Regulation of Adult Neurogenesis 2.0 – Beyond Signaling Pathways and Transcriptional Regulators
The discovery of adult neurogenesis added a new layer of complexity to our understanding of the mechanisms underlying plasticity in the adult mammalian brain. After more than five decades of research, studies in adult rodents combining genetic and pharmacologic manipulations of neurogenesis with behavioral analyses have now convincingly proven that the life-long generation of new neurons in the dentate gyrus of the hippocampus, in the subventricular zone/olfactory bulb system, and potentially in the hypothalamus, is critical for neural circuit plasticity and for adaptation of the organism to a changing environment. Furthermore, analyses of preclinical models for human diseases not only suggest that perturbation of adult neurogenesis contributes to the pathogenesis of cognitive impairment and emotional symptoms in ageing, neurodegenerative and neurodevelopmental diseases but also raise the possibility that ameliorating neurogenesis deficits may be of considerable therapeutic benefit. Boosted by the proof that substantial generation of neurons occurs in some areas of the postnatal and adult human brain [1–4], there is an ever-increasing interest in determining the precise physiological function of the new neurons and in deciphering the molecular and cellular mechanisms underlying their life-long generation in the adult brain.
The origin of new neurons are neural stem/precursor cells (NSPCs) that are either quiescent or slowly proliferating. NSPCs give rise to neurons through a complex sequence of proliferation, differentiation and maturation steps that culminate in the functional integration of the neuron into an existing neural circuit. The adult-born neuron’s ability to powerfully modulate neural circuit function is not solely explained by its addition to the circuit, but also by its electrophysiological properties and connectivity, which are at least for a limited time highly distinct from the properties of its embryonically generated counterparts.
Transplantation experiments conducted around the turn of the millennium were the first to demonstrate that signals provided by the surrounding cells are critical regulators of adult neurogenesis leading to the concept that interaction of NSPCs with a neurogenic microenvironment or neurogenic niche is central for regulating neurogenesis. While the neurogenic niche concept originally focused on the role of the niche to permit neuronal differentiation of NSPCs, later experiments revealed that the environmentally derived signals are involved in the control of virtually every step of the development of the adult-born neuron. The identity of extrinsic signals has been gradually unraveled comprising developmental morphogens, neurotrophins, neurotransmitters, hormones, matrix proteins, and others. The regulation of adult neurogenesis by niche-derived factors has been the subject of a number of previous reviews, to which we would like to refer the reader for further reading [5–7].
As initially predicted by the expression of distinct developmental stage-specific markers and later on confirmed by transcriptomic analyses, developmental progression in the adult neurogenic lineage is accompanied by the initiation and shut-down of gene-expression programs. Consequently, transcription factors (TFs) as the principal regulators of gene expression and potential mediators of extrinsic signals in the regulation of adult neurogenesis have attracted major attention over the past decade and there is now a wealth of knowledge on TFs that are essential for defined stages of adult neurogenesis. The function of TFs and transcriptional networks in the regulation of adult neurogenesis has been extensively reviewed elsewhere [5, 6, 8].
While signaling pathways and transcriptional regulation are arguably central mechanisms in neurogenesis, they are likely part of a more complex and sophisticated regulatory network enabling the controlled generation of neurons from precursor cells in the adult brain. This series of reviews is dedicated to emerging mechanisms in the regulation of neurogenesis.
The first part of the review issue focuses on mechanisms other than signaling pathways and transcriptional regulation that control neurogenesis on the level of gene expression. Gene expression changes arise not only from activities of TFs and signaling pathways but are critically dependent on epigenetic mechanisms such as DNA methylation, histone modifications, chromatin remodeling, imprinting and non-coding RNAs. These mechanisms remain difficult to study in vivo as, e.g., biochemical analysis of epigenetic modifications of DNA or histones still require large amounts of cells, which with regard to cells of specific developmental stages of neurogenesis cannot be easily obtained. At the in vitro level, cell culture systems faithfully modeling all distinct developmental stages of adult neurogenesis are still lacking. Despite these technical obstacles, considerable insight into the role of epigenetics in regulating adult neurogenesis has now been gained through the genetic and pharmacologic manipulation of enzymes that serve as ‘writers’ or ‘erasers’ of epigenetic marks, and of enzymes that ‘read’ these modifications by binding and serving as effectors. In this issue, Jobe and Zhao will review the current knowledge on the function of DNA methylation in the regulation of neurogenesis and how the DNA methylation network connects to other epigenetic regulatory mechanisms such as histone modification and non-coding RNAs. Genomic imprinting has been primarily considered as an important mechanism to control gene-dosage in early embryonic development. There is now evidence that imprinting is critical for the long-term maintenance of the NSPC pool and that aberrations of imprinting in NSPCs may be linked to brain tumor formation. The article by Lozano-Urena and colleagues will highlight new perspectives on this exciting emerging field at the intersection of neurogenesis and cancer biology.
Gene expression is controlled also post-transcriptionally on the level of mRNA stability and mRNA translation. The miRNA-regulated mRNA degradation/translation blockade has long been recognized as a powerful element in the regulation of developmental processes. Bielefeld and colleagues will review the current knowledge of central miRNA players in neurogenesis and will give a glimpse of the therapeutic potential of miRNAs in adult neurogenesis-associated pathologies with a focus on epilepsy. Interestingly, TFs and epigenetic regulators appear to be targets of microRNAs in adult neurogenesis, and vice versa, evidencing a tremendously intricate multilevel control of gene expression. The article by Rolando and Taylor adds to this complexity by highlighting the role of a recently discovered pathway controlling RNA stability in adult NSPCs and provides a perspective on how this novel pathway participates in the regulation of NSPC fate decisions.
Comparative analyses of cellular transcriptomes and proteomes indicate that mRNA levels frequently do not reflect the levels of their corresponding proteins. With regard to adult neurogenesis, robust mRNA expression of neural fate determinants in the absence of the corresponding protein is hypothesized to prime NSPCs for activation and lineage differentiation. In their review, Baser and colleagues will focus on the role of protein synthesis in controlling NSPCs and neuronal differentiation at the post-transcriptional level and will discuss the function of RNA-binding proteins and RNA methylation as emerging regulators of translation in adult neurogenesis. They will also provide a perspective on the energetic advantage of keeping the rate of protein synthesis in NSPCs under tight control, which leads up to the second part of this review issue.
The generation of a functional neuron from a quiescent NSPC requires extensive changes in proliferative activity, cellular growth, and synaptic activity. These changes impose specific demands on the availability of energy equivalents and precursor molecules acting as building blocks for anabolic pathways. The reviews by Knobloch and Beckervordersandforth will focus on the evolving concept that the neuronal development is defined by developmental stage-specific metabolic codes, and that metabolic programs constrain and promote lineage progression in adult neurogenesis. The review by Knobloch will in particular highlight the role of lipid metabolism, which may not only serve as a stage-specific energy source but could provide precursors for membranes and modifications of proteins including histones, thereby linking metabolism to epigenetic regulation of neurogenesis. The article by Beckervordersandforth will review the current knowledge on mitochondria-dependent metabolism and cellular redox state in controlling lineage progression in adult neurogenesis. The review will also discuss the potential role of mitochondrial dysfunction in age-associated impairment of neurogenesis and its promise as therapeutic target to enhance neurogenesis-dependent plasticity in the ageing brain.
Autophagy sits at the crossroads between cellular homeostasis and metabolism. Through its central function in quality control and clearance of functionally compromised cellular components, including dysfunctional mitochondria, autophagy ensures that degradation products are recycled as building blocks for new cellular components or are used as energy source. NSPCs undergo extensive structural and morphological changes en route to their differentiation into a mature neuron. NSPCs, immature neurons, and functionally integrated neurons differ greatly in their cytoskeletal components, their organelle content and shape, cellular growth, and their ability to form and retract contacts through processes and synapses. It is tempting to speculate that autophagic activity and its precise regulation may be of central importance to restructure adult-born neurons during their development. Moreover, NSPCs also experience large metabolic changes when they become activated, requiring an increased biosynthesis that in turn may depend on the cell’s recycling capacity. In this regard, and in agreement with studies performed in other adult tissues, autophagy may be also a crucial checkpoint in the activation of quiescent NSPCs. Indeed, knockout studies of key genes in the autophagic process have been reported to impact on neurogenesis. In their article, Dhaliwal and Lagace will provide a perspective on the emerging role of autophagy in the regulation of adult neurogenesis.
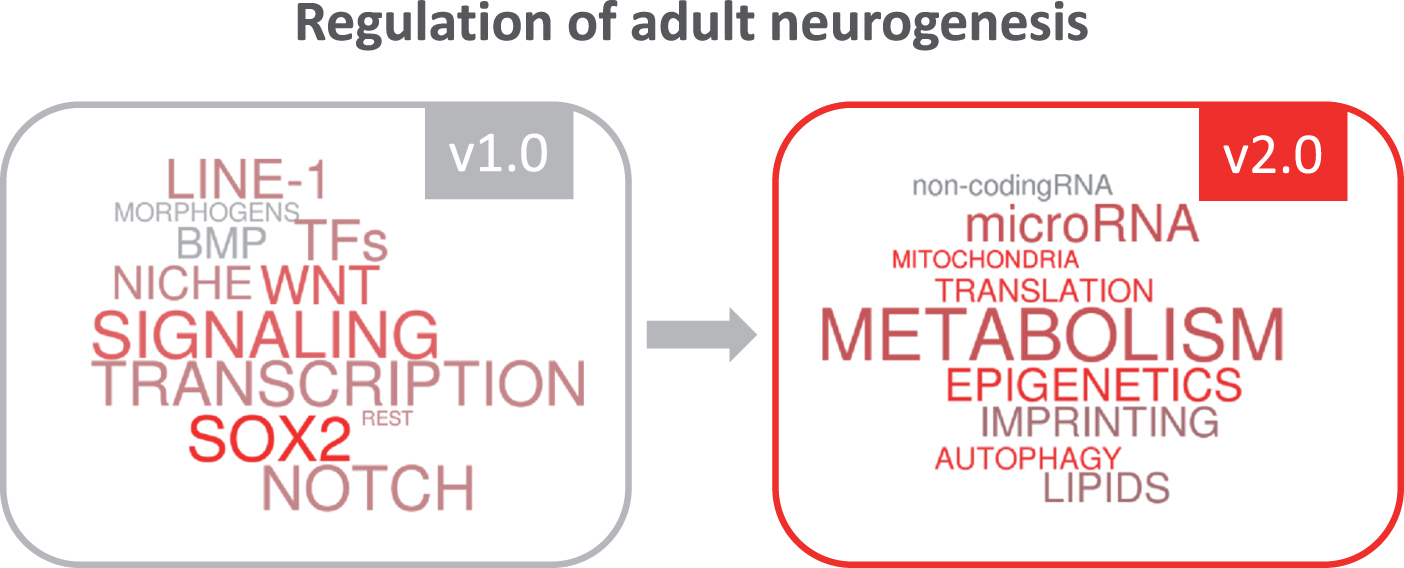
Over the last few years, new actors have gradually emerged on the scene of adult neurogenesis, inspiring us in the elaboration of this special issue. Our vision of the regulation of this complex process of brain plasticity has been turning from a first conceptual view based on “classic players” such as transcription factors or niche signals (version 1.0 of the regulation of adult neurogenesis), to a second more complete and updated view (version 2.0), which now integrates evolving regulatory functions of epigenetic and metabolic mechanisms (Fig. 1). We are aware that some new emerging key players have not been covered in the selection of reviews of the issue and we apologize in advance for that. Given the dizzying speed of the field, it is difficult to compile a volume with up-to-date information that includes last-minute findings. We hope that new and exciting data on metabolism, epigenetics and translational control will come to light as we proceed to the final publication of this v2.0 of the regulation of adult neurogenesis. It is to be expected that our knowledge will soon evolve towards the v3.0, in which we will probably see the crystallization of the concepts emerging today and the flourishing of exciting new ideas.
Fig.1
The growing insight into regulatory mechanisms in adult neurogenesis.
REFERENCES
[1] | Eriksson, et al. Nat Med (1998) ;4: :1313–17. |
[2] | Sanai, et al. Nature (2011) ;478: :382–6. |
[3] | Spalding, et al. Cell (2013) ;153: :1219–27. |
[4] | Ernst, et al. Cell (2014) ;156: :1072–83. |
[5] | Ming and Song. Neuron (2011) ;70: :687–702. |
[6] | Rolando and Taylor. Curr Top Dev Biol (2014) ;107: :183–206. |
[7] | Choe, et al. Cold Spring Harb Perspect Biol. (2015) ;7: (10):a018879. |
[8] | Beckervordersandforth, et al. Cold Spring Harb Perspect Biol. (2015) ;8: (3):a018887. |



