained as Free and/or Open Source Software (FOSS). However, while there is a clear overlap between the objectives of FAIR and FOSS with regard to accessibility and reusability, they are not the same (see also [13]). FOSS is mostly concerned with source code being open and licensed under an open license. Open source and permissive licenses are desirable for FAIR software, but although FAIR has its roots in the “FOSS-loving” research software community, they are not a requirement as such. Indeed FAIR does not require data to be open, as clearly certain data sets (e.g., patients’ electronic health records, genomics sequences) require adequate access control (see go-fair FAQ [14]). However, such privacy and sensitivity concerns are not in the same way valid for research software that relates to published research, where there is an expectation that the methodology is made available. It remains to be discussed how open research software should be in order to meet the intentions behind FAIR.
3.Software quality: Beyond FAIR
Another much-debated relationship is the one between FAIR and software quality. Ultimately, the quality of the content of digital resources is crucial for obtaining valid research results. However, the FAIR Guiding Principles do not cover content-related quality aspects, and it is an ongoing discussion whether software quality considerations are part of FAIR (e.g., [15]). We think that it is important here to distinguish between form (that is, how a software is provided, the code itself) and function (that is, what a software actually does, how it behaves, the algorithm encoded), as different quality considerations apply. This is also in line with how the FAIR principles are interpreted for data: they address the form of providing data sets to the scientific community, but are not concerned with the functional content or quality of the data themselves.
Quality aspects concerning the form of software can be considered as covered by FAIR, in particular by the interoperability and reusability principles. It is important to realise that unlike data, software is not static and can only be (re)used if it is sustainable and evolves along with the continuous development of the entire software ecosystem. The quality of its codebase is decisive for a software’s ability to evolve sustainably. This characteristic is often also referred to as maintainability, and includes aspects like modularity, understandability, changeability, analysability and testability [16]. Following guidelines for good scientific software development, as well as language- and/or community specific coding standards [17] are effective means to making and keeping the code base maintainable. Many of these qualities are measurable/quantifiable and could thus be covered with additional FAIR principles and metrics.
Quality aspects that concern the functionality of software, on the other hand, go beyond what is covered by the FAIR principles. Arguably, the most important quality criterion for research software is functional correctness, i.e., the production of the correct results every time the software is run. Thorough validation of the functional correctness of research software can, however, be significantly more difficult than the testing that is required for code maintainability as discussed above [18,19]. For example, testing the software might require specific resources such as access to high performance computing, validated input/output data pairs to test the implementation of an algorithm might not be available yet (as the purpose of the software is to create them), or require the execution of very long computations. Other important quality criteria related to functionality of research software are security measures (guaranteeing privacy and integrity of research data) and computational efficiency (striving to optimise use of resources and runtime performance). The latter cannot be measured statically and may require systematic scientific benchmarking in order to arrive at meaningful performance estimates [20,21]. Discussion is ongoing to see if for these criteria workable principles and metrics can be developed, but specific training and adequate attention in the development process are certainly key to high functional quality of research software.
4.FAIR principles applied to research software
We understand the original 15 FAIR Guiding Principles as an instantiation of the four foundational FAIR principles in the context of research data. Here we interpret them in the context of research software. We discuss how they apply to software, and suggest rephrased, extended or additional principles when necessary. Table 1 provides an overview of the principles in their original and in the proposed software-specific formulation. Tables 3 (page 53) and 4 (page 55) illustrate the proposed principles by using them for assessing the FAIRness of two exemplary bioinformatics tools.
Table 1
Summary of the proposed FAIR principles for research software and how they relate to the FAIR Guiding Principles for data. It is indicated whether a given FAIR data principle has been simply rephrased to adjust it to software, extended to cover a broader scope, reinterpreted to match the different context, discarded as it does not apply, or newly proposed as it only applies for research software
| FAIR for data | FAIR for software | Operation | |
| F1 | (Meta)data are assigned a globally unique and persistent identifier. | Software and its associated metadata have a global, unique and persistent identifier for each released version. | Rephrased |
| F2 | Data are described with rich metadata. | Software is described with rich metadata. | Rephrased |
| F3 | Metadata clearly and explicitly include the identifier of the data it describes. | Metadata clearly and explicitly include identifiers for all the versions of the software it describes. | Rephrased and extended |
| F4 | (Meta)data are registered or indexed in a searchable resource. | Software and its associated metadata are included in a searchable software registry. | Rephrased |
| A1 | (Meta)data are retrievable by their identifier using a standardized communications protocol. | Software and its associated metadata are accessible by their identifier using a standardized communications protocol. | Rephrased |
| A1.1 | The protocol is open, free, and universally implementable. | The protocol is open, free, and universally implementable. | Remain the same |
| A1.2 | The protocol allows for an authentication and authorization procedure, where necessary. | The protocol allows for an authentication and authorization procedure, where necessary. | Remain the same |
| A2 | Metadata are accessible, even when the data are no longer available. | Software metadata are accessible, even when the software is no longer available. | Rephrased |
| I1 | (Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation. | Software and its associated metadata use a formal, accessible, shared and broadly applicable language to facilitate machine readability and data exchange. | Rephrased and extended |
| I2 | (Meta)data use vocabularies that follow FAIR principles. | – | Reinterpreted, extended and split |
| I2S.1 | – | Software and its associated metadata are formally described using controlled vocabularies that follow the FAIR principles. | Reinterpreted, extended and split |
| I2S.2 | – | Software use and produce data in types and formats that are formally described using controlled vocabularies that follow the FAIR principles. | Reinterpreted, extended and split |
| I3 | (Meta)data include qualified references to other (meta)data. | – | Discarded |
| I4S | – | Software dependencies are documented and mechanisms to access them exist. | Newly proposed |
| R1 | (Meta)data are richly described with a plurality of accurate and relevant attributes. | Software and its associated metadata are richly described with a plurality of accurate and relevant attributes. | Rephrased |
| R1.1 | (Meta)data are released with a clear and accessible data usage license. | Software and its associated metadata have independent, clear and accessible usage licenses compatible with the software dependencies. | Rephrased and extended |
| R1.2 | (Meta)data are associated with detailed provenance. | Software metadata include detailed provenance, detail level should be community agreed. | Rephrased |
| R1.3 | (Meta)data meet domain-relevant community standards. | Software metadata and documentation meet domain-relevant community standards. | Rephrased |
4.1.Findability
Findability is a fundamental principle, since it is necessary to find a resource before any other consideration. The main concern of findability for research software is to ensure software can be identified unambiguously when looking for it using common search strategies. Such strategies include the use of keywords in general-purpose search engines like Google, as well as specialised registries (websites hosting software metadata) and repositories (websites hosting software source code and binaries). Findability can be improved by registering the software in a relevant registry, along with the provision of appropriate metadata, providing contextual information about the software. Registries typically render metadata in a web-findable way and can provide a DOI. Some registries and repositories allow annotating software using domain-agnostic or domain-specific controlled vocabularies, increasing findability via search engines further. In the following we discuss how the original four Findability principles apply to the findability of research software.
F1. (Meta)data are assigned a globally unique and persistent identifier
Persistent identifiers (PIDs) are long-lasting references to documents, web pages, or any other digital objects [22]. Global uniqueness makes PIDs a mechanism that allows for unambiguous identification of the referenced resources. As analysed by project FREYA [23], there are several alternatives for assigning PIDs.
Clearly, research software should have their own PIDs. However, it is not enough to assign a PID to a generic software, but rather needed for all their versions and specific deployments. Software versions should get assigned different PIDs as they represent specific developmental stages of the software. This is important as it will contribute to guaranteeing data provenance and reproducible research processes. Indeed, source code management systems make it easier to track software versions. For example, Git, currently the most popular technology for software source code version control, works with commit hashes (SHA1), which uniquely point to the specific snapshot of the source code. However, this identifier is not globally resolvable and GitHub (one of the most popular repositories for software source code based on Git technologies) does not make any guarantees about the accessibility or sustainability of code on the platform (persistence), and thereby the software published therein. A common community solution to this problem is depositing software releases from GitHub to Zenodo [24], an open publishing platform funded by the European Commision, and developed and hosted by Centre Européen Recherche Nucléaire (CERN). Zenodo mints DOIs for each released version of the software, and also creates a concept DOI which refers to all versions of a given software [25].
To the best of our knowledge, there is no unified mechanism to automatically assign PIDs for research software. Thus, software authors need to actively register their software, and associated versions, at least in one registry or/and repository.
We suggest rephrasing this principle as “Software and its associated metadata have a global, unique and persistent identifier for each released version”.
F2. Data are described with rich metadata
A software’s name alone does not reveal much about it. In order for others to find and use that software, they need information about what it does, what it depends on and how it works. Metadata provide this information. How detailed software metadata has to be and what is considered to be “rich” depends on the concrete context and cannot be answered in general. In the context of FAIR, software metadata should at least describe where to find a specific version of the software, how to cite it, who are the authors, what are the inputs and outputs, and what are dependencies. Furthermore metadata should include elements related to provenance and should follow community agreements (discussed later as part of the Reusability principles).
There are currently multiple projects working on concrete solutions to add structured metadata annotations to software. Examples include the biotoolsSchema [26], a formalised schema (XSD) used by the bio.tools project [27,28]; the CodeMeta set of terms [29] and Bioschemas Tool profile [30]. The latter two work on top of schema.org, a project aiming to make it easier to add structured markup to web pages, and help search engines to index them. Additionally, some programming languages provide a way to add metadata to software sources, i.e., packages; and often require them to be in a specific format and/or adhere to some guidelines. For instance, R packages must include metadata in the DESCRIPTION file [31] while PEP 566 describes metadata for Python software packages.
Regardless of the metadata description approach used, the use of controlled vocabularies provided by community-approved ontologies is recommended. These will vary dependent on the research domain. The Software Ontology [32] is a resource that can be used to describe software, including types, tasks, versions, provenance and associated data. In the case of life sciences, we advise to use elements from ontologies such as EMBRACE Data and Methods ontology (EDAM) [33]. EDAM provides unambiguously defined terms for describing the types of data and data identifiers, data formats, operations and topics commonly used in bioinformatics. In the geosciences, OntoSoft [34,35] is an ontology designed to facilitate the annotation and publication of software with rich metadata.
We suggest rephrasing this principle as “Software is described with rich metadata”.
F3. Metadata clearly and explicitly include the identifier of the data it describes
For reproducibility and reusability purposes, any person and/or system examining the metadata needs to be able to identify which version of the software is described by it. F3 extends F1’s focus regarding the precise identification of versions and/or reference deployments beyond the software itself by including the metadata associated with each version and/or reference deployment of the software. This enables the exact version of a given software to be found when reusing and/or reproducing previously generated scientific results. For example, release metadata files on Zenodo should point to specific releases on software source code repositories such as GitHub, BitBucket or GitLab.
We suggest rephrasing and extending this principle as “Metadata clearly and explicitly include identifiers for all the versions of the software it describes”.
F4. (Meta)data are registered or indexed in a searchable resource
Software and associated metadata should be registered in a suitable, searchable software registry or repository. There are chiefly three classes of registries and repositories: (i) general ones such as Zenodo, GitHub itself, and comprehensive software archives as run by the Software Heritage project [36], (ii) language-specific ones such as CRAN, PyPI, and (iii) domain-specific ones such as the bioinformatics-specific BioConductor [37], bio.tools and BioContainers [38] registries, the Astrophysics Source Code Library (ASCL) [39], swMath [40] for mathematical software, CLARIN [41] for digital humanities software source code and the different science gateways based on the HUBzero [42] open source software platform.
The choice of the registry/repository may be influenced by the programming language used and/or the operating system most used by the respective community. For example, most of the Python packages are registered in PyPI and/or one of the Conda [43] channels. R packages go to CRAN, Bioconductor.org and/or source code repositories like GitHub. Linux distributions have their own package managers with software repositories.
We suggest rephrasing this principle as “Software and associated metadata are included in a searchable software registry”.
4.2.Accessibility
In the original FAIR Guiding Principles, accessibility translates into retrievability through a standardized communication protocol (A1) and accessibility of metadata even when the original resource is no longer accessible (A2). These principles clearly also apply to software. Interpreting accessibility also as the ability to actually use the software (access its functionality), however, we found mere retrievability not enough. In order for anyone to use any research software, a working version of the software needs to be available. This is different from just archiving source code, even in comprehensive and long-term collections like the Software Heritage archive. To use software, a working version (binary or code) has to be either downloadable and/or accessible e.g., via a web interface, along with the required documentation and licensing information. Accessibility requirements depend on the software type, e.g., web-applications, command-line tools, etc. For example, software containers allow the use across different operating systems and environments, e.g., local computers, remote servers, and high-performance computing (HPC) installations. Cloud-based servers can execute existing pieces of code as a service, as software made available through a web interface or via Jupyter Notebooks [44]. Notebooks allow others to see the results and the narrative alongside the code used to generate them.
Furthermore, even for software that can be downloaded or accessed without restrictions, being able to run it might also depend on, for example, data samples, (paid) registration, other (proprietary) software packages, or a non-free operating system like Windows or macOS. For data, the FAIR principles demand that “(Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation” (I1) and in that sense discourage the use of proprietary data formats. This is in our view, however, different from transparent dependencies for running software.
It is worth to re-emphasize that research software are not single, isolated, digital objects. As further discussed for Interoperability, research software interoperate at different levels with other digital objects including other software, and might have different available versions and/or web-based deployments. Still, all implementations should be considered as part of a single entity for the considerations on accessibility with metadata, as to ensure appropriate links among them (see F1, F3). Since accessibility, interoperability and (re)usability are intrinsically connected for research software, we consider aspects of installation instructions (R1.3), software dependencies (I4S), and licensing (R1.1) as part of other principles here, rather than adding another Accessibility principle.
A1. (Meta)data are retrievable by their identifier using a standardized communications protocol
Retrievability of research software and its metadata can be achieved by depositing it in an appropriate repository and/or registry. We will discuss later in this paper that retrieving software source code and/or binaries is however only the first step towards being able to actually use it.
We suggest rephrasing this principle as “Software and associated metadata are retrievable by their identifier using a standardized communications protocol”.
A1.1. The protocol is open, free, and universally implementable. Usually software (and its metadata) can be downloaded directly from the repository and/or website via standard protocols (HTTP/SSH).
There is no need to rephrase this specific item as it generally applies to any digital resource exposed via the web, and thus to both data and software.
A1.2. The protocol allows for an authentication and authorization procedure, where necessary. Authentication and authorization are relevant for accessing research software source code (open and closed source), binaries and/or web applications. Collaborative development platforms like GitHub and BitBucket implement mechanisms to support authentication and authorisation, and control the access to the code base. Similarly, it might be possible that users might need to register, and/or authenticate, before downloading binaries or, in the case of web applications, using the software. In all cases, access conditions should be justified and documented.
There is no need to rephrase this specific item as it generally applies to any digital resource, and thus to both data and software.
A2. Metadata are accessible, even when the data are no longer available
Metadata provides the context for understanding research software, and this should persist even when the software itself is no longer available. To achieve this, metadata should be available separately from research software objects, here understood as either the source code, binaries and/or web servers hosting the deployed software. For example, GitHub can host the software source code and can be connected to Zendo, FigShare, bio.tools or FAIRsharing.org [45] for hosting additional copies of the research software metadata. Zenodo promises metadata, and a snapshot of the software release, to be available for the upcoming 20 years, even when the versioned source code on GitHub may not be accessible any more. Metadata should follow community agreements. In this way, it will contribute towards the findability of the metadata as well as the software it references and provide details on how the software interoperates with other digital objects and how it may be (re)used.
We suggest rephrasing this principles as “Software metadata are accessible, even when the software is no longer available”.
4.3.Interoperability
The IEEE Standard Glossary of Software Engineering Terminology [46] defines interoperability as the “ability of two or more systems or components to exchange information and to use the information that has been exchanged”. This definition is further complemented by semantic interoperability, ensuring “that these exchanges make sense – that the requester and the provider have a common understanding of the ‘meanings’ of the requested services and data.” [47]. When examining the FAIR data principles from a research software perspective, interoperability turns out to be the most challenging among the four high-level principles. This is not surprising given the complexity of the software interoperability challenges that form a research area of its own [48–52].
Already for data and its associated metadata, interoperability has been found to be “the most challenging of the four FAIR principles. This, in part, is due to interoperability not being well understood” [53]. In contrast to the rather static nature of data, research software are live digital objects that interact at different levels with other objects, e.g., other software, managed data, execution environments; and either directly and/or indirectly, as scripts or as part of a workflow (see Fig. 1). The interoperability principles are therefore even more challenging to apply to software, some are not directly applicable, others need to be rephrased and even new principles need to be defined to appropriately address the dynamic nature of software.
Fig. 1.
Interoperability for research software can be understood in two dimensions: as part of workflows (horizontal dimension) and as stack of digital objects that need to work together at compilation and execution times (vertical dimension). Importantly, workflows do not need to use the same physical hardware or the same operating system, as long as there are agreed mechanisms for software to interoperate with one another.
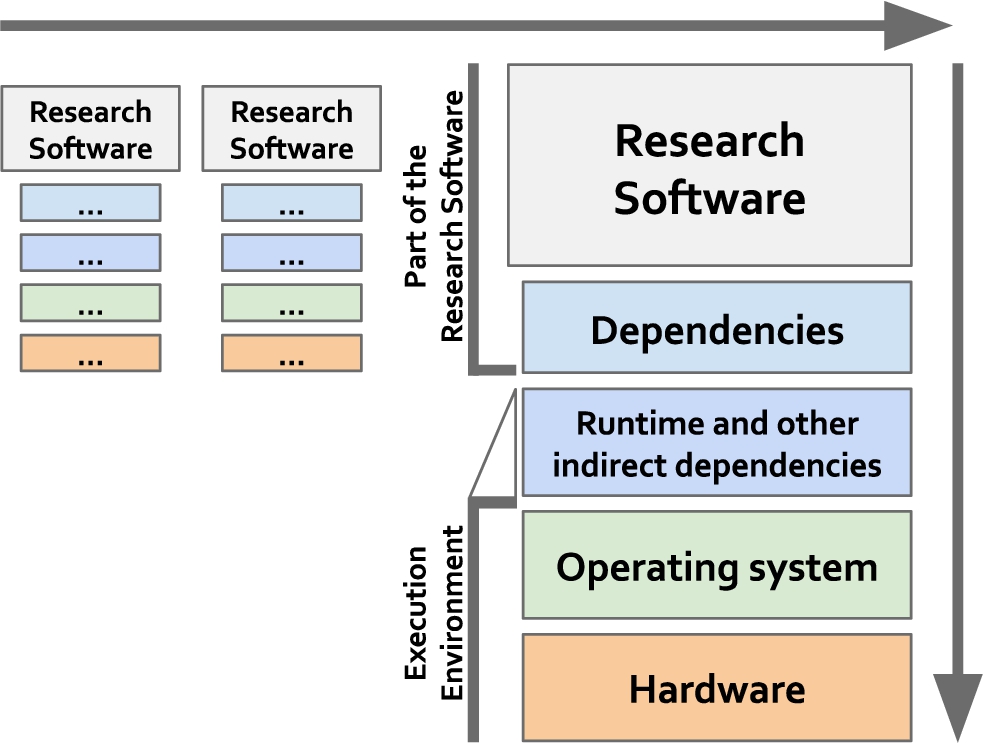
Software interoperability can be defined from three different angles:
1. for a set of independent but interoperable objects to produce a runnable version of the software, including libraries, software source code, APIs and data formats, and any other resources for facilitating that task;
2. for a stack of digital objects that should work together for being able to execute a given task including the software itself, its dependencies, other indirect dependencies, the whole execution environment including runtime dependencies and the operating system, the execution environment, dependencies, and the software itself; and
3. for workflows, which interconnect different standalone software tools for transforming one or more data sets into one or more output data sets through agreed protocols and standards.
Thus, interoperability for software can be considered both for individual objects, which are the final product of a digital stack, and as part of broader digital ecosystems, which includes complex processes and workflows as well as their interaction [6,54,55]. Different pieces of software can also work together independent of programming languages, operating systems and specific hardware requirements through the use of APIs and/or other communication protocols.
Software metadata are a necessity for interoperability. They provide the context in which the software is used and contributes towards provenance, reproducibility and reusability. However, a balance is needed between the detail level and its generation cost. Depending on whether research software is considered as an individual product or as part of an ecosystem, the associated metadata might differ [28,56,57], with workflows having specific mechanisms to capture it through their specifications, e.g., using Common Workflow Language (CWL) [58,59] and/or Workflow Description Language (WDL) [60], among others. This metadata should include software version, dependencies (including which version), input and output data types and formats (preferably using a controlled vocabulary), communication interfaces (specified using standards like OpenAPI), and/or deployment options.
Another aspect associated with interoperability is the ability to run the software in different operating systems, i.e. software portability. Software portability strongly depends on the availability of the full execution stack in other operating systems (vertical axis in Fig. 1), which may not always be given. This dependency on other digital objects to have a working software is further extended in the newly introduced FAIR principle I4S. The present tendency to package software and its dependencies, in software containers e.g., Docker, Singularity, Rocket, contributes to enhanced software portability. Although these differences are not negligible, given that these terms are often used interchangeably, we will be considering both under the FAIR principle of interoperability, highlighting any issues that arise due to this divergence.
I1. (Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation
In contrast to data, which can be represented in very informal ways, software source code is written in a programming language, and is thus formal by design. The use of openly accessible, shared and broadly applicable programming languages facilitates the interaction with the execution environment. When considering research software as part of a workflow, software should be able to share input and/or output data sets with other software. The proper specification and use of formally defined, shared and broadly applicable data types, models and formats for the data consumed and/or produced by the software are key to facilitating the syntactically and semantically correct interconnection of different pieces of software via their associated metadata.
We suggest rephrasing and extending this principles as “Software and its associated metadata use a formal, accessible, shared and broadly applicable language to facilitate machine readability and data exchange”.
I2. (Meta)data use vocabularies that follow the FAIR principles
Following on from the previous interoperability principle, and considering the differences between data and software, we consider two different cases here: the software itself and the data that it operates on (i.e. inputs and outputs). In both cases, ontologies and controlled vocabularies that are themselves FAIR should be used for the formal description. FAIR software should operate on FAIR data, and not undermine the principles by, e.g., producing outputs only in proprietary data formats. Without using FAIR vocabularies, it might become impossible to understand, for both machines and humans, what is described (software) and/or to what it refers (software metadata). Whenever possible, those descriptions should be generated, agreed and maintained by communities as a mechanism towards the sustainability of such resources, keeping metadata understandable even if the resources disappear for unforeseen reasons. A registry of the available controlled vocabularies, data types, formats and schemas that may be used by the software can be found at FAIRsharing.org.
Thus, we propose to reinterpret and extend I2 by splitting it into two sub-principles to account for such differences:
I2S.1 “Software and its associated metadata are formally described using controlled vocabularies that follow the FAIR principles”.
I2S.2 “Software use and produce data in types and formats that are formally described using controlled vocabularies that follow the FAIR principles”.
I3. (Meta)data include qualified references to other (meta)data
I3 aims to interconnect data sets by semantically meaningful relationships. This approach is useful to prevent information silos and to facilitate machine interpretability of existing relationships between data sets, enabling the automated combination and reasoning over data, and even the inference of new knowledge. However, such relationships are difficult to translate to the case of research software. We found the closest resemblance of this principle to be in software dependencies.
Dependencies are a key element for building working software. Building software usually requires a number of additional modules, libraries and/or other research software that are not included in the original software distribution (see Fig. 1). Such dependencies include not only those modules directly used within the software, but also (recursively) dependencies to additional libraries used by the imported modules. The scenario often builds a complex network of interconnected modules that precludes the software building. Despite all the complexity associated with software dependencies, the semantically meaningful information required is rather limited, essentially boiling down to the “dependsOn” relationship. Although there are additional concepts involved, such as “relatedTo” and “derivedFrom”, they are not utilized in a software dependency context.
This leads us to propose a new FAIR principle I4S for research software: “Software dependencies are documented and mechanisms to access them exist”.
The present tendency to package software and its dependencies, either in virtual environments and/or software containers, alleviates the practical concerns for the final user, and simply moves the issue to the generation of those packages. Software deployment systems (PyPI, Conda, CRAN, …) provide solutions for this, and this information can be aggregated by services such as Libraries.io. In order to follow this principle, software dependencies need to be clearly documented in a formal, accessible, machine-readable, and shared way, and formally described following each programming language format.
4.4.Reusability
Reusability in the context of software has many dimensions. At its core, reusability aims for someone to be able to re-use software reproducibly as described by Benureau and Rougier 2018 [61]. The context of this usage can vary and should cover different scenarios: (i) reproducing the same outputs reported by the research supported by the software, (ii) (re)using the code with data other than the test one provided to obtain compatible outputs, (iii) (re)using the software for additional cases other than those stated as supported, or (iv) extending the software in order to add to its functionality.
Software reusability depends to a high degree on software maintainability (see also Section Software quality: beyond FAIR), including proper documentation at various levels of detail. The legal framework, e.g., software licenses, is also important in terms of reusability as it determines how software can be built, modified, used, accessed and distributed. Furthermore, as research software is an integral part of the scientific process, credit attribution (citation) is another important aspect to consider with regard to (re)usability.
R1. Meta(data) are richly described with a plurality of accurate and relevant attributes
Based on the revision of principles R1.1, R1.2 and R1.3 as detailed below, we suggest rephrasing principle R1 as “Software and its associated metadata are richly described with a plurality of accurate and relevant attributes”.
R1.1. (Meta)data are released with a clear and accessible data usage license. Licenses are useful to protect intellectual property. Software licenses let others know what they are allowed to do, e.g., using the software for free with their own data, and how they are restricted, e.g., not modifying or redistributing it. Without a license, others cannot legally use software in any way as the usage rules are not even defined. Metadata should have separate (data) usage licenses. Licenses for metadata are mainly needed to establish mechanisms on how the software is referenced via its metadata by third parties. A clear example is the indexing of software metadata by registries.
Proper management of software licenses is a challenging task considering the multi-faceted nature of research software, which is often the product of combining libraries, modules, and execution environments with the software itself. The legal implications of misusing software by not considering dependencies and incompatibilities between the associated licenses can be severe. Therefore, it is necessary that licenses for research software are included as part of the available documentation and are structured to facilitate its machine-readability. For example, the Software Package Data Exchange standard [62] facilitates that software licenses becomes machine readable. This is important because licenses go beyond the software itself and have to take into account limitations established by the licenses of all of its dependencies. If every piece of software has made available its license information, then it should be possible to automatically derive potential incompatibilities on software usage as well as to establish whether licenses between software components and dependencies are compatible at the build stage.
Metadata usage licenses are independent of software licenses. Data usage licenses for metadata establish how the metadata can be consumed by third-parties for purposes of indexing, citing and/or referencing software. As there are no dependencies between pieces of software in terms of metadata, there is no need to propagate the data usage licenses among them. Similarly to metadata, if any data is distributed with the software, e.g. as demo input data, it should have its own data usage licenses, where the terms for third parties to make use of it are clearly stated.
We suggest rephrasing and extending this principle as “Software and its associated metadata have independent, clear and accessible usage licenses compatible with the software dependencies”.
R1.2. (Meta)data are associated with detailed provenance. Provenance refers to the origin, source and history of software and its metadata. It is recommended to use well-known provenance vocabularies, for instance PROV-O [63], that are FAIR themselves. There are some elements commonly presented on any provenance data, including a person or organization providing the resource and how to contact them, published date, location and other resources used to produce the one described.
There are standards to capture how software is being used while transforming a given data set. To track the provenance of the software itself, specific software versions is the minimum required information. Software versions reference specific algorithmic implementations which might change over time and/or be included/removed among major software releases. This aspect connects with principles F1 and F3 on identifying specific software versions and its associated metadata. Software provenance also incorporates aspects of how the software is produced. Specifically referring to executable compiled software, provenance should include information on how the software has been compiled and which dependencies it incorporates. This is in line with the newly proposed interoperability principle I4S.
Furthermore, information on how to cite software and how to contribute to it [64] are related to provenance, as they provide information about the people involved in creating the software. Citation information should be included in the metadata, since it makes it easier for others (re)using the software to acknowledge the developers. Although there is no standard way to cite software currently, the Software Sustainability Institute provides more information and discussions on this topic, and there are guidelines developed for particular domains e.g. earth sciences [65] and mathematics [66], as well as generic guidelines defined by the FORCE11 Software Citation Working Group based on the Software Citation Principles [64].
We suggest rephrasing this principle as “Software metadata include detailed provenance information”.
R1.3. (Meta)data meet domain-relevant community standards. Community standards are important as they provide guidelines on what is the minimum expected information about certain kinds of research object. These standards might even influence reviewing and certification processes. Given the multi-faceted nature of software and their strong dependencies on other pieces of software, non-compliance with community standards might render software unable to be reused. Indeed, non-compliance with standards will also prevent to integrate research software within other applications. The software’s documentation should provide information on how to install, run and use a software. In order to make it easier for users, it should include examples with inputs and expected outputs. Any dependency should be clearly stated as it contributes not only to (re)usability but also accessibility and interoperability.
Initiatives such as CodeMeta [29], Bioschemas [67] and the RDA Research Schemas [68] have made preliminary recommendations for community-agreed software descriptors, but broader work, especially regarding the mapping across different vocabularies, is still needed.
We suggest rephrasing this principle as “Software metadata and documentation meet domain-relevant community standards”.
5.Conclusions
Software has become an essential constituent of scientific research. It is therefore desirable to apply the FAIR Guiding Principles, which have so far mostly been interpreted as principles for scientific data management and stewardship, also to research software. As we have discussed in this work, many of the FAIR principles can be directly applied to research software, where software and data can be treated as the same kind of digital research objects. However, when specific characteristics of software are involved, such as their executability, composite nature, and continuous evolution accompanied by frequent versioning, it is necessary to revise and extend the original principles. Furthermore, it can be argued that considerations about the functionality of software (as opposed to the form in which it is provided) are by definition out of the scope of FAIR, and thus need to be addressed by other guiding principles, for example based on best practices for (research) software development [17].
This work aims to become the starting point for further community-led discussions and proposals on how to effectively apply FAIR principles to research software, and eventually the development of specific FAIR principles for research software. In addition to the work on the principles, the development of community-specific metadata schemes for software has to play an important role, as defined metadata standards are key to the successful application of many of the principles. There are groups within the wider research software community beginning to address these issues. For example, recently the Software Source Code Identification Working Group [69] has been initiated in the scope of the Research Data Alliance (RDA) and FORCE11 to produce an initial collection of software identification use cases and corresponding schemas as well as to give an overview of the different contexts in which software artifact identification is relevant. Results from this working group can assist in the definition of principles related to software annotation.
Another important aspect discussed during the work on this paper is the need of a governance model for the FAIR principles. A governance model is crucial to enable an open and transparent process for updating the FAIR principles and should be defined in the scope of the community discussions for each of the domains where they are applied e.g., research data, workflows, research software, etc.
Finally, the aim of this work is to set the foundations to develop metrics and associated maturity models that can ultimately inform software users and developers how FAIR their software is. Making software FAIR comes with a cost due to the required efforts. Hence, software developed to be used by others, such as libraries, can be expected to reach a higher degree of FAIRness than software that has not been implemented with reuse as a primary goal, for example a script that has been created as a side-effect of demonstrating an algorithm. Based on FAIR software metrics, communities will be able to agree on degrees of FAIRness that the different kinds of software should comply to, in order to reflect their Open Science ideals.
Acknowledgements
We are grateful to the numerous people who contributed to the discussions around FAIR research software at different occasions preceding the work on this paper. Making no claims to completeness, these include Peter Doorn, Michel Dumontier, Chris Erdmann, José María Fernández, Rafael C. Jimenez, Katrin Leinweber, Jason Maassen, Mustapha Mokrane, Jurriaan Spaaks, Mark Wilkinson and Amrapali Zaveri. We also thank the Japan BioHackathon for sponsoring a FAIR software related project for the 2018 edition. Furthermore, we would like to thank Stian Soiland-Reyes for his valuable comments on earlier versions of this manuscript.
NCH and CAG were supported by EP/N006410/1 and EP/S021779/1 for the UK Software Sustainability Institute. AV, JLG, SCG and CAG were supported by ELIXIR-EXCELERATE 676559. EM, AV, JLG and SCG has been additionally supported by PT17/0009/0001. EM and SCG has been additionally supported by IMI2 FAIRplus 802750.
Appendices
Appendix
Table 2
Recent events with discussions around FAIR and software. This paper is a result of the lessons learned from these discussions
| Event | Main outcomes | Publications |
| “FAIR principles for Software” at 2019 Workshop on Sustainable Software Sustainability (WOSSS19) (https://www.software.ac.uk/wosss19) | FAIR principles can serve as a baseline for enabling software sustainability. However, in contrast to data, software is dynamic, depends on the environment it is executed in, and needs to evolve along with the changing research software ecosystem. It is unclear if and how such software sustainability principles are covered by FAIR. | [70] |
| “FAIR Software” Birds of a Feather meeting (https://www.derse.org/en/conf2019/talk/S8T8HW/) at deRSE 2019 | FAIR and FOSS are not the same, but they do overlap with regard to their intentions about reusability. For discussing the relationship between FAIR and software sustainability, we should distinguish between the code/implementation and the provided functionality. FAIR provides a useful baseline for research software engineering, but developers should be more concerned about software quality. | |
| Top 10 FAIR Data & Software Global Sprint, including “10 easy things to make your software FAIR” (https://librarycarpentry.org/Top-10-FAIR/2018/12/01/research-software/) | Identification and description of 10 “low hanging fruits” that help developers of research software to make their software FAIRer. | [71] |
| “Sharing Your Software – What is FAIR?” at the 2018 American Geophysical Union (AGU) Fall Meeting | A lot of support for making software FAIR is already there. People seem to find writing metadata much more difficult than writing software. Better community software development practices will help here. | [72] |
| “FAIRness assessment for software” (https://github.com/dbcls/bh18/wiki/FAIRnessassessment-for-software) at the ELIXIR 2018 BioHackathon | Applying the general FAIR principles to software is mainly a question of suitable metadata. Community efforts are needed to define these. | |
| “Making Software FAIR” (https://www.aanmelder.nl/dtl2018/makingsoftware-fair) at the DTL Communities@Work 2018 Conference | Generally the FAIR principles are applicable also to research software, the F and A however more directly than the I and R. Re-interpretation and/or extension of the original principles seems to be required. | |
| TIB Training workshops on FAIR Data and Software | Practical tips on improving the citation ability and long-term usability of software as part of making it FAIR. | |
| “Applying FAIR Principles to Software” at the 2017 Workshop on Sustainable Software Sustainability (WOSSS17) | Criteria for evaluating software and software sustainability already exist. It is desirable, but not trivial, to align and combine them with FAIR. | [73] |
| CodeMeta Workshop (https://codemeta.github.io/workshop/) 2016 on The Future of Software Metadata | Community-driven consensus for software metadata. Developed before FAIR, but absolutely applicable to it. |
Table 3
FAIRness assessment of the Fastme tool
| Principle | Description | Fulfilled | Comment |
| F1 | Software and its associated metadata have a global, unique and persistent identifier for each released version. | YES (partially) | Identifier is ‘fastme’ or ‘FastMe’ plus version in X.x in all metadata sources. Sources: bio.tools, webpage, FastMe 2.0’s publication, GitLab and Galaxy. It complies with this principle from version 2.0. It does not have a specific PID, but it can be easily found across different repositories and registries including version information. |
| F2 | Software is described with rich metadata. | YES | Metadata covers the description, usage and accessibility of the software. Regarding the degree of structure and content formalization, metadata in bio.tools is available in biotoolsSchema format and makes use of EDAM terms as well as others that belong to the project’s own vocabulary. Galaxy’s metadata is structured (XML), but does not use any controlled vocabulary. In the remaining sources, metadata is unstructured and does not use any controlled vocabulary. |
| F3 | Metadata clearly and explicitly include identifiers for all the versions of the software it describes. | YES (partially) | All metadata include the version they apply to. However, it is unclear if that same metadata apply to posterior releases of the software. There is a lot of metadata about FastMe 2.0 (webpage, FastMe 2.0’s publication), but not so much for posterior ones. Observation: bio.tools metadata, which refers to FastMe 2.1.4, contains cross-references to metadata belonging to FastMe 2.0. |
| F4 | Software and its associated metadata are included in a searchable software registry. | YES | bio.tools. |
| A1 | Software and its associated metadata are accessible by their identifier using a standardized communications protocol. | YES | Both software and metadata are accessible through HTTP/S: bio.tools, webpage, FastMe 2.0’s publication, GitLab, Galaxy. |
| A1.1 | The protocol is open, free, and universally implementable. | YES | All software and associated metadata are available using HTTP/s across various sites: bio.tools (open), webpage (open), FastMe 2.0’s publication (open access), GitLab (public source code repository) and Galaxy (Galaxy instance at Institut Pasteur, open). |
| A1.2 | The protocol allows for an authentication and authorization procedure, where necessary. | NA | Not necessary. |
| A2 | Software metadata are accessible, even when the software is no longer available. | YES | Metadata is independent of software accessibility in the cases of bio.tools, webpage, FastMe 2.0’s publication and GitLab. |
| I1 | Software and its associated metadata use a formal, accessible, shared and broadly applicable language to facilitate machine readability and data exchange. | YES | Software: This program is written in C (source code available at the src folder in GitLab), a formal, machine readable and widely used language. Metadata: bio.tools metadata is available in JSON and XML formats (following biotoolsSchema). Galaxy fastme metadata is also available in XML. Metadata at webpage and GitLab are not machine readable. |
| I2S.1 | Software and its associated metadata are formally described using controlled vocabularies that follow the FAIR principles. | YES | Software: bio.tools metadata use EDAM terms as well as others that belong to the project’s own vocabulary. In the remaining cases, used terms do not belong to any controlled vocabulary. Metadata: bio.tools metadata is described in an XML schema. |
| I2S.2 | Software use and produce data types and formats that are formally described using controlled vocabularies that follow the FAIR principles. | YES | Software uses and produces phylogenetic trees in NEWICK format, consumes multiple sequence alignment files in PHYLIP format and produce distance matrices in PHYLIP format as well. All these formats have been formally described using EDAM (edamontology.org/format_1910, edamontology.org/format_1997, edamontology.org/format_1445). NETWICK is also contained in the Eagle-I Research Resource Ontology (ebi.ac.uk/efo/swo/SWO_0000634) and Ensembl Glossary (ensembl.org/glossary/ENSGLOSSARY_0000289). |
| I4S | Software dependencies are documented and mechanisms to access them exist. | NO | Dependencies are not mentioned anywhere in the metadata. An exception is Galaxy, but it regards ‘galaxy dependencies’. |
| R1 | Software and its associated metadata are richly described with a plurality of accurate and relevant attributes. | YES | See comments for R1.1 and R1.2. |
| R1.1 | Software and its associated metadata have independent, clear and accessible usage licenses compatible with the software dependencies. | YES | Software: GNU General Public License as published by the Free Software Foundation, either version 3 of the license or later versions, in webpage, bio.tools, GitLab. No conditions of use in Galaxy. Metadata: bio.tools: Attribution 4.0 International (CC BY 4.0) license. GitLab: not stated. Webpage: Creative Commons Attribution 4.0 International license. Galaxy: not stated. |
| R1.2 | Software metadata include detailed provenance, detail level should be community agreed. | YES (partially) | To some extent in metadata at GitLab, as there is not available the whole history of releases, the first ones are missing. No provenance in the remaining metadata. |
| R1.3 | Software metadata and documentation meet domain-relevant community standards. | YES (partially) | Authors do provide documentation but do not follow any community-agreed standard for doing that. Metadata do not follow any community-agreed standard at the authors’ repository and webpage. Metadata registered in bio.tools and Galaxy do follow the standards established by those initiatives. |
Table 4
FAIRness assessment of the ChIPseeker tool
| Principle | Description | Fulfilled | Comment |
| F1 | Software and its associated metadata have a global, unique and persistent identifier for each released version. | YES | Bioconductor: 10.18129/B9.bioc.ChIPseeker. It resolves to the latest stable version. Each version in X.x.x form. This DOI is mentioned in neither bio.tools nor the GitHub repository. |
| F2 | Software is described with rich metadata. | YES | Metadata covers the description, usage and accessibility of the software. Regarding the degree of structure and content formalization, metadata in bio.tools follows the structure set by the biotoolsSchema and makes use of EDAM terms as well as others that belong to the project own vocabulary. Bioconductor and GitHub DESCRIPTION follow Bioconductor’s metadata standardized structure and vocabulary. In the remaining sources, metadata is unstructured and does not make use of any controlled vocabulary. |
| F3 | Metadata clearly and explicitly include identifiers for all the versions of the software it describes. | YES | All metadata include the version they apply to. |
| F4 | Software and its associated metadata are included in a searchable software registry. | YES | Bioconductor and bio.tools. |
| A1 | Software and its associated metadata are accessible by their identifier using a standardized communications protocol. | YES | Both software and metadata are accessible through HTTP/S: bio.tools, GitHub and webpage, Bioconductor. |
| A1.1 | The protocol is open, free, and universally implementable. | YES | All software and associated metadata are available using HTTP/s across various sites: bio.tools (open), webpage (open), Bioconductor (open), and GitHub (public source code repository). |
| A1.2 | The protocol allows for an authentication and authorization procedure, where necessary. | YES | Not necessary. |
| A2 | Software metadata are accessible, even when the software is no longer available. | YES | Available in bio.tools, webpage, Bioconductor, and GitHub. GitHub contains DESCRIPTION files since version 0.99.6. |
| I1 | Software and its associated metadata use a formal, accessible, shared and broadly applicable language to facilitate machine readability and data exchange. | YES | Software: The software is written in R, a formal, machine readable and widely used language. Metadata: bio.tools metadata is available in JSON and XML (following biotoolsSchema). Galaxy metadata is available in XML format. Bioconductor and GitHub DESCRIPTION follow Bioconductors metadata standardized structure, which is not a widely used format and thus less interoperable than the previous ones. Metadata at the software’s webpage is not machine readable. |
| I2S.1 | Software and its associated metadata are formally described using controlled vocabularies that follow the FAIR principles. | YES | Software: bio.tools metadata uses EDAM terms as well as others that belong to the project own vocabulary. In the remaining cases, used terms do not belong to any controlled vocabulary. Metadata: bio.tools metadata is described in an XML schema. Bioconductor and GitHub DESCRIPTION use Bioconductor’s metadata accepted terms. |
| I2S.2 | Software use and produce data types and formats that are formally described using controlled vocabularies that follow the FAIR principles. | YES | The software accepts BED files as an input and produce GRanges objects as output. Sources: In manual, referenced in Bioconductor and bio.tools. |
| I4S | Software dependencies are documented and mechanisms to access them exist. | YES | Stated in Bioconductor and GitHub DESCRIPTION. Automatically downlodable and installable through Bioconductor. |
| R1 | Software and its associated metadata are richly described with a plurality of accurate and relevant attributes. | YES | See comments for R1.1 and R1.2. |
| R1.1 | Software and its associated metadata have independent, clear and accessible usage licenses compatible with the software dependencies. | YES | Software: Artistic-2.0, coherent across metadata sources. Metadata: Depends on the resource used to gather it. bio.tools: Attribution 4.0 International (CC BY 4.0) license. Bioconductor: not stated. Webpage: Artistic-2.0 license. GitHub: not stated. |
| R1.2 | Software metadata include detailed provenance, detail level should be community agreed. | YES | Commits on GitHub since version 0.99. No provenance in the remaining metadata. |
| R1.3 | Software metadata and documentation meet domain-relevant community standards. | YES | Authors follow the standards by Bioconductor, bio.tools and GitHub DESCRIPTION to structure metadata and documentation. |
References
[1] | M.D. Wilkinson, M. Dumontier, I.J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg et al., The FAIR guiding principles for scientific data management and stewardship, Scientific Data 3: ((2016) ), 160018. doi:10.1038/sdata.2016.18. |
[2] | M.D. Wilkinson, R. Verborgh, L. Olavo Bonino da Silva Santos, T. Clark, M.A. Swertz, F.D.L. Kelpin, A.J.G. Gray et al., Interoperability and FAIRness through a novel combination of web technologies, PeerJ Computer Science 3: ((2017) ), e110. doi:10.7717/peerj-cs.110. |
[3] | RTD (Directorate-general for research), Turning FAIR into reality: Final report and action plan from the European Commission Expert Group on FAIR Data. Publications Office of the European Union, 2018. https://publications.europa.eu/en/publication-detail/-/publication/7769a148-f1f6-11e8-9982-01aa75ed71a1/language-en (accessed August 16, 2019). |
[4] | D.S. Katz, K.E. Niemeyer, A.M. Smith, W.L. Anderson, C. Boettiger, K. Hinsen, R. Hooft et al., Software vs. data in the context of citation, PeerJ Preprints, e2630v1, 2016. doi:10.7287/peerj.preprints.2630v1. |
[5] | M.G. Atkinson, S. Montagnat and I. Johan Taylor, Scientific workflows: Past, present and future, Future Generation Computer Systems 75: ((2017) ), 216–227. doi:10.1016/j.future.2017.05.041. |
[6] | C. Goble, S. Cohen-Boulakia, S. Soiland-Reyes, D. Garijo, Y. Gil, M.R. Crusoe, K. Peters and D. Schober, FAIR computational workflows, Data Intelligence, (2019) . doi:10.1162/dint_a_00033. |
[7] | S. Hettrick, M. Antonioletti, L. Carr, N. Chue Hong, S. Crouch, D. De Roure, I. Emsley et al., UK Research Software Survey 2014, Zenodo, 2014. doi:10.5281/zenodo.14809. |
[8] | T. Schlauch, M. Meinel and C. Haupt, DLR software engineering guidelines (version 1.0.0), Zenodo, 2018. doi:10.5281/zenodo.1344612. |
[9] | J.-F. Abramatic, R. Di Cosmo and S. Zacchiroli, Building the universal archive of source code, Communications of the ACM 61: (10) ((2018) ), 29–31. doi:10.1145/3183558. |
[10] | CRAN, The comprehensive R archive network. https://cran.r-project.org/ (accessed August 16, 2019). |
[11] | CPAN, The comprehensive Perl archive network. https://www.cpan.org/ (accessed August 16, 2019). |
[12] | Warehouse Project, PyPI · The Python package index. https://pypi.org/ (accessed August 16, 2019). |
[13] | R. Higman, D. Bangert and S. Jones, Three camps, one destination: The intersections of research data management, FAIR and Open, Insights 32: ((2019) ), 18. doi:10.1629/uksg.468. |
[14] | GO FAIR, What is the difference between ‘FAIR Data’ and ‘Open Data’ if there is one? https://www.go-fair.org/faq/ask-question-difference-fair-data-open-data/ (accessed August 16, 2019). |
[15] | P. Doorn, Does it make sense to apply the FAIR data principles to software?, SlidePlayer, 2017. https://slideplayer.com/slide/12849777/. |
[16] | Object Management Group, Automated source code maintainability measure. https://www.omg.org/spec/ASCMM/1.0/PDF (accessed August 16, 2019). |
[17] | R.C. Jiménez, M. Kuzak, M. Alhamdoosh et al., Four simple recommendations to encourage best practices in research software, F1000Research 6: ((2017) ), 876. doi:10.12688/f1000research.11407.1. |
[18] | D. Heaton and J.C. Carver, Claims about the use of software engineering practices in science: A systematic literature review, Informaion and Software Technology 67: ((2015) ), 207–219. doi:10.1016/j.infsof.2015.07.011. |
[19] | U. Kanewala and J.M. Bieman, Testing scientific software: A systematic literature review, Information and Software Technology 56: (10) ((2014) ), 1219–1232. doi:10.1016/j.infsof.2014.05.006. |
[20] | S. Capella-Gutierrez, D. de la Iglesia, J. Haas, A. Lourenco, J.M. Fernández, D. Repchevsky, C. Dessimoz et al., Lessons learned: Recommendations for establishing critical periodic scientific benchmarking, 2017. doi:10.1101/181677. |
[21] | S. Mangul, L.S. Martin, B.L. Hill, A. Ka-Mei Lam, M.G. Distler, A. Zelikovsky, E. Eskin and J. Flint, Systematic benchmarking of omics computational tools, Nature Communications 10: (1) ((2019) ), 1393. doi:10.1038/s41467-019-09406-4. |
[22] | Wikimedia Foundation, Inc., Persistent identifier. https://en.wikipedia.org/wiki/Persistent_identifier (accessed August 16, 2019). |
[23] | C. Ferguson, J. McEntrye, V. Bunakov, S. Lambert, S. van der Sandt, R. Kotarski, S. Stewart et al., D3.1 Survey of current PID services landscape, Zenodo, 2018. doi:10.5281/zenodo.1324296. |
[24] | GitHub, Making your code citable. https://guides.github.com/activities/citable-code/ (accessed August 16, 2019). |
[25] | Zenodo, Frequently asked questions | DOI versioning. https://help.zenodo.org/ (accessed August 16, 2019). |
[26] | bio.tools, biotoolsSchema – resource description model for bioinformatics software. https://github.com/bio-tools/biotoolsSchema (accessed August 16, 2019). |
[27] | J. Ison, H. Ienasescu, P. Chmura, E. Rydza, H. Ménager, M. Kalaš, V. Schwämmle et al., The Bio.tools registry of software tools and data resources for the life sciences, Genome Biology 20: (1) ((2019) ), 1–4. doi:10.1186/s13059-019-1772-6. |
[28] | J. Ison, H. Ménager, B. Brancotte et al., Community curation of bioinformatics software and data resources, Briefings in Bioinformatics ((2019) ), bbz075. doi:10.1093/bib/bbz075. |
[29] | Codemeta, The CodeMeta Project user guide. https://codemeta.github.io/user-guide/ (accessed August 16, 2019). |
[30] | Bioschemas community, Bioschemas – Tools, vr 0.1. https://bioschemas.org/specifications/Tool/ (accessed August 16, 2019). |
[31] | H. Wickham, Package metadata, R Packages. http://r-pkgs.had.co.nz/description.html (accessed August 16, 2019). |
[32] | The Software Ontology. http://theswo.sourceforge.net/ (accessed August 16, 2019). |
[33] | J. Ison, M. Kalas, I. Jonassen, D. Bolser, M. Uludag, H. McWilliam, J. Malone, R. Lopez, S. Pettifer and P. Rice, EDAM: An ontology of bioinformatics operations, types of data and identifiers, topics and formats, Bioinformatics 29: (10) ((2013) ), 1325–1332. doi:10.1093/bioinformatics/btt113. |
[34] | OntoSoft. http://www.ontosoft.org/ (accessed August 16, 2019). |
[35] | Y. Gil and R. Varun, OntoSoft: Capturing scientific software metadata. http://www.ontosoft.org/ (accessed August 16, 2019). |
[36] | Software Heritage, The Software Heritage archive. https://www.softwareheritage.org/archive/ (accessed August 16, 2019). |
[37] | Bioconductor. https://www.bioconductor.org/ (accessed August 16, 2019). |
[38] | F. da Veiga Leprevost, B.A. Grüning, S. Alves Aflitos, H.L. Röst, J. Uszkoreit, H. Barsnes, M. Vaudel et al., BioContainers: An open-source and community-driven framework for software standardization, Bioinformatics 33: (16) ((2017) ), 2580–2582. doi:10.1093/bioinformatics/btx192. |
[39] | ASCL.net, Astrophysics source code library. http://ascl.net (accessed August 16, 2019). |
[40] | swMATH, An information service for mathematical software. https://swmath.org/ (accessed August 16, 2019). |
[41] | CLARIN-NL, CLARIN NL resource list. https://dev.clarin.nl/clarin-resource-list-fs (accessed August 16, 2019). |
[42] | HUBzero. https://hubzero.org (accessed August 16, 2019). |
[43] | Anaconda Inc., Conda documentation. https://docs.conda.io/en/latest/ (accessed August 16, 2019). |
[44] | Jupyter Project and Community, Project Jupyter. https://www.jupyter.org (accessed August 16, 2019). |
[45] | S.-A. Sansone, P. McQuilton, P. Rocca-Serra, A. Gonzalez-Beltran, M. Izzo, A.L. Lister and M. Thurston, FAIRsharing as a community approach to standards, repositories and policies, Nature Biotechnology 37: (4) ((2019) ), 358–367. doi:10.1038/s41587-019-0080-8. |
[46] | IEEE, Standard glossary of software engineering terminology, IEEE Std 610.12-1990, 1990. doi:10.1109/IEEESTD.1990.101064. |
[47] | S. Heiler, Semantic interoperability, ACM Computing Surveys (CSUR) 27: (2) ((1995) ), 271–273. doi:10.1145/210376.210392. |
[48] | Y. Charalabidis, R.J. Gonçalves and K. Popplewell, Towards a scientific foundation for interoperability, in: Interoperability in Digital Public Services and Administration: Bridging E-Government and E-Business, (2011) , pp. 355–373. doi:10.4018/978-1-61520-887-6.ch019. |
[49] | G. da Silva Serapião Leal, W. Guédria and H. Panetto, Interoperability assessment: A systematic literature review, Computers in Industry 106: ((2019) ), 111–132. doi:10.1016/j.compind.2019.01.002. |
[50] | Y. Naudet, T. Latour, W. Guedria and D. Chen, Enterprise interoperability with SOA: A survey of service composition approaches, Computers in Industry 61: (2) ((2010) ), 176–185. doi:10.1016/j.compind.2009.10.014. |
[51] | R. Rezaei, T. Chiew and S. Lee, A review of interoperability assessment models, Journal of Zhejiang University SCIENCE C 14: (9) ((2013) ), 663–681. doi:10.1631/jzus.C1300013. |
[52] | R. Rezaei, T.K. Chiew, S.P. Lee and Z.S. Aliee, Interoperability evaluation models: A systematic review, Computers in Industry 65: (1) ((2014) ), 1–23. doi:10.1016/j.compind.2013.09.001. |
[53] | R. Allen and D. Hartland, FAIR in practice – Jisc report on the findable accessible interoperable and reuseable data principles, Zenodo, 2018. doi:10.5281/zenodo.1245568. |
[54] | E. Elmroth, F. Hernández and J. Tordsson, Three fundamental dimensions of scientific workflow interoperability: Model of computation, language, and execution environment, Future Generation Computer Systems 26: (2) ((2010) ), 245–256. doi:10.1016/j.future.2009.08.011. |
[55] | R. Mantovaneli Pessoa, E. Silva, M. van Sinderen et al., Enterprise interoperability with SOA: A survey of service composition approaches, in: 12th Enterprise Distributed Object Computing Conference Workshops, (2008) , pp. 238–251. doi:10.1109/EDOCW.2008.32. |
[56] | M. Palmblad, A.-L. Lamprecht, J. Ison and V. Schwämmle, Automated workflow composition in mass spectrometry-based proteomics, Bioinformatics 35: (4) ((2019) ), 656–664. doi:10.1093/bioinformatics/bty646. |
[57] | C. Wroe, C. Goble, M. Greenwood, P. Lord, S. Miles, J. Papay, T. Payne and L. Moreau, Automating experiments using semantic data in a bioinformatics grid, IEEE Intelligent Systems 19: (1) ((2004) ), 48–55. doi:10.1109/MIS.2004.1265885. |
[58] | Common Workflow Language, v1.0, figshare, 2016. doi:10.6084/m9.figshare.3115156.v2. |
[59] | F. Khan, S. Soiland-Reyes, R.O. Sinnott, A. Lonie, C. Goble and M.R. Crusoe, Sharing interoperable workflow provenance: A review of best practices and their practical application in CWLProv, GigaScience 8: (11) ((2018) ), giz095. doi:10.1093/gigascience/giz095. |
[60] | Open WDL. http://www.openwdl.org/ (accessed August 16, 2019). |
[61] | F.C.Y. Benureau and N.P. Rougier, Re-run, repeat, reproduce, reuse, replicate: Transforming code into scientific contributions, Frontiers in Neuroinformatics 11: ((2018) ), 69. doi:10.3389/fninf.2017.00069. |
[62] | SPDX Workgroup – a Linux Foundation Project, Software package data exchange. https://spdx.org/ (accessed August 16, 2019). |
[63] | T. Lebo, S. Sahu and D. MacGuiness, PROV-O: The PROV ontology. https://www.w3.org/TR/prov-o/ (accessed August 16, 2019). |
[64] | A.M. Smith, D.S. Katz and K.E. Niemeyer, Software citation principles, PeerJ Computer Science 2: ((2016) ), e86. doi:10.7717/peerj-cs.86. |
[65] | J. Hausman, S. Stall, J. Gallagher and M. Wu, Software and services citation guidelines and examples ver 1, ESIP, 2019. doi:10.6084/m9.figshare.7640426.v4. |
[66] | D.S. Katz and N.P. Chue Hong, Software citation in theory and practice, 2018. doi:10.1007/978-3-319-96418-8_34. |
[67] | A.J.G. Gray, C.A. Goble and R. Jimenez, Bioschemas: From potato salad to protein annotation, in: International Semantic Web Conference (Posters, Demos & Industry Tracks), (2017) . https://iswc2017.semanticweb.org/paper-579/ (accessed August 16, 2019). |
[68] | RDA Research Metadata Schemas WG. https://www.rd-alliance.org/groups/research-metadata-schemas-wg (accessed August 16, 2019). |
[69] | RDA Software Source Code Identification WG. https://rd-alliance.org/groups/software-source-code-identification-wg. |
[70] | P.J.C. Aerts, C. Hof, S. Sufi and C. Martinez-Ortiz, Sustainable software sustainability – Workshop report, DANS, SSI, Netherlands eScience Center, 2019. https://www.software.ac.uk/wosss19 (accessed August 16, 2019). |
[71] | P.A. Martinez, C. Erdmann, N. Simons et al., Top 10 FAIR data & software things, Zenodo, 2019. doi:10.5281/zenodo.2555498. |
[72] | N. Chue Hong and D.S. Katz, FAIR enough? Can we (already) benefit from applying the FAIR data principles to software?, figshare, 2018. doi:10.6084/m9.figshare.7449239.v2. |
[73] | P.J.C. Aerts, Sustainable software sustainability – Workshop report, DANS, 2017. doi:10.17026/dans-xfe-rn2w. |




