Association of Histone H3 Trimethylation in Circulating Monocytes with Lack of Early Recurrence in Patients with Bladder Cancer following BCG Induction Therapy
Abstract
BACKGROUND:
The mode of action of Bacillus Calmette-Guérin (BCG) in the treatment of patients with non-muscle invasive bladder cancer (NMIBC) is incompletely understood, but recent studies support an association between BCG-induced trained immunity in circulating monocytes and disease-free survival.
OBJECTIVE:
We compared epigenetic profiles in monocytes from NMIBC patients with early disease recurrence with those from recurrence-free patients.
METHODS:
We conducted chromatin immunoprecipitation and DNA sequencing (ChIP-seq) on monocytes from seven patients treated with BCG (four with early recurrences and three recurrence-free after one year) to determine genome-wide distribution and abundance of histone 3 lysine 4 trimethylation (H3K4me3) prior to and after five weeks of induction therapy.
RESULTS:
Genome-wide H3K4me3 profiles before or after BCG induction distinguished patients with early recurrences from those remaining recurrence-free. Furthermore, H3K4me3 levels at genes involved in specific pathways were increased in the recurrence-free group. Independent quantification showed increased H3K4me3 levels in elements of the Wnt and AMPK signaling pathways in the recurrence-free group before BCG initiation, while elements of the MAPK showed increased levels after five weeks of induction in the same group. Validation of these genes on an independent cohort of four additional patients that remained recurrence-free after one year and three with early recurrences revealed consistent increases in H3K4me3 levels associated with MAPK pathway genes after five weeks of BCG treatment in the recurrence-free group.
CONCLUSIONS:
Recurrence-free survival following BCG immunotherapy for NMIBC is associated with the accumulation of H3K4me3 at specific gene loci, and could lead to identification of prognostic biomarkers.
BACKGROUND
Up to 50% of patients with non-muscle invasive bladder cancer (NMIBC) suffer from early recurrence after tumour resection and adjuvant immunotherapy with Bacillus Calmette-Guérin (BCG), with some eventually progressing to muscle-invasive disease [1, 2]. Furthermore, the majority of patients treated with BCG suffer local and systemic side effects such as bladder irritation, malaise, and fever [3]. Thus, there is a need for biomarkers that can be used for the early identification of patients who are unlikely to benefit from BCG therapy.
We recently reported results of a longitudinal study that provided evidence of an association between BCG-mediated acquisition of trained immunity in peripheral blood monocytes and disease-free survival in patients with NMIBC [4]. Trained immunity, a form of innate immune memory, is acquired upon stimulation of pattern recognition receptors (PRRs) on innate immune cells with microbial ligands called pathogen-associated molecular patterns (PAMPs), or with damage-associated molecular patterns (DAMPs), which are endogenous non-infectious molecules released after tissue injury. Trained immunity is characterized by enhanced non-specific Th1 inflammatory responses following a secondary inflammatory challenge [5, 6].
At the molecular level, the acquisition of trained immunity involves metabolic reprogramming and modifications of histones associated with inflammatory genes. Combinations of methylation at specific lysine residues of histone 3 are important determinants of the transcriptional permissiveness of chromatin at corresponding gene loci. While di- and tri-methylation of lysine 9 (H3K9me2/3) generally leads to condensed chromatin and inhibition of transcriptional activity, mono- and tri-methylation of lysine 4 (H3K4me1 and H3K4me3) results in decondensation of chromatin at enhancer and promoter regions respectively. In the case of trained immunity, these latter modifications facilitate enhanced and rapid transcriptional activation of inflammatory genes [7]. To explore a potential prognostic value of H3K4me3 in monocytes from patients with NMIBC before and after BCG therapy, we performed chromatin immunoprecipitation (ChIP) followed by TLR-4 and TNFα promoter-specific PCR amplification of co-precipitated DNA as well as high-throughput DNA sequencing (ChIP-seq) to determine genome-wide distribution and abundance of modified H3K4me3-bound targets. We selected TLR-4 and TNFα promoters for the first experimental approach because increased H3K4me3 presence at these promoters is a feature of trained immunity in monocytes [8, 9]. ChIP-seq was performed to further identify potential genes and biological pathways associated with early recurrence or remission. We present here putative genes and biological pathways that are associated with early recurrence or remission, and we suggest that these may underpin trained immunity in NMIBC.
MATERIALS AND METHODS
Study participants
A subset of seven patients from our previous study [4], recruited between September 2018 and March 2019, was selected for ChIP-seq analysis. Patients were recruited following informed consent and protocol approval by the Queen’s University Health Sciences and Affiliated Hospitals Research Ethics Board (approval number UROL-282-13). All resected tumors were at the Ta stage but varied in grade and American Urological Association risk (see Table 1). Four of these participants suffered neoplastic recurrence within a year while three remained in remission at the time of this study. For ChIP-qPCR validations, seven additional patients with Ta or T1 stage tumors (recruited between March 2019 and October 2019; Table 2) were selected. Three of these patients suffered recurrences within a year of therapy initiation. All patients were BCG naïve prior to enrollment in this study.
Table 1
Characteristics of patients enrolled in the ChIP-seq study
| Patient # | Sex | Age | Stage | Grade | AUA risk | Days to recurrence |
| 29 | M | 69 | Ta | high | 3 | 83 |
| 38* | M | 88 | Ta | low | 2 | n/a |
| 41* | F | 86 | Ta | high | 3 | 154 |
| 42 | M | 71 | Ta | high | 2 | 305 |
| 43* | M | 80 | Ta | high | 2 | 73 |
| 46* | M | 84 | Ta | high w/ CIS | 3 | n/a |
| 54* | M | 70 | Ta | high | 2 | n/a |
Participants used for ChIP-qPCR confirmation (*).
Table 2
Characteristics of patients enrolled for validation of H3K4me3 target genes
| Patient # | Sex | Age | Stage | Grade | AUA risk | Days to recurrence |
| 55 | M | 71 | Ta | high w/ CIS | 3 | n/a |
| 76 | M | 84 | T1 | high | 3 | n/a |
| 79 | M | 81 | Ta | high | 2 | n/a |
| 80 | M | 70 | Ta | high | 3 | 201 |
| 81 | M | 80 | T1 | high w/ CIS | 3 | 91 |
| 87 | M | 75 | T1 | high w/ CIS | 3 | n/a |
| 88 | M | 78 | Ta | high | 2 | 104 |
Blood collection and cell isolations
Peripheral blood (20 mL) was collected during patient clinical visits at two time-points (Fig. 1A): before the first BCG instillation (week one) and before the sixth BCG instillation (week six). Blood was collected in Vacutainer K2-EDTA-K tubes (Becton Dickinson, Mississauga, ON) and transferred to the laboratory within two hours for isolation of peripheral blood mononuclear cells (PBMCs) by density gradient centrifugation using Lymphoprep (STEMCELL Technologies, Vancouver, Canada) according to the manufacturer’s instructions. Cells were frozen in 8% DMSO/fetal bovine serum at a concentration of 1×107 cells per vial (1 mL) and stored in liquid nitrogen. Monocytes were subsequently isolated from thawed PBMCs using an EasySepTM Human Monocyte Isolation Kit (STEMCELL Technologies).
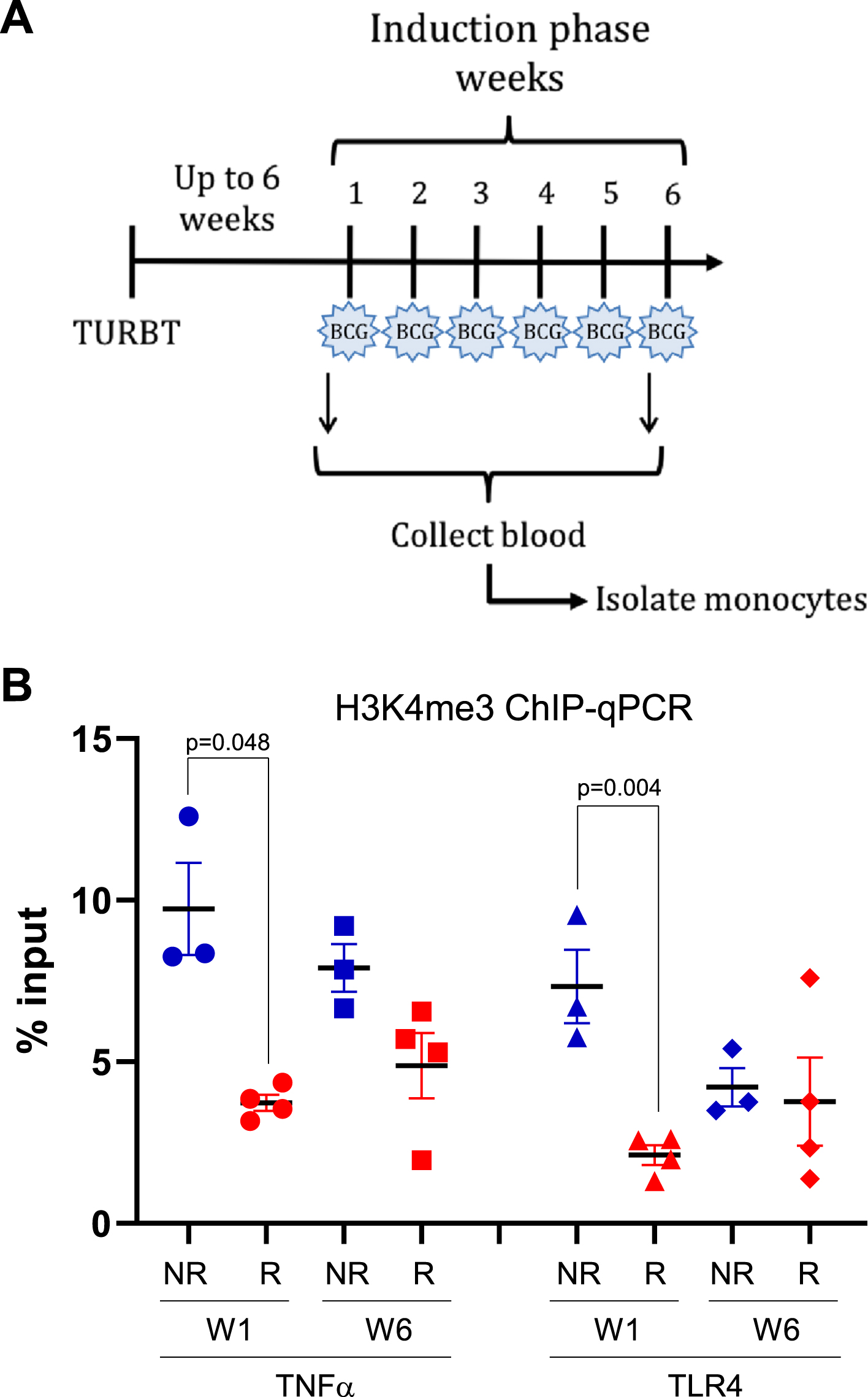
Fig. 1
H3K4me3 methylation at promoters of innate immune memory-related genes. A) peripheral blood from patients with NMIBC was collected before the first (week 1) and last (week 6) intravesical BCG instillation of the first induction phase. TURBT: transurethral resection of bladder tumor; BCG: bacillus Calmette-Guérin. B) H3K4me3 in monocytes at promoters of TNFα and TLR4 genes before the first BCG instillation (week 1) and the last (week 6). Blue solid shapes: no-early recurrence (NR) patients used for ChIP-seq; red solid shapes: early recurrence (R) patients used for ChIP-seq; circles: week 1 (pre-BCG); squares: week 6 (before 6th BCG instillation).
Chromatin immunoprecipitation
For each chromatin immunoprecipitation (ChIP) reaction, one million freshly purified monocytes were pelleted, and their chromatin was cross-linked in 1% paraformaldehyde for 10 minutes at room temperature. Cross-linking of chromatin was stopped by adding glycine to a final concentration of 125 mM and incubating under rotation for five minutes at room temperature. Cell pellets were washed twice with ice-cold PBS and stored at –80°C until sonication. Before sonication, nuclei were prepared according to the truChIP Chromatin Shearing Kit (Covaris, Woburn, MA) low cell protocol. Sonication was performed using a Covaris M220 focused ultrasonicator under the following conditions: PIP = 75, duty factor = 5%, CPB = 200, time = 10 minutes, temperature = 7°C. Immunoprecipitation was performed on fragmented chromatin using a rabbit monoclonal anti-H3K4me3 antibody (Cell Signaling Technology, Danvers, MA) and SimpleChIP Plus Sonication IP Kit reagents (Cell Signaling Technology), from which antibody and protein G magnetic beads amounts were adjusted for the number of cells used.
DNA amplification
Immunoprecipitated DNA was analyzed by qPCR using SYBR Green PCR Master Mix (Applied Biosystems – Fisher Scientific, Ottawa, ON, Canada) on a Roche Lightcycler 480. Amplification conditions included one cycle of 10 minutes at 95°C followed by 40 cycles of amplification (20 seconds at 95°C and one minute at 60°C per cycle). Percent input values were calculated to quantify the relative levels of immunoprecipitated histone. Primers used are listed in Table 3.
Table 3
Primers used for ChIP-qPCR
| Primer name | Sequence | Genome coordinates |
| AKT3-F | gtgcccaatgaggtagggac | chr1:243851071-243851090 |
| AKT3-R | tcttccccgaggtgcagg | chr1:243851145-243851128 |
| FZD5-F | gaccacagcggactcacg | chr2:207769868-207769885 |
| FZD5-R | cgggcattagaagcaggtga | chr2:207769941-207769922 |
| IRS2-F | agtgagtaacacatcgcgca | chr13:109786574-109786593 |
| IRS2-R | gcgtaacgccgagtcacat | chr13:109786644-109786626 |
| LEPR-F | cggggctctgcgtgg | chr1:65420605-65420619 |
| LEPR-R | gcagcctgcccaagcc | chr1:65420674-65420659 |
| MET-F | gactaggggacggacagca | chr7:116672165-116672183 |
| MET-R | caggcgaccagactgagg | chr7:116672242-116672225 |
| TLR4-F | cctttagcccagaactgctttg | chr9:117704314-117704335 |
| TLR4-R | ggctcgctatcaccgtctg | chr9:117704420-117704402 |
| TNFα-F | atgattctttccccgccctc | chr6:31575499-315755518 |
| TNFα-R | ctggtcctctgctgtccttg | chr6:31575601-315755582 |
Sequencing of immunoprecipitated DNA and analysis
Chromatin immunoprecipitation sequencing (ChIP-seq) libraries were constructed using SimpleChIP ChIP-seq DNA Library Prep Kit for Illumina and SimpleChIP ChIP-seq Multiplex Oligos for Illumina (Cell Signaling Technology) according to the manufacturer’s instructions. ChIP-seq libraries were subjected to quality control and the size of each library was quantified so that library concentrations (nM) could be calculated. Libraries were pooled and purified prior to dilution to 4 nM and confirmed in triplicate using a Qubit 4 Fluorometer (ThermoFisher Scientific, Mississauga, ON, Canada). The library pool was denatured and diluted to 1.8 pM in 1.3 mL of HT1 solution and paired-end sequenced with the Illumina NextSeq550 system using the HIGH output v2.5 flowcell resulting in >25 million PE reads per sample. Data were stored in the secure server of the Queen’s University Centre for Advanced Computing. Demultiplexing and Fastq generation were processed using bcl2fastq conversion software (Illumina). Fastq files were aligned using Bowtie against HG38 and duplicates were filtered using Samtools. Peak calling was performed using HOMER [10] with length between peaks and length of a peak set to 370 and 250, respectively.
To identify peaks that best discriminate between patients who suffered from early disease recurrence and patients with no early recurrence at different time points, we used a machine learning-based feature selection algorithm (MFeaST) [11]. The algorithm relies on a set of machine learning techniques to rank the features based on their ability to discriminate between two groups and is highly effective at identifying molecular patterns among samples. The following pair-wise comparisons were performed: early recurrence week one (pre-BCG) versus no early recurrence week one (post-BCG), and early recurrence week six versus no early recurrence week six. To prepare for the MFeaST, the peaks data were segregated by chromosome. For each chromosome, a data matrix was generated where the intensity of a peak was recorded for each sample and for each genomic location block of 400 base pairs. Blocks with no peaks in more than one sample were filtered out. For each comparison category, the processed data were run twice through MFeaST with leave-one-out validation and all available algorithms. First, for each chromosome, the top 5% of the ranked blocks in each chromosome were selected and combined into a single data matrix. The combined data were then passed through MFeaST to generate the final combined ranking. From the top 5% of combined top ranked peaks, only the promoter and exon peaks were used in the subsequent analysis. For each peak, a median-based fold-change value was calculated in each comparison category (e.g., week one versus week six, etc.). Peaks with log2 fold change of ≥2 were used in pathway analysis to identify enriched peaks (2625 genes for week one and 2735 genes for week six). The aggregate output files from these data are presented in the following location (DOI: 10.17632/vgt4jzvfg4.1). Raw data are available upon request to the corresponding authors.
Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed by applying the “clusterprofiler” package [12]. GO terms were prioritized using a corrected false discovery rate (FDR;<0.05). The significantly enriched KEGG signaling pathways were displayed in the form of a dot plot [13]. Genes involved in Wnt, AMPK and MAPK signaling pathways were selected for validation. The Integrative Genomics Viewer software (IGV, v2.9.4) [14] was used to visualize the selected candidates to confirm the genomic locations of the peaks to be used in the validation.
Validation of differentially active pathways
To validate the newly identified pathways enriched in patients with no early recurrences, we performed an independent H3K4me3 immunoprecipitation experiment on monocytes from the same patients as well as an additional independent cohort using the monoclonal antibody (Cell Signaling Technology) used earlier for ChIP-seq. Primers within the identified differential peaks were used to quantify H3K4me3 in immunoprecipitated chromatin by qPCR on the co-precipitated DNA (see Table 2 for primers).
Quantification of transcript levels
RNA was extracted from flash frozen monocytes of NMIBC patients using a PureLinkTM RNA Mini Kit (Invitrogen). One hundred nanograms of RNA was used for reverse transcription using an AMV 1st Strand cDNA Synthesis Kit (Roche). qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) on a Roche Lightcycler 480 system with the following amplification conditions: one cycle of 10 minutes at 95°C followed by 40 cycles of amplification (20 seconds at 95°C and one minute at 60°C per cycle). GAPDH was used as a reference for relative quantitation by ΔCt. The sequences for primers used were as follows: TNFα sense 5’-CCTCTCTCTAATCAGCCCTCTG-3’; TNFα antisense 5’-GAGGACCTGGGAGTAGATGAG-3’; TLR4 sense 5’-AGACCTGTCCCTGAACCCTAT-3’; TLR4 antisense 5’-CGATGGACTTCTAAACCAGCCA-3’; GAPDH sense 5’-GGAGCGAGATCCCTCCAAAAT-3’; GAPDH antisense 5’-GGCTGTTGTCATACTTCTCATGG-3’.
Statistical analysis
Data processing and visualization were performed in MATLAB (Mathworks, Inc., MA, USA, v. R2020a) and GraphPad Prism (GraphPad Software Inc., MA, USA, v. 9.5.1). Clustering results were generated using the t-distributed stochastic neighbor embedding (tSNE) [15]. A two-sided t-test was used to assess differences between no-early recurrence and early recurrence groups at weeks one and six. A one-sided t-test was used for the analysis of the validation results, since the direction of the effect was known. Assumptions of all statistical tests were checked and satisfied. A p-value <0.05 was considered statistically significant.
RESULTS
H3K4me3 at inflammatory gene promoters
Since BCG-induced trained immunity leads to the accumulation of histone 3 lysine 4 trimethylation at pro-inflammatory gene promoters [16], we performed ChIP on chromatin extracted from monocytes using an H3K4me3-specific antibody followed by gene promoter-specific amplification of immunoprecipitated DNA. Specifically, we amplified DNA associated with the promoters of the TNFα and TLR4 genes (known to play a role in trained immunity [17]) in monocytes isolated before treatment (week one) and prior to the last weekly instillation of BCG (week six). Patients who remained recurrence-free for one year (no-early recurrence) had significantly higher H3K4me3 modifications at gene promoters for both TNFα and TLR4 locations in monocytes isolated prior to BCG induction, compared to patients who suffered early recurrences (Fig. 1B, Supplementary Table 1). However, the significance of these differences weakened after five rounds of BCG instillations (Supplementary Table 1). This indicates that, for some NMIBC patients, inflammatory genes may be more transcriptionally accessible even before BCG induction therapy, and could indicate a favorable response to treatment.
Epigenomic profiles of monocytes from patients with and without early recurrence
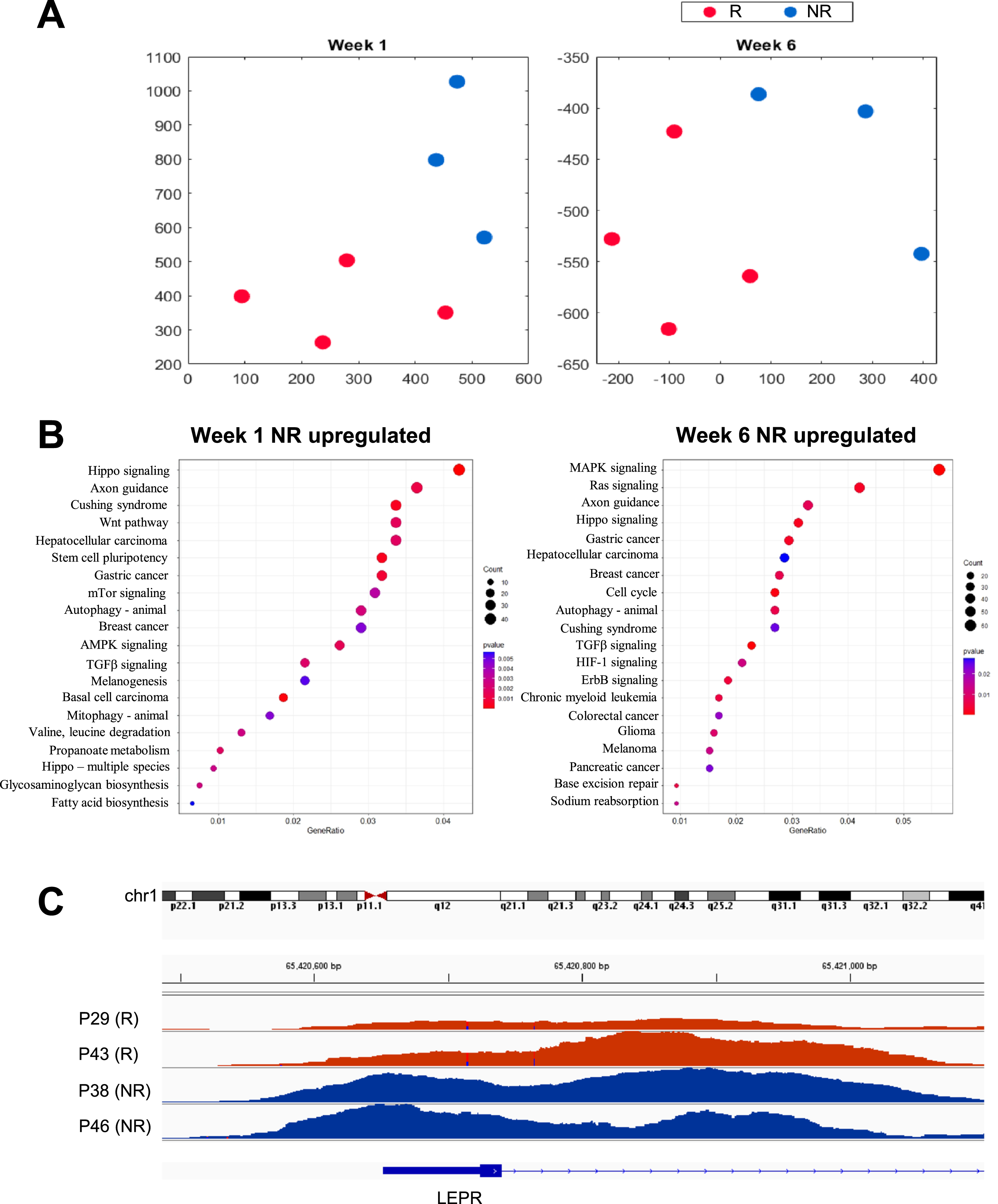
To determine whether monocytes from patients without early recurrences had a specific epigenomic profile, we performed ChIP-seq on DNA associated with chromatin regions immunoprecipitated with the anti-H3K4me3 antibody. We selected monocytes from the three patients without early recurrences previously identified as showing higher H3K4me3 histone modifications at inflammatory gene promoters as well as the four patients who suffered high-grade early recurrences (Table 1). ChIP-seq profiles were generated using monocytes from these patients at week one and week six. Initial analysis of the peak-calling data showed that the patient group without recurrence at one year had more peaks with average higher scores at the promoter and exon locations compared to the patient group with early recurrence at week one and week six (differences not statistically significant, Supplementary Table 2). To identify genomic locations that are most discriminating between the early recurrence and no early recurrence groups, we performed feature selection using MFeast [11]. The number of peaks at the selected locations was significantly higher in the no early recurrence group compared with the early recurrence group at both week-one and week-six time points (p = 0.02, <0.001, respectively, Supplementary Table 2). Consequently, on average, the peak scores at the selected locations were also higher in the group with no early recurrence at week one and week six (p = 0.04, Supplementary Table 2, Supplementary Figure 1B). Clustering analysis of the selected peak locations revealed that the H3K4me3 epigenomic profiles of patients with and without early recurrence localized to distinct clusters before BCG treatment was initiated (Fig. 2A), suggesting the presence of a distinct chromatin accessibility profile in patients responding successfully to BCG immunotherapy.
GO and KEGG pathway analyses of H3K4me3-associated genes enriched in the groups with and without early recurrence revealed that, at week one, the most significantly enriched pathways in the group without early recurrence included genes involved in Wnt signaling (overlapping multiple GO pathways), as well as in AMPK signaling. At week six, elements of the MAPK signaling pathway were the most enriched in the group without early recurrence (Fig. 2B).
Fig. 2
High-dimensional arrangement and pathway enrichment from H3K4me3 epigenomic profiles of monocytes from patients with NMIBC. A) Two-dimensional t-SNE embeddings of data from no early recurrence (NR, blue) and early recurrence (R, red) samples at week 1 (left panel) and week 6 (right panel). B) Gene Ontology (GO) terms mostly enriched among genes upregulated in recurrence-free patients at week1 one (left panel) and week 6 (right panel). C) Example of the LEPR promoter element that shows noticeable differences in monocytes from recurrence-free (blue) versus early recurrence (red) patients.
Validation of H3K4me3 target genes enriched in monocytes from patients with no early recurrence
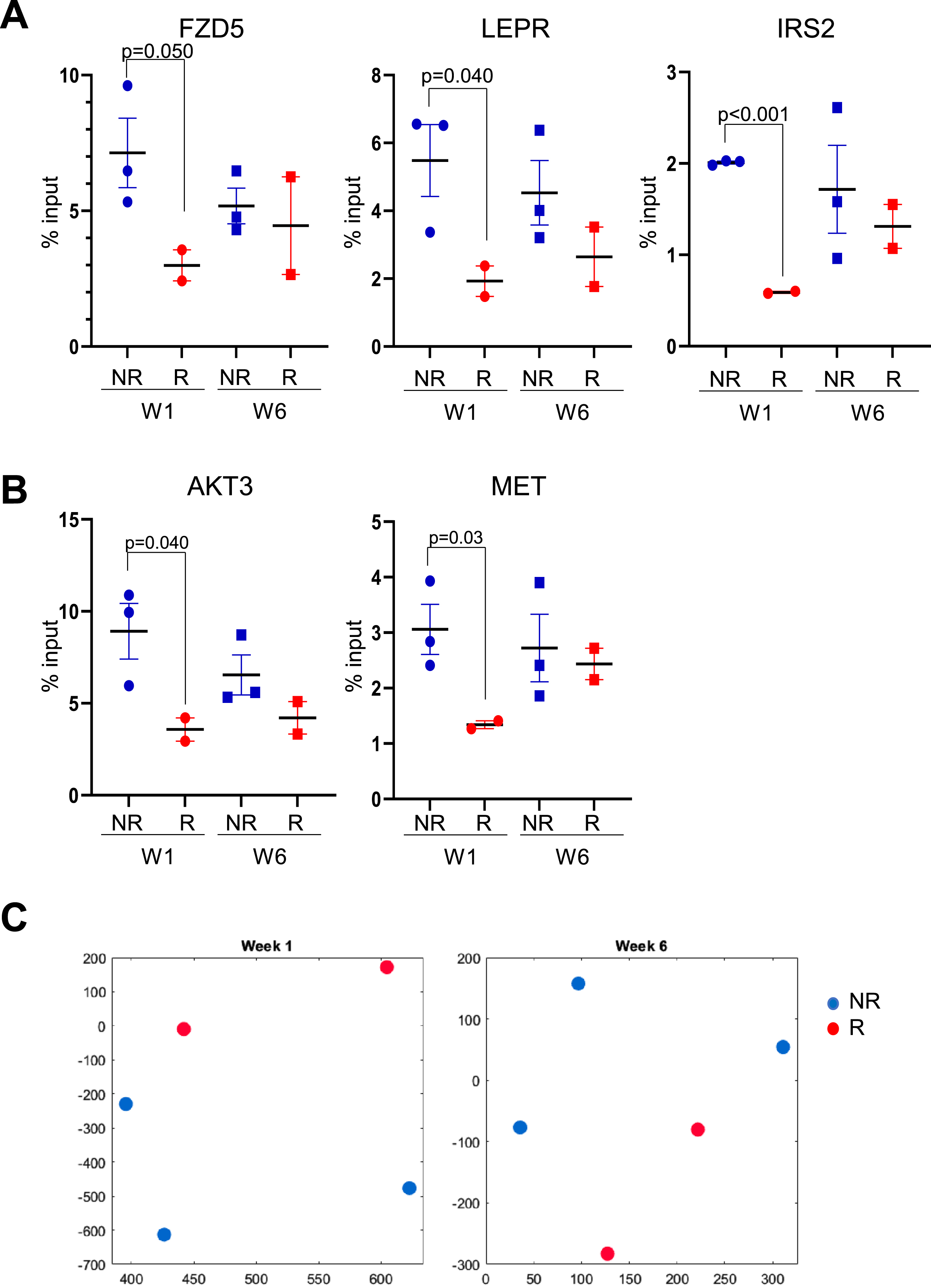
To confirm that these identified pathways were associated with monocytes from patients without early recurrent disease, we first performed an independent ChIP on a sample taken from the same population of monocytes used for ChIP-seq. Due to limitations in the number of monocytes isolated from each patient, we were only able to perform this confirmation experiment using monocytes from five of the seven patients used for the ChIP-seq study (set 1, two patients with early recurrence and three without early recurrence, indicated by * in Table 1). We first evaluated the H3K4me3 enrichment at promoter sequences from a gene involved in Wnt signaling (FZD5) that was repeatedly identified in the pathways upregulated in monocytes from week-one patients without early recurrence, as well as promoter sequences of genes from the AMPK signaling pathway (LEPR, IRS2; LEPR peaks visualized in Fig. 2C). Results showed that the levels of H3K4me3 associated with these three gene promoter regions were higher in recurrence-free patients than in patients with early recurrence at week one (Fig. 3A, Supplementary Table 3). We also found that the levels of H3K4me3 enriched for two genes involved in MAPK signaling (AKT3, MET) initially identified following ChIP-seq pathway analysis were higher at week six in monocytes from recurrence-free patients when compared with monocytes from patients with early recurrence, although the difference was not significant (Fig. 3B, Supplementary Table 3). These results show that the candidate genes identified in the ChIP-seq study were representative of H3K4me3 enrichment present in the patients’ monocytes. The levels of H3K4me3 enrichment obtained for the genes involved in the identified pathways were used to cluster the patients before and at the end of treatment by t-SNE. We found that the patients evaluated by qPCR for enriched pathways genes were clustering distinctly between early recurrence and no early recurrence (Fig. 3C). This showed that the clustering observed from the epigenomic profiles was maintained when using a subset of enriched genes quantified independently.
Fig. 3
Validation of gene promoter elements identified as enriched in patients without early recurrence. A) H3K4me3 in monocytes at the promoter sequences of FZD5, LEPR and IRS2. B) H3K4me3 in monocytes at promoter sequences of AKT3 and MET. Circles: week 1 (pre-BCG); squares: week 6 (before 6th BCG instillation) C) Two-dimensional t-SNE embeddings of ChIP-qPCR data at week 1 (left panel) and week 6 (right panel) obtained from monocyte samples of patients without early recurrence (NR, blue) and patients with early recurrence (R, red). Blue solid shapes: monocytes from patients with no early recurrence (NR) used for ChIP-seq; red solid shapes: monocytes from patients with early recurrence (R) used for ChIP-seq.
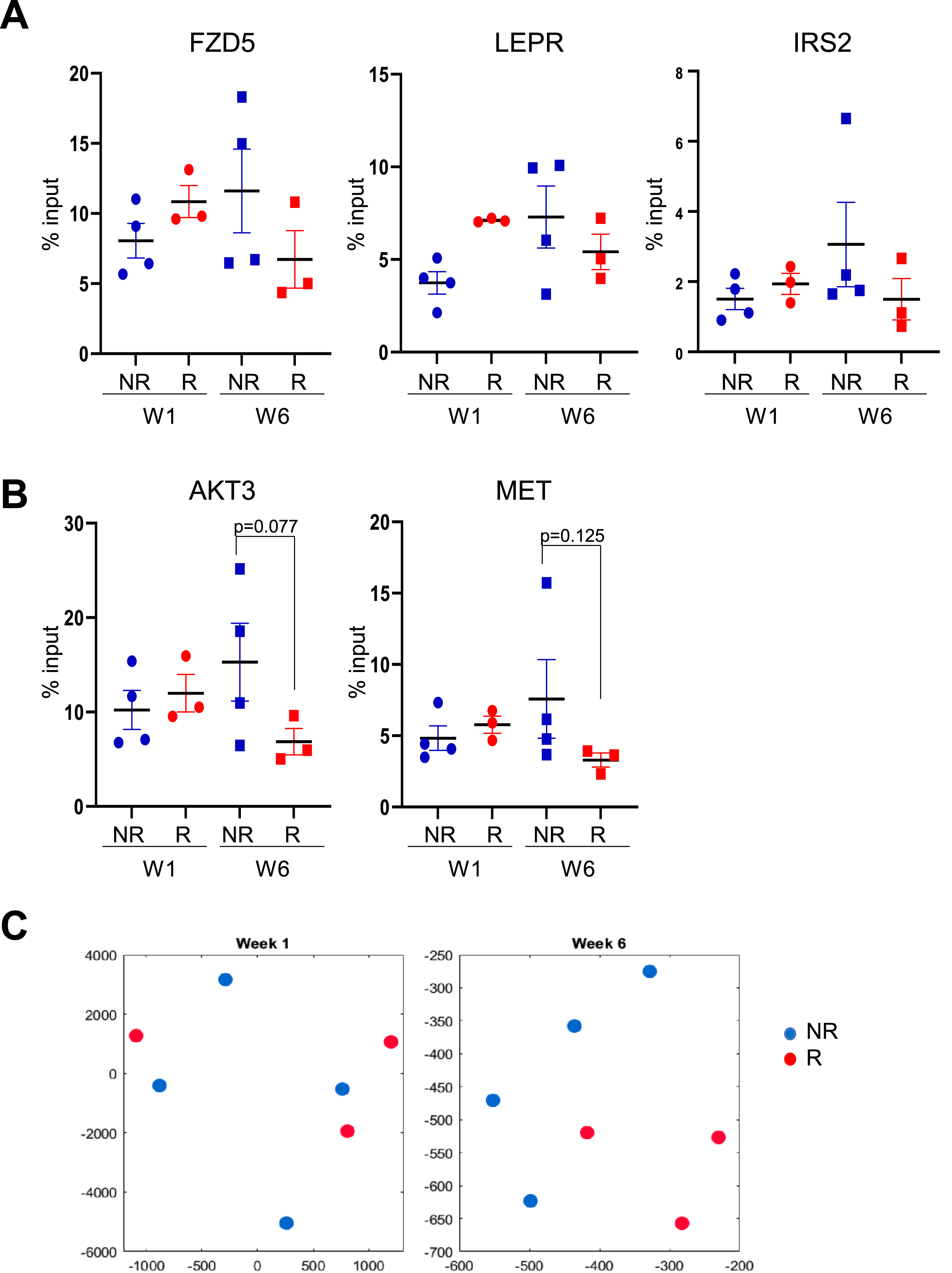
Further ChIP validation experiments using monocytes from seven independent patients with NMIBC (set 2, three with early recurrences and four without early recurrences) revealed that the higher enrichment of genes (AKT3 and MET) from the MAPK pathway at week six was maintained in this new subset of recurrence-free patients when compared to the new subset of patients who suffered early recurrence (Fig. 4B, Supplentary Table 3). However, the enrichment of pathways associated with recurrence-free status at week one (FZD5 for Wnt pathway, LEPR and IRS2 for AMPK pathway) was not maintained (Fig. 4A). These week-one pathway genes were more enriched at week six in patients without early recurrence when compared with week-six patients with early recurrence (Fig. 4B). Clustering of the candidate genes’ enrichment showed separation according to recurrence status at week six, but not at week one (Fig. 4C). Together, these results indicate that when expanded to patients not profiled in our H3K4me3 study, patients who remain recurrence-free for at least one year may not be identified through a single type of epigenomic H3K4me3 profile at week one of BCG induction therapy, but their profiles at week six reveal a stronger association with patients that did suffer early disease recurrence.
Fig. 4
Methylation levels on monocytes from an independent validation cohort of patients at gene promoters previously identified as enriched in the original group of patients without early disease recurrence. A) H3K4me3 in monocytes at gene promoters for FZD5, LEPR and IRS2 genes. B) H3K4me3 in monocytes at gene promoters for AKT3 and MET genes. C) Two-dimensional t-SNE embeddings of ChIP-qPCR data from monocytes of a validation cohort at week one (left panel) and week six (right panel). Blue solid shapes: monocytes from patients with no early recurrence (NR) used for ChIP-seq; red solid shapes: monocytes from patients with early recurrence (R) used for ChIP-seq.
DISCUSSION
In this study, we compared monocyte epigenomic profiles of NMIBC patients undergoing BCG immunotherapy before treatment initiation and just prior to the end of the induction treatment course. Results revealed a distinct pattern of H3K4me3 target genes associated with disease-free survival. Genes involved in the Wnt and AMPK pathways were enriched prior to BCG treatment in patients who remained recurrence-free for at least one year, whereas genes involved in the MAPK pathway were enriched after five instillations of BCG in these patients. While ChIP-qPCR performed on monocytes from additional patients did not show the enrichment of the few representative genes from the Wnt and AMPK pathways observed in monocytes from recurrence-free patients prior to BCG induction therapy, their enrichment following five instillations of BCG was maintained. Since these two pathways are known to be involved in myeloid cell differentiation [18–20], it is possible that the chromatin status of their associated genes reflects the ability of monocytes from patients to respond to the BCG stimulus and trigger an effective anti-cancer response. Also, the enrichment of MAPK genes associated with another pathway active during myeloid differentiation [21, 22] was maintained following five BCG instillations in monocytes from patients without early recurrence.
Although the number of patient samples analyzed in this study was small and some of the identified changes were not statistically significant, the effect size (Cohen’s d) was large, and the results suggest that activation of these specific signaling pathways in circulating monocytes is associated with patients that remained disease-free for at least a year. Pathways identified as being significantly different in the gene set enrichment analysis can result from a combination of genes that do not exhibit statistically significant differences when analyzed individually. Therefore, the maintenance of the differences at single genes, although not always statistically significant, is most likely indicative of a representative feature of monocytes from patients who remained free of recurrence when compared to patients with early disease recurrence. Moreover, the increased peak intensity observed in monocyte samples from disease-free patients indicates a more relaxed chromatin state that would lead to an increased response to a therapeutic stimulus.
Although the pathways enriched in monocytes from disease-free patients are not the expected pro-inflammatory pathways characteristic of trained immunity, they point to other properties of monocytes that may be important for their anti-tumorigenic functions. For example, it has recently been demonstrated that monocytes respond to leptin secreted by surrounding adipocytes to regulate skin wound healing following bacterial infection [23]. The identification of the leptin receptor (LEPR) promoter as one epigenetic enrichment associated with disease-free survival in our study indicates that a similar function could mediate the immunotherapeutic benefit of BCG. As for the enrichment of the H3K4me3 epigenetic mark on genes from the Wnt and MAPK pathways, it is possible that it is a consequence of reprogramming of bone marrow myeloid progenitors following exposure to DAMPs released into the circulation by the treated bladder.
While we validated gene sets that may be involved in disease-free survival of patients with higher risk NMIBC, further identification and validation of genes differentially marked prior to BCG induction therapy is important. Since H3K4me3 is only one of several histone modifications that regulate the response to BCG, epigenomic analysis of additional modifications, e.g., H3K4me1 and H3K9me2, would be informative. It is also possible that only small subpopulations of monocytes mediate the response to BCG in NMIBC and, consequently, their epigenetic marks may remain undetected when analyzed in pooled populations of monocytes. This potential limitation could be overcome by performing epigenomic analysis at the single-cell level, such as scATAC-seq analysis. Finally, one should consider the possibility that other innate immune cell populations, e.g., neutrophils and NK cells which are also known to acquire memory, may contribute to the immunotherapeutic benefit of BCG.
In conclusion, we have identified pathways whose genes transcriptional accessibility during treatment is linked to successful BCG immunotherapy in NMIBC patients. Monitoring these pathways early after the induction of BCG treatment could help identify patients who are more likely to respond successfully and, consequently, reduce the number of unnecessary BCG treatments and recurrences by switching the less responsive patients to alternative interventions.
ACKNOWLEDGMENTS
This research was supported by grants from the Southeastern Ontario Medical Organization (SEAMO) AHSC AFP Innovation Fund and the Canadian Institutes of Health Research (grant number PJT-173383).
AUTHOR CONTRIBUTIONS
JFP, DRS and CHG contributed to the conception of this study. JFP, MT, AG, AA, CCTH and KT contributed to performance of work. JFP, MT, CCTH, KT, and CGH contributed to writing of the article. All authors contributed to interpretation of the data. All authors had access to the data.
CONFLICTS OF INTEREST
JFP, MT, AG, AA, CCTH, KT, DRS and CG have no conflict of interest to report.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/BLC-230028.
REFERENCES
[1] | Zlotta AR , Fleshner NE , Jewett MA . The management of BCG failure in non-muscle-invasive bladder cancer: An update. Can Urol Assoc J. (2009) ;3: (6 Suppl 4):S199–205. |
[2] | Witjes JA . Management of BCG failures in superficial bladder cancer: A review. Eur Urol. (2006) ;49: (5):790–7. |
[3] | Decaestecker K , Oosterlinck W . Managing the adverse events of intravesical bacillus Calmette-Guerin therapy. Res Rep Urol. (2015) ;7: :157–63. |
[4] | Graham CH , Pare JF , Cotechini T , Hopman W , Hindmarch CCT , Ghaffari A , et al. Innate immune memory is associated with increased disease-free survival in bladder cancer patients treated with bacillus Calmette-Guerin. Can Urol Assoc J. (2021) ;15: (8):E412–E7. |
[5] | Netea MG , Joosten LA , Latz E , Mills KH , Natoli G , Stunnenberg HG , et al. Trained immunity: A program of innate immune memory in health and disease. Science. (2016) ;352: (6284):aaf1098. |
[6] | Crisan TO , Netea MG , Joosten LA . Innate immune memory: Implications for host responses to damage-associated molecular patterns. Eur J Immunol. (2016) ;46: (4):817–28. |
[7] | Netea MG , Dominguez-Andres J , Barreiro LB , Chavakis T , Divangahi M , Fuchs E , et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. (2020) ;20: (6):375–88. |
[8] | Bhattarai S , Li Q , Ding J , Liang F , Gusev E , Lapohos O , et al. TLR4 is a regulator of trained immunity in a murine model of Duchenne muscular dystrophy. Nat Commun. (2022) ;13: (1):879. |
[9] | Bekkering S , Arts RJW , Novakovic B , Kourtzelis I , van der Heijden C , Li Y , et al. Metabolic induction of trained immunity through the mevalonate pathway. Cell. (2018) ;172: (1-2):135–46e9. |
[10] | Heinz S , Benner C , Spann N , Bertolino E , Lin YC , Laslo P , et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. (2010) ;38: (4):576–89. |
[11] | Gerolami J , Wong JJM , Zhang R , Chen T , Imtiaz T , Smith M , et al. A computational approach to identification of candidate biomarkers in high-dimensional molecular data. Diagnostics (Basel). (2022) ;12: (8). |
[12] | Yu G , Wang LG , Han Y , He QY . clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. (2012) ;16: (5):284–7. |
[13] | Wickham H Ggplot2: Elegant graphics for data analysis (2nd ed): Springer International Publishing; 2016. |
[14] | Robinson JT , Thorvaldsdottir H , Winckler W , Guttman M , Lander ES , Getz G , et al. Integrative genomics viewer. Nat Biotechnol. (2011) ;29: (1):24–6. |
[15] | van der Maaten L , Hinton, G . Visualizing data using t-SNE. Journal of Machine Learning Research. (2008) ;9: :2579–805. |
[16] | Arts RJW , Moorlag S , Novakovic B , Li Y , Wang SY , Oosting M , et al. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. (2018) ;23: (1):89–100e5. |
[17] | Owen AM , Fults JB , Patil NK , Hernandez A , Bohannon JK . TLR agonists as mediators of trained immunity: Mechanistic insight and immunotherapeutic potential to combat infection. Front Immunol. (2020) ;11: :622614. |
[18] | Blumenthal A , Ehlers S , Lauber J , Buer J , Lange C , Goldmann T , et al. The Wingless homolog WNT5A and its receptor Frizzled-5 regulate inflammatory responses of human mononuclear cells induced by microbial stimulation. Blood. (2006) ;108: (3):965–73. |
[19] | Lento W , Congdon K , Voermans C , Kritzik M , Reya T . Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb Perspect Biol. (2013) ;5: (2). |
[20] | Jacquel A , Luciano F , Robert G , Auberger P . Implication and regulation of AMPK during physiological and pathological myeloid differentiation. Int J Mol Sci. (2018) ;19: (10). |
[21] | Zhang W , Liu HT . MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. (2002) ;12: (1):9–18. |
[22] | Geest CR , Coffer PJ . MAPK signaling pathways in the regulation of hematopoiesis. J Leukoc Biol. (2009) ;86: (2):237–50. |
[23] | Kratofil RM , Shim HB , Shim R , Lee WY , Labit E , Sinha S , et al. A monocyte-leptin-angiogenesis pathway critical for repair post-infection. Nature. (2022) ;609: (7925):166–73. |




