Systematic Review: Characteristics and Preclinical Uses of Bladder Cancer Cell Lines
Abstract
Background:
Bladder cancer (BC) cell lines are indispensable in basic and preclinical research. Currently, an up-to-date and comprehensive overview of available BC cell lines is not available.
Objective:
To provide an overview and resources on the origin, pathological and molecular characteristics of commonly used human, murine and canine BC cell lines.
Methods:
A PubMed search was performed for relevant articles published between 1980 and 2017 according to the following MeSH terms: cell line; cell line, tumor; urinary bladder neoplasms; carcinoma, transitional cell. The Cellosaurus database was searched, using the term “bladder” and/or “urothelial carcinoma”. We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines.
Results:
We provide information on 157 human, murine and canine BC cell lines. 103 human BC cell lines have molecular data available, of which 69 have been profiled by at least one “omic” technology. We outline how these cell lines are currently being used for in vitro and in vivo experimental models. These results allow direct comparison of BC cell lines to patient samples, providing information needed to make informed decisions on the most genomically appropriate cell line to answer research questions. Furthermore, we show that cross-contamination remains an issue and describe guidelines for prevention.
Conclusions:
In the BC field, multiple human, murine and canine BC cell lines have been developed and many have become indispensable for in vitro and in vivo research. High-throughput -omic technologies have dramatically increased the amount of molecular data on these cell lines. We synthesized a comprehensive overview of these data as a resource for the BC scientific community.
INTRODUCTION
In 2017, an estimated 17.000 deaths are caused by bladder cancer (BC) in the United States. Worldwide, BC is the ninth most common cancer, with a prevalence of more than 1.300.000 patients [1, 2]. Urothelial carcinoma is the predominant histological subtype with 70–80% being non-muscle invasive disease (NMIBC) at diagnosis. NMIBC patients have a high 5-year overall survival, but tumors frequently recur and ∼30% of these patients will have progression to muscle invasive bladder cancer (MIBC), which is associated with a high morbidity and mortality [3, 4]. Important goals for BC research include the discovery of (urine) biomarkersfor non-invasive surveillance of NMIBC patients, biomarkers to predict response to treatment or progression and development of novel therapeutics for both NMIBC and MIBC.
To address these goals, human tissue and cell lines have been the primary tools used in preclinical studies [5]. BC cell lines reflect some of the genetic aberrations found in human BC and thus provide the opportunity to investigate basic tumor biology and preclinical pharmacological responses [6–11]. For example, urothelial cell lines containing Fibroblast Growth Factor Receptor-3 (FGFR3) mutations and cell lines with FGFR3-transforming acid coiled coil 3 (FGFR3-TACC3) fusions, which are highly sensitive to FGFR-selective agents, have been used to evaluate the efficacy of these new targeted treatments and are the foundation of currently running clinical trials [NCT01004224, NCT01732107, NCT02278978 and NCT02529553] [12, 13]. Furthermore, the COXEN (co-expression extrapolation) bioinformatics algorithm was used to generate multigene predictive models for various malignancies by using in vitro chemotherapy and radiation sensitivity data of transcriptionally profiled cancer cell lines [14]. This work also led to the identification of novel drugs for BC as well as potential drug targets that could enhance radiosensitivity of cancercells [15, 16].
In recent years, large-scale cell line characterization by major research consortia has increased significantly [17–19]. For example, project “Achilles” is aimed at identifying and cataloguing genes that are essential for cell survival [20]. In this project, genome-scale RNAi and CRISPR-Cas9 knockout is used in multiple human cancer cell lines to silence individual genes and data is available online as a resource. The Cancer Cell Line Encyclopedia (CCLE) has so far catalogued 1457 cell lines and performed multi-omic profiling of the majority of these lines [17]. Genomic characterization has led to opportunities for new in vitro and in vivo studies [21, 22]. Combined, these models offer different and, in many cases, complementary benefits. To fully exploit their potential, detailed information on cell line characteristics and preclinical models is essential for experimental design by every BC researcher. At the same time, concern about adequate selection, cross-contamination and verification of cell lines has not yet been resolved [23]. Here, we present a systematic review on the origin and molecular characteristics of currently available human, murine and canine BC cell lines for preclinical BC research. We discuss in vitro and in vivo BC models using these cell lines, together with their advantages, shortcomings and points of interest.
EVIDENCE ACQUISITION
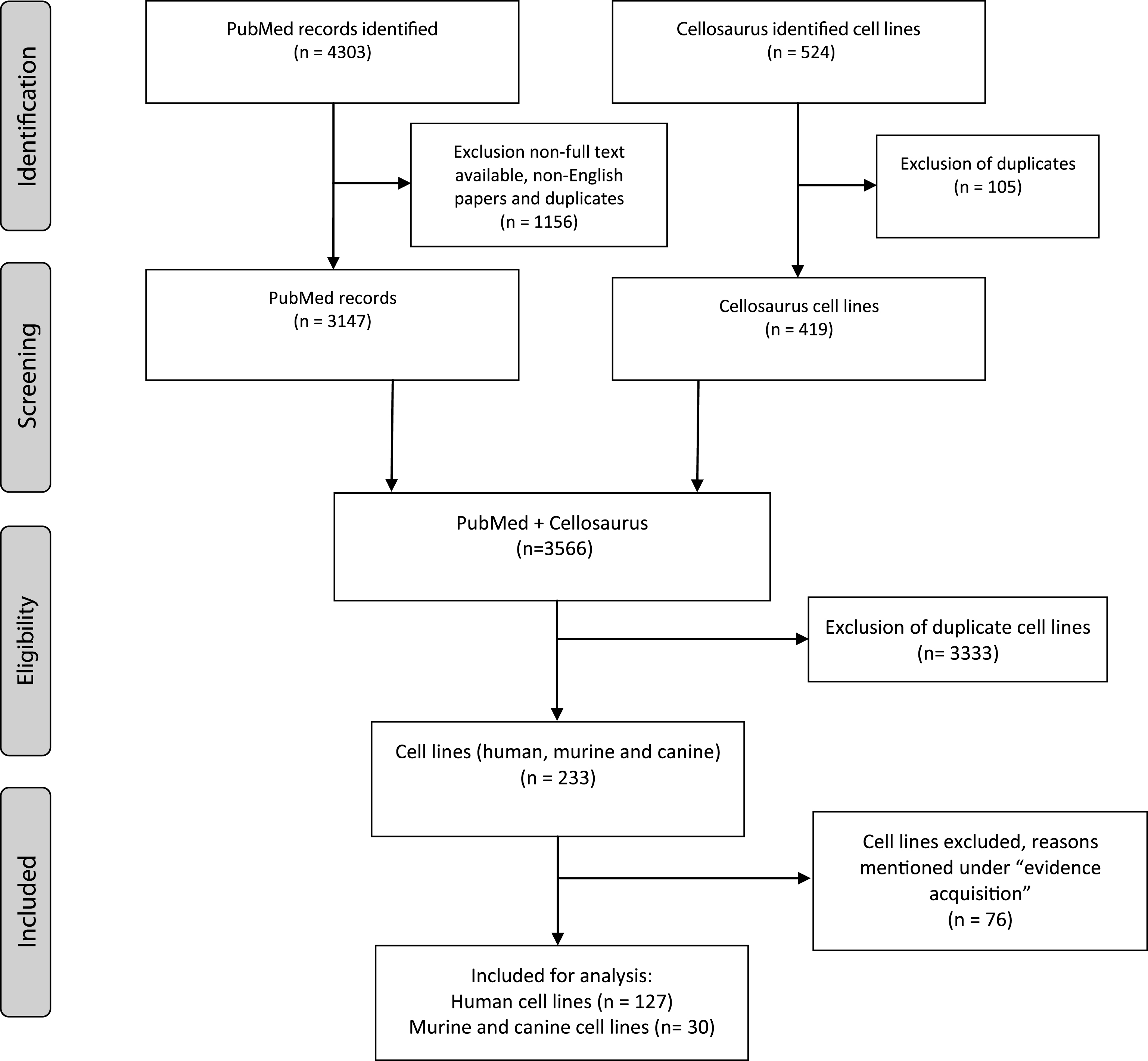
A literature search was conducted using the PubMed, Medline, Embase, Web of Science OvidSP, Google Scholar and Cellosaurus databases for all published results on BC cell lines from 1980 to 2017. The PubMed archive was searched for BC cell lines (November 2017) and the search strategy was as follows: (cell line[MeSH Major Topic] OR cell line, tumor[MeSH Terms]) AND (Urinary Bladder Neoplasms[MeSH Terms] OR Carcinoma, Transitional Cell[MeSH Terms]). The search resulted in 3147 English language, full text articles. BC cell lines were included if they were published at least twice since 1980, of which at least one publication since 2000. Incidentally found urothelial cancer cell lines of ureter or renal pelvis and a single urachal adenocarcinoma cell line were included as well. Contaminated BC cell lines, genetically engineered BC cell lines and patient-derived xenografts (PDX) were excluded. A search on Cellosaurus for ‘bladder’ and ‘urothelial’ provided 387 and 137 hits respectively (December 2017). After excluding duplicates, 419 remained. All cell lines were investigated and eligible cell lines were verified on PubMed and/or Google Scholar and added to the search results. Eventually, a total of 157 human, murine and canine bladder cancer cell lines were included in this systematicreview (Fig. 1).
Fig.1
Summary of evidence search and selection. Selection process for studies to be included is in compliance with PRISMA.
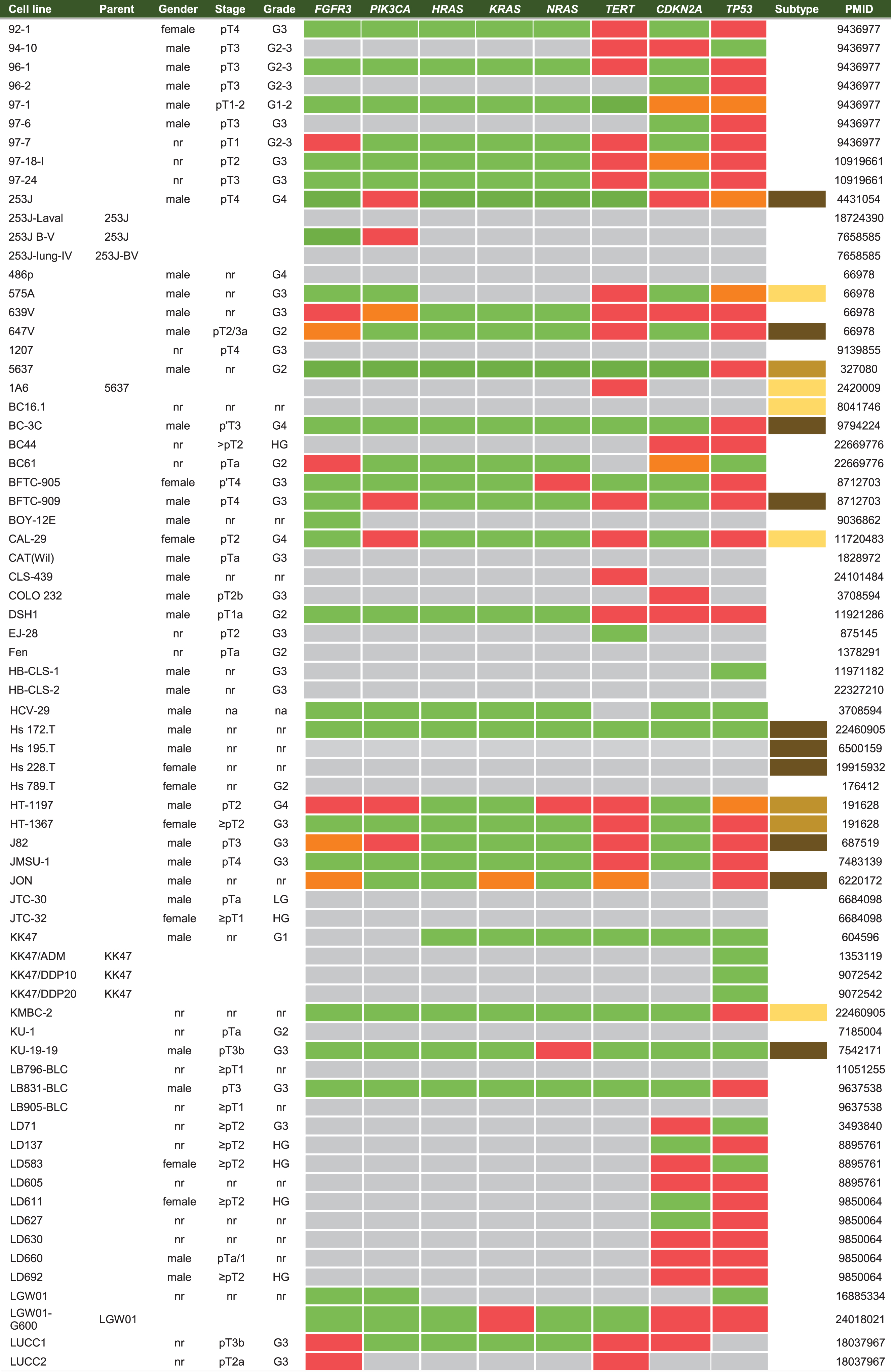
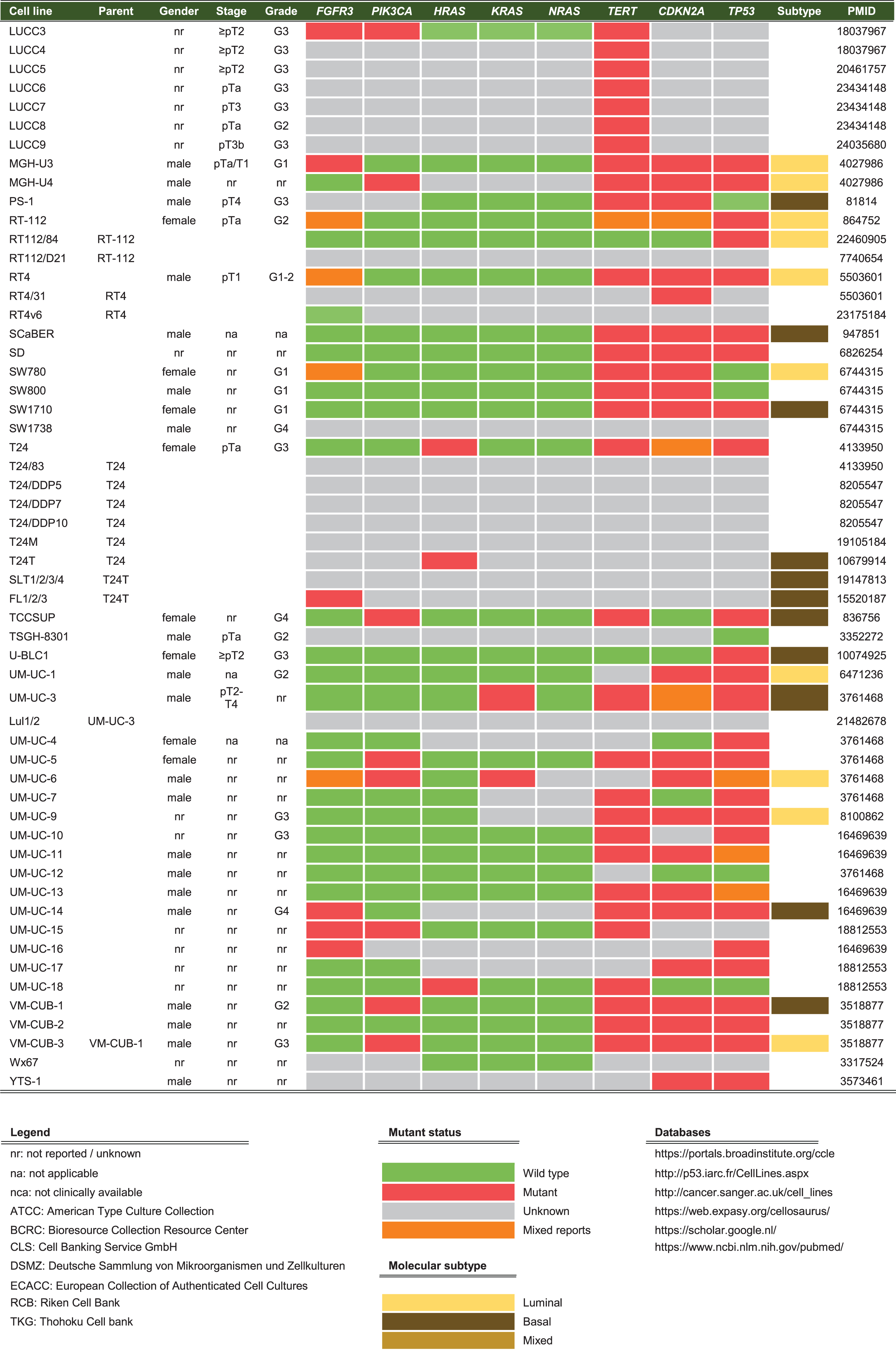
Historical cell line data like origin, gender, development, stage and grade were collected for each cell line. Thereafter, databases of the Catalogue of Somatic Mutations in Cancer (COSMIC), CCLE, the International Agency for Research on Cancer (IARC) TP53, Earl et al. (2015) and Nickerson et al. (2017) were used to report on the molecular characterization of the reviewed cell lines [17–19, 24]. These large databases incorporate molecular data from multiple international research projects working with cell lines, while in the past characterization was limited to low-throughput techniques [25, 26]. An overview of widely-used online databases for BC cell line research is reported inTable 1.
Table 1
Overview of available online resources for bladder cancer cell lines
| Database | Name | Resource | Website |
| Achilles | Genome scale RNAi and CRISPR-Cas9 silencing and knockdown of individual genes to identify genes that affect cell survival. | https://portals.broadinstitute.org/achilles | |
| BC-BET | Bladder Cancer Biomarker Evaluation Tool | Rapid evaluation of candidate gene expression across 40 commonly used human bladder cancer cell lines. | https://gdancik.github.io/bioinformatics/BCBET |
| CCLE | Cancer Cell Line Encyclopedia | Genetic characterization of human cell lines | https://portals.broadinstitute.org/ccle |
| Cellosaurus | Database for cell lines | https://web.expasy.org/cellosaurus/ | |
| COSMIC | Catalogue of Somatic Mutations in Cancer | Somatic mutations in human cancer cell lines | http://cancer.sanger.ac.uk/cosmic |
| CTRP | Cancer Therapeutics Response Portal | Identifying and targeting cancer dependencies with small molecules | https://portals.broadinstitute.org/ctrp/ |
| GDSC | Genomics of Drug Sensitivity in Cancer | Identifying molecular features of cancers that predict response to anti-cancer drugs | http://www.cancerrxgene.org/ |
| IARC TP53 | International Agency for Research on Cancer (IARC) TP53 Database | Compiles various types of data and information on human TP53 gene variations related to cancer | http://p53.iarc.fr/ |
| ICLAC | International Cell Line Authentication Committee | Resource lists cell lines that are currently known to be cross-contaminated or misidentified | http://iclac.org/ |
| TCPA | MD Anderson Cancer Cell Line Project | Proteomic database of cancer cell lines | http://tcpaportal.org/ |
| UC-25 | Exome sequencing of bladder cancer cell lines | http://www.synapse.org/UC25 |
IN VITRO MODELS: CELL LINE ORIGIN, DERIVATION AND MOLECULAR CHARACTERISTICS
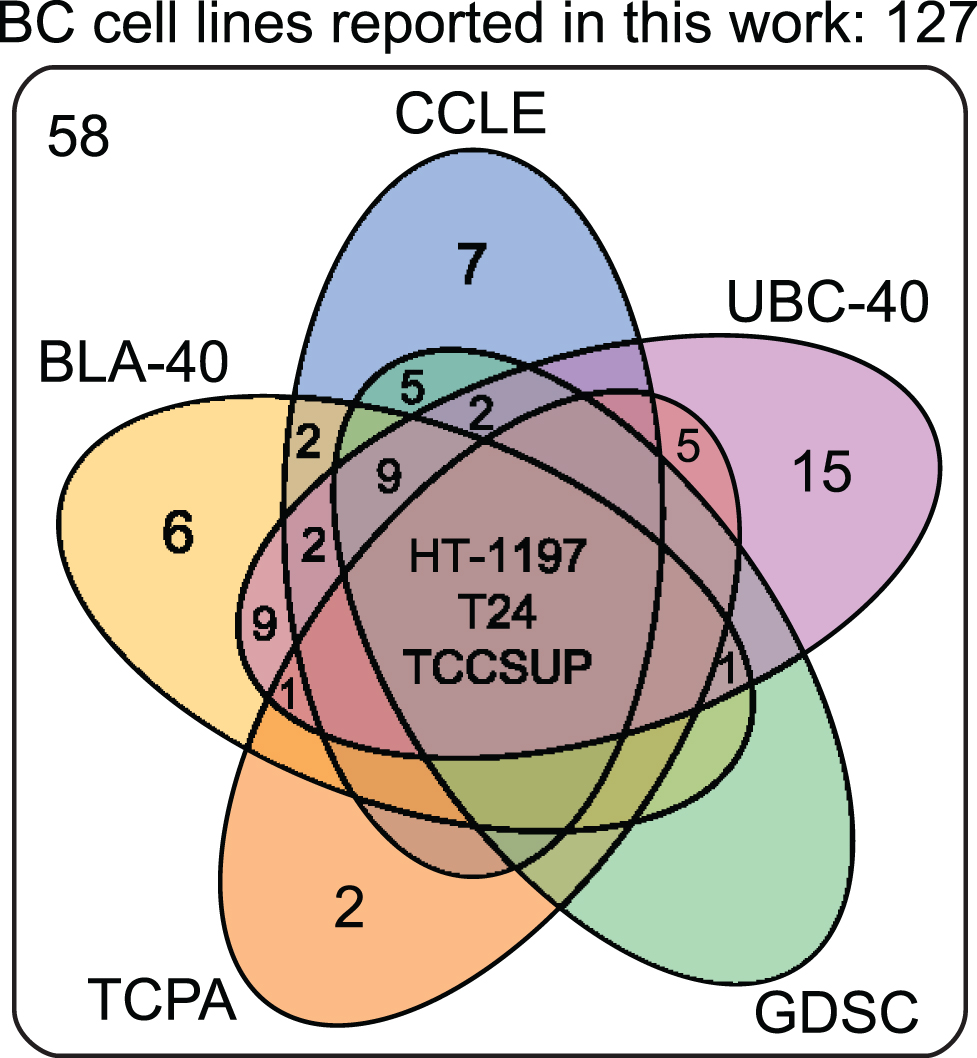
Cell lines are produced by either direct in vitro culture of patient tumor cells or by implantation of a patient’s tumor cells in immunodeficient mice and passaging the tumor multiple times before taking cells into culture. Currently, approximately 103 human BC cell lines of different stages and grades have molecular data available, of which 69 have been profiled by at least one – omic technology [18]. In general, molecular characterization of BC cell lines revealed that most genetic aberrations found in BC cell lines resemble those of the original tumor [19, 27–29]. Condensed historical and molecular data of the reviewed human BC cell lines is summarized in Table 2 (Complete information available in Supplementary Table 1). Many cell lines are now part of panels that have been characterized by different research groups. For example, Genomics of Drug Sensitivity in Cancer (GDSC) determined molecular alterations by using 20 BC cell lines to predict response to anti-cancer therapeutics [30]. Similarly, The Cancer Proteome Atlas (TCPA), included 11 BC cell lines in their effort to measure expression of cancer related proteins and how this may impact drug sensitivity [31]. More recently, whole exome sequencing on a subset of the UBC-40 panel (UC-25) confirmed the detection of mutations in TP53, TERT, PIK3CA and ERBB2 and other cancer-associated genes, in line with earlier TCGA findings in patients [18, 19, 32]. Also, 12 cell lines had homozygous deletions of chromosome 9p21, resulting in significant loss of expression of MTAP, CDKN2A and CDKN2B, concordant with previous findings [19, 33]. Because cell lines now overlap in different panels, we provide a full report of which cell lines were profiled in each panel in Table 2 and Fig. 2. In total, 69 of 127 human BC cell lines are represented in at least one of the five different panels. The most frequently characterized cell lines that have been profiled in all five panels are HT-1197, T24, and TCCSUP (Fig. 2). Table 2 shows that a relatively small number of human BC cell lines are of low stage (12/127) and low-grade (8/127). This is mainly caused by the difficulty of culturing low-grade tumors in vitro and the clinical need for late-stage and high-grade disease models. However, NMIBC are the bulk of all BC tumors worldwide and the high costs associated with bladder cancer mainly originates from these patients [2]. Thus, development of low-grade cell lines should not be neglected by the BC community. Finally, molecular subtyping of BC has shown promising results to predict clinical outcomes in BC patients [34]. Luminal and basal molecular subtypes have also been demonstrated in BC cell lines [11, 35]. Our data shows that 39/127 human cell lines currently have been analysed; 15 luminal cell lines, 21 basal cell lines and 3 cell lines have a mixed report (Table 2 and Supplementary Table 2). Overall, data in Table 2 show that cell line mutational frequencies correspond to frequencies reported in in patients [32]. These findings could be useful in selecting the optimal cell line for specific research questions.
Fig.2
Venn diagram of human bladder cancer cell lines analyzed in five panels. 127 human BC cell lines are present in five panels. The overlap between different molecular panels is visualized. Only HT-1197, T24 and TCCSUP have been profiled in all five panels. CCLE: The Cancer Cell Line Encyclopedia; UBC-40: Urothelial Bladder Cancer-40; GDSC: Genomics of Drug Sensitivity in Cancer; TCPA: The Cancer Proteome Atlas; BLA-40:Bladder-40.
MURINE AND CANINE BLADDER CANCER CELL LINES
As in vivo use of human cell lines requires immunodeficient models, multiple BC cell lines from different species have been developed to study aspects of tumor immunity in vivo. Studies in the 1960s first explored chemically induced BC in mice by exposure to different carcinogens [36]. After exposure, most mice developed poorly differentiated tumors, which were heterogeneous and resembled human MIBC [37]. For instance, murine BC can be induced by exposure to carcinogenic chemicals like, 7, 12-dimethylenzanthacene (DMBA), N-methyl-N-nitrosourea (MNU), (N-butyl-N-(4-hydroxybutyl) nitrosamine (BBN), or N-[4-(5-nitro-2-furyl)-2thiazolyl]-formamide (FANFT) [36]. DMBA is a mutagen that causes DNA damage after activation. MNU is a genotoxic compound, which is administered intravesically and acts directly on urothelium with tumor formation after ∼8 weeks. BBN is administered orally to mice, with tumors developing after a ∼16-week diet of 0.05% BBN in drinking water [38–40]. Like BBN, FANFT can also be administered orally, resulting in carcinomas of both urothelial and squamous cell origin after ∼8 months [41]. After tumor formation in the bladder, tumor cells are harvested, cultured and passaged in vitro to develop a novel murine BC cell line. Currently, multiple carcinogen-induced BC cell lines are available. However, two murine BC cell lines are predominantly used in BC research: MB49 and MBT2 [42, 43]. MB49 was developed by orthotopic transplantation of mouse bladder urothelial cells, which were transformed by exposure to DMBA for 24 hours [42]. Although a male mouse was the original donor, karyotypic exploration showed MB49 has lost the Y-chromosome after in vitro culture [44]. MBT2 was produced by serial transplantation of a FANFT-induced tumor in mice [43]. Recently, Fantini et al. reported that BBN tumors had an overexpression of markers of the MIBC basal subtype [45]. Moreover, BBN tumors showed a high mutational burden similar to MIBC patients. Interestingly, the predominantly used MB49 (DMBA) and MBT2 (FANFT) cell lines in research showed important differences in their mutational profiles compared to human and BBN-induced tumors [45]. These findings demonstrate the importance of molecular profiling to select the appropriate research model and to ultimately improve translational research. BTT-T739 is another murine cell line that causes multiple visceral metastases, but is less frequently used in practice [46]. A fourth murine line is AY-27, generated after feeding FANFT to Fisher 344 rats. AY-27 causes in vivo high-grade NMIBC with progression to MIBC after ∼6 days [47]. A condensed overview of all murine cell lines is given in Table 3 (complete information available in Supplementary Table 3).
Table 3
Summary of murine & canine BC cell lines (see Supplementary Table 3 for complete overview)
| Cell line | Parent | Gender | Species | Origin | Stage/Grade | Development | PMID |
| BTT-T739 | female | Mouse | T739 | MIBC/HG | Cultured bladder tumor cells from T739 mice fed with BBN | 23691487 | |
| MB49 | male | Mouse | C57BL/6J | MIBC | 24 hr DBMA treatment in vitro, transformed transplanted into cells in syngeneic mice | 107359 | |
| MB49-I | MB49 | MIBC | MB49 tumor passeged 13x subcutaneously | 19539312 | |||
| MBT-2 | female | Mouse | C3H/He | MIBC/HG | Serially transplant FANFT-induced tumor in syngeneic mice, used at 20th transplant | 19388981 | |
| MBT-2V | MBT-2 | nr | Cultured lung metastasis cells from i.m. injected MBT-2 cells | 16140114 | |||
| NCI-HPN-F1G | male | Mouse | C57BL/lcrf-a | nr | 24 hr DBMA treatment in vitro, transformed transplanted into cells in syngeneic mice | 22161874 | |
| NCI-HPN-F2G | female | Mouse | C57BL/6 | nr | Spontaeous transformation by in vitro culture | 22161874 | |
| NCI-HPN-M1C | male | Mouse | C57BL/6 | nr | Spontaeous transformation by in vitro culture | 22161874 | |
| NCI-HPN-M1I | male | Mouse | C57BL/6 | nr | Spontaeous transformation by in vitro culture | 22161874 | |
| NCI-HPN-M2C | Mouse | C57BL/6 | nr | Spontaeous transformation by in vitro culture | 22161874 | ||
| 804 G | female | Rat | ACI | nr | Exposure of normal bladder epithelial cells to BCPN for 23 months in vitro | 7448785 | |
| AY-27 | male | Rat | Fischer 344 | NMIBC/G2-3 | Cultured bladder tumor cells from rats fed with FANFT | 7298281 | |
| BC31 | male | Rat | ACl/N | nr | Cultured bladder tumor cells from rats fed with BBN | 381090 | |
| BC47 | male | Rat | ACl/N | nr | Cultured bladder tumor cells from rats fed with BBN | 3918910 | |
| BC50 | male | Rat | ACl/N | nr | Cultured bladder tumor cells from rats fed with BBN | 381090 | |
| NBT-II | male | Rat | Wistar | nr | Cultured bladder tumor cells from rats fed with BBN | 4941696 | |
| M-NBT-II | NBT-II | male | Rat | Wistar | nr | Cultured from lymphnode metastasis from nude mouse injected s.c. with NBT-II | 7929634 |
| NBT-L2b | NBT-II | male | Rat | Wistar | nr | NTB-II cultured in vitro; colonies with distinct morphology selected for expansion | 9369930 |
| NBT-T2 | NBT-II | Rat | Wistar | nr | NTB-II cultured in vitro; colonies with distinct morphology selected for expansion | 9369930 | |
| Bliley | nr | Canine | nr | nr | nr | 27257868 | |
| K9TCC | Female | Canine | Mixed breed | nr | nr | 7653891 | |
| K9TCC#1Lillie | Female | Canine | English Pointer | Invasive | Cultured cells from bladder tumor biopsy | 24964787 | |
| K9TCC#2Dakota | Female | Canine | Bichon Fries | Invasive | Cultured cells from bladder tumor biopsy | 24964787 | |
| K9TCC#4Molly | Female | Canine | Maltese | Invasive | Cultured cells from bladder tumor biopsy | 24964787 | |
| K9TCC-PU-AxA | Female | Canine | nr | Invasive G3 | Cultured cells from bladder tumor biopsy (same dog as K9TCC-PU-AxC) | 18562222 | |
| K9TCC-PU-AxC | Female | Canine | nr | Invasive G3 | Cultured cells from bladder tumor biopsy (same dog as K9TCC-PU-AxA) | 18562222 | |
| K9TCC-PU-In | Female | Canine | German Shephard | Invasive G3 | Cultured cells from bladder tumor biopsy | 18562222 | |
| K9TCC-PU-Mx | Female | Canine | German Shephard | Invasive G3 | Cultured cells from bladder tumor biopsy | 18562222 | |
| K9TCC-PU-Nk | Female | Canine | nr | Invasive G3 | Cultured cells from bladder tumor biopsy | 18562222 | |
| K9TCC-PU-Sh | Female | Canine | Collie | Invasive G3 | Cultured cells from bladder tumor biopsy | 18562222 |
Legend: nr = not reported/unknown. Databases: https://portals.broadinstitute.org/ccle; http://p53.iarc.fr/CellLines.aspx; http://cancer.sanger.ac.uk/cell_lines; https://web.expasy.org/cellosaurus/; https://scholar.google.nl/; https://www.ncbi.nlm.nih.gov/pubmed/.
Canine cell lines are increasingly available and used in BC research. In dogs, BC is a naturally occurring disease, with some breeds (Scottish Terriers) even having a life-time risk of BC than other breeds [48]. Unfortunately, BC is typically diagnosed when the tumor is already muscle-invasive or metastasized, leaving most dogs incurable [49]. Therefore, canine BC offers a unique opportunity to study advanced BC and as a result characterization of canine BC has increased in the last years [50]. Invasive BC in dogs is similar to human MIBC both histologically and molecularly and therefore provides us with a potential BC model [51]. Knapp et al. generated one of the first canine lines (K9TCC) and thereafter more canine BC cell lines followed [52, 53]. Recently, extensive molecular characterization of a canine tumor cell line panel was done by the Flint Animal Cancer Center (FACC) [54]. FACC data will prove a valuable resource for BC research. An overview of our search for commonly used canine BC cell lines is presented in Table 3. Of note is the lack of high-throughput molecular characterization in both murine and canine cell lines. As these cell lines are frequently used, it would be of great benefit to the research community to expand the molecular characterization of murine and canine cell lines so the correct model can be chosen accordingly.
BLADDER CANCER CELL LINES FOR IN VIVO RESEARCH
The use of immunocompromised mouse models for human xenografts limits investigation of interactions between the tumor and its host. In vivo immunocompetent mouse tumor models have the advantage of an intact host immune system and tumor microenvironment (TME) and thus provide a platform to study tumor-host interactions and immunotherapeutics [6, 40, 55, 56]. In vivo models can be divided in two main groups: 1) spontaneous or autochthonous tumor models (subdivided in carcinogen-induced and genetically engineered/transgenic models) and 2) engraftment or non-autochthonous models (subdivided in allograft/syngeneic and xenograft/xenogeneic models). In contrast to tumor heterogeneity of carcinogen-induced models, BC in genetically engineered mouse models (GEM) originates due to fixed experimentally applied genetic changes found to be associated with BC and complement carcinogen-induced mouse models. Recently published TCGA data will be used to aid the development of novel GEMs [32]. More detailed information on GEMs and carcinogen-induced BC models has been published elsewhere [57–59]. Syngeneic models are generated by implantation of murine BC cell lines into immunocompetent mice and provide insight into the biology of TME interactions and are excellent tools to evaluate immunotherapy such as intravesical Bacillus Calmette-Guérin or checkpoint inhibitors [60–62]. Xenogeneic models are generated by implantation of human BC cell lines (xenografts) or PDXs into immunodeficient mice [63–65]. PDXs are molecularly more related to the original tumor, show greater intratumoral heterogeneity and histological similarity to the primary tumor, even after several passages in mice [65–67]. PDXs can therefore be used to directly evaluate a patient’s response to treatment [65–67]. Here, we only focus on immortalized, tumor-derived cell lines and reviews on PDX models have been published elsewhere [58, 63, 68].
In vivo models can also be classified according to the site of tumor implantation; orthotopic (in the bladder) or heterotopic (elsewhere, not in the bladder) [58, 69, 70]. Orthotopic models can either be spontaneous (i.e. orthotopic carcinogen-induced) or engrafted [71]. Damage to the bladder wall is generally used to increase orthotopic tumor engraftment [69], yet the “tumor take rate” varies considerably between studies [69, 72, 73]. Multiple procedures exist to damage the bladder wall, for example transurethral mechanical injuring, intravesical exposure to corrosive chemicals, laparotomy or minimal invasive percutaneous surgery [73–76]. The main advantage of inoculating tumor cells inside the bladder, is a more realistic simulation of the TME found in BC patients. Challenges with orthotopic models include the requirement for imaging methods to measure tumor growth or procedures of sequential cohort euthanasia and necropsy [77]. Non-invasive assessment of tumors can be accomplished through magnetic resonance imaging, intravesical ultrasonography and in vivo fluorescence of tumors [61, 78, 79]. Common heterotopic tumor inoculation sites include subcutaneous or subrenal capsule models [61, 65]. Subcutaneous models allow easy tumor size measurements but may have different responses to drugs and a different TME compared to the orthotopic location [80, 81]. Alternatively, subrenal capsule models have a higher tumor take than orthotopic engraftment models, probably due to elimination of orthotopic tumor cells by voiding or because subrenal capsules tumors benefit from the rich blood supply of the subrenalarea [82].
METASTATIC BLADDER CANCER MODELS
Patients with metastases have a poor clinical outcome and contribute to the bulk of mortality in BC. Experimental models are invaluable to investigate the complex multistep metastatic process that enables invasive growth of the primary tumor, detachment from the primary site, intravasation, migration, extravasation and colonization of distant organs. This multistep process is difficult to replicate in vivo, because mice often die before metastases develop. Moreover, kidney failure occurs frequently, as both engrafted and carcinogen-induced orthotopic tumors cause post-renal obstruction due to a large bladder tumor. Another limitation is the lack of available tissue from highly metastatic human tumors and thus limited availability of metastatic BC models. Therefore, researchers use in vivo serial passaging to acquire more aggressive cell lines. Serial passaging can be orthotopic, by portal vein injections, tail vein injections or intracardiac injections. The different routes determine the metastatic pattern of spreading [83]. For example, metastatic cell line Lul1/2 was produced from the UM-UC-3 parent cell line by tail vein injection, causing tumor formation in the lungs. Next, lung nodules were harvested and re-injected into the tail vein and the cycle was repeated multiple times, generating Lul1/2 cells with greater capacity to proliferate and aggressively expand in a metastatic site [84]. Two models from the parental cell line T24 have been developed by using the same approach and are currently available for studying lung and liver tropism in BC [85, 86]. Lung metastasis model T24T/FL1-3 was developed by repeated tail vein injections of T24T followed by harvesting of lung nodules to develop cell lines with increased capabilities to colonize distant organs [85]. T24T/SLT1-3 liver metastatic lines were developed by intrasplenic injections of T24T followed by harvesting of liver nodules and repeated intrasplenic injections [86]. These models allowed identification of molecular changes associated with the metastatic phenotype to liver and lung.
For syngeneic murine models, MBT-2 and MB49 cell lines are frequently used. MBT-2 has a low frequency of metastatic spreading to the lungs when injected intravenously, but metastatic potential increases when tumor cells are injected intramuscularly [87]. To improve the metastatic potential of MB49, cells were passaged multiple times subcutaneously, causing cells to acquire a more invasive phenotype (MB49-I). After orthotopic inoculation of MB49-I cells, mice developed multiple lung metastasis [88]. In summary, there are limited metastatic BC cell lines and models available. Expanding the number of metastatic BC lines and optimizing current metastatic models is an area that would benefit the BC research community.
CAVEATS OF CANCER CELL LINES
Despite their advantages and contributions to the field, researchers should be aware of cell line limitations. With each passage, cells experience genetic changes due to selective pressure, which may lead to a change in phenotype [89]. After multiple passages, BC cell lines showed loss of heterozygosity, microsatellite instability and changes in ploidy compared to the original tumor [90–92]. Genetic changes and selection are also induced by passaging cells for long time periods under suboptimal culture conditions (e.g. lack of nutrients or passaging cells at low density) [89, 93]. It is therefore important to be aware of how a cell line is developed (single clone or heterogeneous tumor cell culture), culture conditions and to realize that cell lines may represent a subset of cellular features of the original tumors [6, 70, 94].
Cross-contamination
Another challenging aspect when using cancer cell lines is cross-contamination. Cross-contamination is the result of contamination of an original cell line with a contaminant cell line that has a dominant growth pattern, eventually outgrowing the original cell line. Cross-contamination was discovered as early as 1967, when Gartler et al. accidently found that a panel of cell lines of mixed gender all shared a common X-linked genetic polymorphism (G6PD allele) that was almost exclusive for people of African descent. Further analysis showed that his cell lines were likely contaminated with HeLa cells from donor Henrietta Lacks, who is of African descent [95, 96]. Unfortunately, the process of cross-contamination can be gradual and insidious, especially if visual morphology of the contaminating cells is similar to that of the host line. HeLa cells have often been associated with cross-contamination of other cultures, because HeLa cells grow rapidly and are widely used [97, 98]. Genetic fingerprinting of various BC cell lines suggests some are now HeLa cells [99, 100]. For example, cell line KU-7 was derived from a LG papillary tumor, yet acquired invasive growth characteristics after multiple passages [99]. KU-7 turned out to be contaminated with HeLa cells, likely even before 1984, thereby raising questions about several studies [99, 101, 102]. Another example is T24, which has contaminated a variety of cell lines [103–106]. Recent estimates suggest that up to a third of all documented cell lines are contaminated and no longer represent the original tumor [107, 108]. Given the magnitude of the cross-contamination problem, the National Institute of Health (NIH) requires authentication of cell lines, for instance by using short tandem repeat profiling (STR-profiling) when cell lines are used in NIH funded research [23, 109–111]. Additionally, multiple tests are available to determine which species a cell line originated from, like isoenzymology and DNA-barcoding [112]. Last, consulting overviews of cross-contaminated cell lines before usage is strongly recommended. An overview of published cross-contaminated BC cell lines is given in Table 4 [113]. Verification of cell lines before manuscripts are accepted will hopefully reduce the frequency of cross-contamination and improve the quality of cell line-related research in the future[114–116].
Table 4
Overview of documented cross-contaminated BC cell lines
| Cell line | Cross-contaminant | Reference/PMID |
| ACCS | T24 | 19557180 |
| BIU-87 | Non-human | 26116706 |
| BIU-87/ADMR | Child BIU-87 | na [26116706] |
| BT-B | HeLa subclone | DSMZ |
| ECV-304 | T24/umbilical vein | 12197766 |
| EJ-1 | T24 | 11416159 |
| Ej138 | T24 | 17254797 |
| EJ-30 (A3/B3) | T24, child Ej138 | na [17254797] |
| EJ-30tgr | T24, grandchild Ej138 | na [17254797] |
| GHR3 | T24, grandchild Ej138 | na [17254797] |
| GHE | T24/astrocytoma | 10508494 |
| hAG | T24/thyroid | 10508494 |
| HBMEC-2 | T24 | ATCC |
| HCV-29Tmv | T24 | 8105864 |
| Hu456 | T24 | 3708594 |
| Hu549 | T24 | 8105864 |
| Hu609 | J82 | 8105864 |
| Hu609T | J82, child Hu609T | na [8105864] |
| Hu609Tmv | T24 | 8105864 |
| Hu961a (-t) | T24 | 8105864 |
| Hu1734 | HCV-29 | 20143388 |
| JCA-1 | T24 | 11522622 |
| KS Y-1 | T24 | 22987579 |
| KU-7 | Hela | 23500642 |
| MGH-U1 | T24 | 6823318 |
| MGH-U1/OCI(-m/F1) | T24, grandchild MGH-U1 | na [6823318] |
| MGH-U2 | T24 | 3355767 |
| RAMAK-1 | T24 | 10508494 |
| RO-H85-1 | 647V/thyroid | 23472229 |
| SBC-2 | Hela | 10508494 |
| SBC-7 | Hela | 10508494 |
| SNK-57 | XY, should be female | 9183630 |
| SW 850 | Bladder/pancreatic | 25997541 |
| TSU-Pr1 | T24/prostatic | 11522622 |
| TSU-Pr1-B1 (-B2) | T24, child TSU-Pr1 | na [11522622] |
| TsuGK27 | T24, child TSU-Pr1 | na [11522622] |
| UM-UC-2 | T24 | 19375735 |
| UM-UC-3-GFP | Unknown | 19375735 |
| UROtsa (GEM; SV40) | Partial T24, incl. children | 23691160 |
| VM-CUB-3 | VM-CUB-I | 25877200 |
Legend: GEM: Genetically Engineered Model; DSMZ: Deutsche Sammlung von Mikroorganismen und Zellkulturen; na: cells derived from parental cells found to be cross-contaminated. Database: http://iclac.org/wp-content/uploads/Cross-Contaminations-v7_2.pdf; https://web.expasy.org/cgi-bin/cellosaurus/search - ‘Problematic cell line bladder’.
CONCLUSIONS
Many cell lines have been developed for BC research, yet a relatively small selection of these lines is currently being used. Important considerations for cell line research and selection of preclinical models are; 1) each model has unique advantages and limitations to take into account; 2) cell line authentication is considered mandatory before use; 3) orthotopic inoculation more accurately represents clinical BC, however these models are more challenging in use;4) human BC cell lines implanted in immunodeficient mice cannot be used to explore the tumor microenvironment in vivo and 5) extensive genomic and molecular characterization of cell lines allow investigators to select lines with desired genetic characteristics. While there is no flawless model, cell lines are an indispensable part of ongoing research. Optimization of current models is needed, as well as development of new metastasis models, especially from highly metastatic tumors. Finally, current in vitro and in vivo models using cell lines can be used in conjunction with new GEMs and PDXs to create a wide range of modelling options for the BC research community.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/BLC-180167.
REFERENCES
[1] | Siegel RL , Miller KD , Jemal A . Cancer statistics, 2016. CA Cancer J Clin. (2016) ;66: (1):7–30. |
[2] | Ferlay JSI , Dikshit R , Eser S , Mathers C , Rebelo M , Parkin DM , Forman D , Bray F . Estimated cancer incidence, mortality and prevalance worldwide in 2012 by WHO international agency for research on cancer. Int J Cancer. (2015) . |
[3] | Sylvester RJ , van der Meijden AP , Oosterlinck W , Witjes JA , Bouffioux C , Denis L , et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: A combined analysis of 2596 patients from seven EORTC trials. Eur Urol. (2006) ;49: (3):466–5; discussion 75–7. |
[4] | Fonteyne V , Ost P , Bellmunt J , Droz JP , Mongiat-Artus P , Inman B , et al. Curative treatment for muscle invasive bladder cancer in elderly patients: A systematic review. Eur Urol 2017. |
[5] | Bernardo C , Costa C , Palmeira C , Pinto-Leite R , Oliveira P , Freitas R , et al. What we have learned from urinary bladder cancer models. Journal of Cancer Metastasis and Treatment. (2016) ;2: (2):51–8. |
[6] | Eiján AM , Lodillinsky C, Sandes E . Animal Models for Basic and Preclinical Research in Bladder Cancer. In: Canda A , editor. Bladder Cancer - From Basic Science to Robotic Surgery: InTech (2012) . |
[7] | Li C , Yang Z , Du Y , Tang H , Chen J , Hu D , et al. BCMab1, a monoclonal antibody against aberrantly glycosylated integrin alpha3beta1, has potent antitumor activity of bladder cancer in vivo. Clin Cancer Res. (2014) ;20: (15):4001–13. |
[8] | van Kessel KE , Zuiverloon TC , Alberts AR , Boormans JL , Zwarthoff EC . Targeted therapies in bladder cancer: An overview of in vivo research. Nat Rev Urol. (2015) ;12: (12):681–94. |
[9] | Zhang X , Godbey WT . Preclinical evaluation of a gene therapy treatment for transitional cell carcinoma. Cancer Gene Ther. (2011) ;18: (1):34–41. |
[10] | Zhao F , Lin T , He W , Han J , Zhu D , Hu K , et al. Knockdown of a novel lincRNA AATBC suppresses proliferation and induces apoptosis in bladder cancer. Oncotarget. (2015) ;6: (2):1064–78. |
[11] | Goodspeed A , Heiser LM , Gray JW , Costello JC . Tumor-derived celllines as molecular models of cancer pharmacogenomics. Mol Cancer Res. (2016) ;14: (1):3–13. |
[12] | Lamont FR , Tomlinson DC , Cooper PA , Shnyder SD , Chester JD , Knowles MA . Small molecule FGF receptor inhibitors blockFGFR-dependent urothelial carcinoma growth in vitro and in vivo . Br J Cancer. (2011) ;104: (1):75–82. |
[13] | Williams SV , Hurst CD , Knowles MA . Oncogenic FGFR3 gene fusions in bladder cancer. Human molecular genetics. (2013) ;22: (4):795–803. |
[14] | Williams PD , Cheon S , Havaleshko DM , Jeong H , Cheng F , Theodorescu D , et al. Concordant gene expression signatures predict clinical outcomes of cancer patients undergoing systemic therapy. Cancer Res. (2009) ;69: (21):8302–9. |
[15] | Smith SC , Havaleshko DM , Moon K , Baras AS , Lee J , Bekiranov S , et al. Use of yeast chemigenomics and COXEN informatics in preclinical evaluation of anticancer agents. Neoplasia. (2011) ;13: (1):72–80. |
[16] | Williams PD , Owens CR , Dziegielewski J , Moskaluk CA , Read PW , Larner JM , et al. Cyclophilin B expression is associated with in vitro radioresistance and clinical outcome after radiotherapy. Neoplasia. (2011) ;13: (12):1122–31. |
[17] | Barretina J , Caponigro G , Stransky N , Venkatesan K , Margolin AA , Kim S , et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. (2012) ;483: (7391):603–7. |
[18] | Earl J , Rico D , Carrillo-de-Santa-Pau E , Rodriguez-Santiago B , Mendez-Pertuz M , Auer H , et al. The UBC-40 Urothelial Bladder Cancer cell line index: A genomic resource for functional studies. BMC Genomics. (2015) ;16: :403. |
[19] | Nickerson ML , Witte N , Im KM , Turan S , Owens C , Misner K , et al. Molecular analysis of urothelial cancer cell lines for modeling tumor biology and drug response. Oncogene. (2017) ;36: (1):35–46. |
[20] | Tsherniak A , Vazquez F , Montgomery PG , Weir BA , Kryukov G , Cowley GS , et al. Defining a cancer dependency map. Cell. (2017) ;170: (3):514–76 e16. |
[21] | Zeng S , Zhu Y , Ma AH , Yu W , Zhang H , Lin TY , et al. The Phosphatidylinositol 3-kinase Pathway as a Potential Therapeutic Target in Bladder Cancer. Clin Cancer Res. (2017) . |
[22] | Ler LD , Ghosh S , Chai X , Thike AA , Heng HL , Siew EY , et al. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci Transl Med. (2017) ; 9: (378) . |
[23] | Korch C . Line of Attack. In: Neimark N, editor. Science Magazine: AAAS; (2015) . pp. 938–40. |
[24] | Landry JJ , Pyl PT , Rausch T , Zichner T , Tekkedil MM , Stutz AM , et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda). (2013) ;3: (8):1213–24. |
[25] | Kim SK , Yun SJ , Kim J , Lee OJ , Bae SC , Kim WJ . Identification of gene expression signature modulated by nicotinamide in a mouse bladder cancer model. PLoS One. (2011) ;6: (10):e26131. |
[26] | Kang MR , Kang JS , Yang JW , Kim BG , Kim JA , Jo YN , et al. Gene expression profiling of KBH-A42, a novel histone deacetylase inhibitor, in human leukemia and bladder cancer cell lines. Oncol Lett. (2012) ;3: (1):113–8. |
[27] | Hoffmann MJ , Koutsogiannouli E , Skowron MA , Pinkerneil M , Niegisch G , Brandt A , et al. The new immortalized uroepithelial cell line HBLAK contains defined genetic aberrations typical of early stage urothelial tumors. Bladder Cancer. (2016) ;2: (4):449–63. |
[28] | Chen F , Zhang G , Cao Y , Hessner MJ , See WA . MB49 murine urothelial carcinoma: Molecular and phenotypic comparison to human cell lines as a model of the direct tumor response to bacillus Calmette-Guerin. J Urol. (2009) ;182: (6):2932–7. |
[29] | Lin CW , Lin JC , Prout GRJr . Establishment and characterization of four human bladder tumor cell lines and sublines with different degrees of malignancy. Cancer Res. (1985) ;45: (10):5070–9. |
[30] | Yang W , Soares J , Greninger P , Edelman EJ , Lightfoot H , Forbes S , et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Research. (2013) ;41: (Database issue):D955–D61. |
[31] | Li J , Zhao W , Akbani R , Liu W , Ju Z , Ling S , et al. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell. (2017) ;31: (2):225–39. |
[32] | Robertson AG , Kim J , Al-Ahmadie H , Bellmunt J , Guo G , Cherniack AD , et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. (2017) ;171: (3):540–56 e25. |
[33] | Stadler WM , Sherman J , Bohlander SK , Roulston D , Dreyling M , Rukstalis D , et al. Homozygous deletions within chromosomal bands 9p21-22 in bladder cancer. Cancer Res. (1994) ;54: (8):2060–3. |
[34] | Seiler R , Ashab HAD , Erho N , van Rhijn BWG , Winters B , Douglas J , et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol. (2017) ;72: (4):544–54. |
[35] | Warrick JI , Walter V , Yamashita H , Chung E , Shuman L , Amponsa VO , et al. FOXA1, GATA3 and PPAR cooperate to drive luminal subtype in bladder Cancer: A molecular analysis of established human cell lines. Sci Re. (2016) ;6: :38531. |
[36] | Soloway MS . Single and combination chemotherapy for primary murine bladder cancer. Cancer. (1975) ;36: (2):333–40. |
[37] | Williams PD , Lee JK , Theodorescu D . Molecular credentialing of rodent bladder carcinogenesis models. Neoplasia. (2008) ;10: (8):838–46. |
[38] | Overdevest JB , Knubel KH , Duex JE , Thomas S , Nitz MD , Harding MA , et al. CD24 expression is important in male urothelial tumorigenesis and metastasis in mice and is androgen regulated. Proc Natl Acad Sci U S A.E. (2012) ;109: (51):3588–96. |
[39] | Oliveira PA , Vasconcelos-Nobrega C , Gil da Costa RM , Arantes-Rodrigues R . The N-butyl-N-4-hydroxybutyl nitrosamine mouse urinary bladder cancer model. Methods Mol Biol. (2018) ;1655: 155–67. |
[40] | Hori S , Miyake M , Tatsumi Y , Onishi S , Morizawa Y , Nakai Y , et al. Topical and systemic immunoreaction triggered by intravesicalchemotherapy in an N-butyl-N-(4-hydroxybutyl) nitorosamine inducedbladder cancer mouse model. PLoS One. (2017) ;12: (4):e0175494. |
[41] | Cohen SM , Yang JP , Jacobs JB , Arai M , Fukushima S , Friedell GH . Transplantation and cell culture of rat urinary bladder carcinoma. Invest Urol. (1981) ;19: (3):136–41. |
[42] | Summerhayes IC , Franks LM . Effects of donor age on neoplastic transformation of adult mouse bladder epithelium in vitro. J Natl Cancer Inst. (1979) ;62: (4):1017–23. |
[43] | Soloway MS . Intravesical and systemic chemotherapy of murinebladder cancer. Cancer Res. (1977) ;37: (8 Pt 2):2918–29. |
[44] | Fabris VT , Lodillinsky C , Pampena MB , Belgorosky D , Lanari C , Eijan AM . Cytogenetic characterization of the murine bladdercancer model MB49 and the derived invasive line MB49-I. Cancer Genet. (2012) ;205: (4):168–76. |
[45] | Fantini D , Glaser AP , Rimar KJ , Wang Y , Schipma M , Varghese N , et al. A Carcinogen-induced mouse model recapitulates the molecular alterations of human muscle invasive bladder cancer. Oncogene. (2018) . |
[46] | Yang XH , Ren LS , Wang GP , Zhao LL , Zhang H , Mi ZG , et al. A new method of establishing orthotopic bladder transplantable tumor in mice. Cancer Biology & Medicine. (2012) ;9: (4):261–5. |
[47] | Hendricksen K , Molkenboer-Kuenen J , Oosterwijk E , Hulsbergen-van de Kaa CA , Witjes JA . Evaluation of an orthotopic rat bladder urothelial cell carcinoma model by cystoscopy. BJU Int. (2008) ;101: (7):889–93. |
[48] | Knapp DW , Glickman NW , Denicola DB , Bonney PL , Lin TL , Glickman LT . Naturally-occurring canine transitional cell carcinoma of theurinary bladder A relevant model of human invasive bladder cancer. Urol Oncol. (2000) ;5: (2):47–59. |
[49] | KE. KDR-VJMGDDBPY. Animal models: Naturally Occurring Canine Urinary Bladder Cancer. In: CN. LSSMS, editor. Textbook of Bladder Cancer: Taylor and Francis, Oxon, UK; (2006) . pp. 171–5. |
[50] | Dhawan D , Ramos-Vara JA , Stewart JC , Zheng R , Knapp DW . Canine invasive transitional cell carcinoma cell lines: In vitro tools to complement a relevant animal model of invasive urinary bladder cancer. Urol Oncol. (2009) ;27: (3):284–92. |
[51] | Knapp DW , Ramos-Vara JA , Moore GE , Dhawan D , Bonney PL , Young KE . Urinary bladder cancer in dogs, a naturally occurring model for cancer biology and drug development. Ilar J. (2014) ;55: (1):100–18. |
[52] | Knapp DW , Chan TC , Kuczek T , Reagan WJ , Park B . Evaluation of in vitro cytotoxicity of nonsteroidal anti-inflammatory drugs against canine tumor cells. Am J Vet Res. (1995) ;56: (6):801–5. |
[53] | Shapiro SG , Knapp DW , Breen M . A cultured approach to canine urothelial carcinoma: Molecular characterization of five cell lines. Canine Genetics and Epidemiology. (2015) ;2: :15. |
[54] | Fowles JS , Dailey DD , Gustafson DL , Thamm DH , Duval DL . The FlintAnimal Cancer Center (FACC) Canine Tumour Cell Line Panel: Aresource for veterinary drug discovery, comparative oncology andtranslational medicine. Vet Comp Oncol. (2017) ;15: (2):481–92. |
[55] | van Hooren L , Sandin LC , Moskalev I , Ellmark P , Dimberg A , Black P , et al. Local checkpoint inhibition of CTLA-4 as a monotherapy or in combination with anti-PD1 prevents the growth of murine bladder cancer. European Journal of Immunology. (2017) ;47: (2):385–93. |
[56] | Le Magnen C , Dutta A , Abate-Shen C . Optimizing mouse models for precision cancer prevention. Nature Reviews Cancer. (2016) ;16: (3):187–96. |
[57] | Zhang ZT , Pak J , Shapiro E , Sun TT , Wu XR . Urothelium-specific expression of an oncogene in transgenic mice induced the formation of carcinoma in situ and invasive transitional cell carcinoma. Cancer Res. (1999) ;59: (14):3512–7. |
[58] | John BA , Said N . Insights from animal models of bladder cancer: Recent advances, challenges, and opportunities. Oncotarget. (2017) ;8: (34):57766–81. |
[59] | Kobayashi T , Owczarek TB , McKiernan JM , Abate-Shen C . Modelling bladder cancer in mice: Opportunities and challenges. Nat Rev Cancer. (2015) ;15: (1):42–54. |
[60] | Kates M , Nirschl T , Sopko NA , Matsui H , Kochel CM , Reis LO , et al. Intravesical BCG induces CD4+ T-cell expansion in an immune competent model of bladder cancer. Cancer Immunol Res. (2017) ;5: (7):594–603. |
[61] | van Hooren L , Sandin LC , Moskalev I , Ellmark P , Dimberg A , Black P , et al. Local checkpoint inhibition of CTLA-4 as a monotherapy or in combination with anti-PD1 prevents the growth of murine bladder cancer. Eur J Immunol. (2017) ;47: (2):385–93. |
[62] | Zhu YT , Pang SY , Lei CY , Luo Y , Chu QJ , Tan WL . Development of a therapy against metastatic bladder cancer using an interleukin-2 surface-modified MB49 bladder cancer stem cells vaccine. Stem Cell Res Ther. (2015) ;6: :224. |
[63] | Inoue T , Terada N , Kobayashi T , Ogawa O . Patient-derived xenografts as in vivo models for research in urological malignancies. Nat Rev Urol. (2017) ;14: (5):267–83. |
[64] | Tentler JJ , Tan AC , Weekes CD , Jimeno A , Leong S , Pitts TM , et al. Patient-derived tumour xenografts as models for oncology drug development. Nature reviews Clinical oncology. (2012) ;9: (6):338–50. |
[65] | Jäger W , Xue H , Hayashi T , Janssen C , Awrey S , Wyatt AW , et al. Patient-derived bladder cancer xenografts in the preclinical development of novel targeted therapies. Oncotarget. (2015) ;6: (25):21522–32. |
[66] | Gao H , Korn JM , Ferretti S , Monahan JE , Wang Y , Singh M , et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response ((2015) ;21: (11):1318–25. |
[67] | Hay JH , Busuttil A , Steel CM , Duncan W . The growth and histological characteristics of a series of human bladder cancer xenografts. Radiother Oncol. (1986) ;7: (4):331–40. |
[68] | Bernardo C , Santos LL . Patient-derived bladder cancer xenografts. Methods Mol Biol. (2018) ;1655: :169–75. |
[69] | Chan E , Patel A , Heston W , Larchian W . Mouse orthotopic models for bladder cancer research. BJU Int. (2009) ;104: (9):1286–91. |
[70] | Zhang N , Li D , Shao J , Wang X . Animal models for bladder cancer: The model establishment and evaluation (Review). Oncol Lett. (2015) ;9: (4):1515–9. |
[71] | Chan ES , Patel AR , Smith AK , Klein JB , Thomas AA , Heston WD , et al. Optimizing orthotopic bladder tumor implantation in a syngeneic mouse model. J Urol. (2009) ;182: (6):2926–31. |
[72] | Horinaga M , Harsch KM , Fukuyama R , Heston W , Larchian W . Intravesical interleukin-12 gene therapy in an orthotopic bladder cancer model. Urology. (2005) ;66: (2):461–6. |
[73] | Larchian WA , Horiguchi Y , Nair SK , Fair WR , Heston WD , Gilboa E . Effectiveness of combined interleukin 2 and B7.1 vaccination strategy is dependent on the sequence and order: A liposome-mediated gene therapy treatment for bladder cancer. Clin Cancer Res. (2000) ;6: (7):2913–20. |
[74] | Dobek GL , Godbey WT . An orthotopic model of murine bladder cancer. J Vis Exp. (2011) (48). |
[75] | Jäger W , Moskalev I , Janssen C , Hayashi T , Gust KM , Awrey S , et al. Minimally invasive establishment of murine orthotopic bladder xenografts. Journal of Visualized Experiments: JoVE. (2014) )84 51123. |
[76] | Werthman PE , Drazan KE , Rosenthal JT , Khalili R , Shaked A . Adenoviral-p53 gene transfer to orthotopic and peritoneal murine bladder cancer. J Urol. (1996) ;155: (2):753–6. |
[77] | Wu Y , McRoberts K , Berr SS , Frierson HFJr , Conaway M , Theodorescu D . Neuromedin U is regulated by the metastasis suppressor RhoGDI2 and is a novel promoter of tumor formation, lung metastasis and cancer cachexia. Oncogene. (2007) ;26: (5):765–73. |
[78] | John BA , Xu T , Ripp S , Wang HR . A real-time non-invasive auto-bioluminescent urinary bladder cancer xenograft model. Mol Imaging Biol. (2017) ;19: (1):10–4. |
[79] | Scheepbouwer C , Meyer S , Burggraaf MJ , Jose J , Molthoff CF . A multimodal imaging approach for longitudinal evaluation of bladder tumor development in an orthotopic murine model. PLoS One. (2016) ;11: (8):e0161284. |
[80] | Black PC , Dinney CPN . Bladder cancer angiogenesis and metastasis–translation from murine model to clinical trial. Cancer Metastasis Reviews. (2007) ;26: (3-4):623–34. |
[81] | Wilmanns C , Fan D , Obrian C , Radinsky R , Bucana C , Tsan R , et al. Modulation of Doxorubicin sensitivity and level of p-glycoprotein expression in human colon-carcinoma cells by ectopic and orthotopic environments in nude-mice. Int J Oncol. (1993) ;3: (3):413–22. |
[82] | Cutz JC , Guan J , Bayani J , Yoshimoto M , Xue H , Sutcliffe M , et al. Establishment in severe combined immunodeficiency mice of subrenal capsule xenografts and transplantable tumor lines from a variety of primary human lung cancers: Potential models for studying tumor progression-related changes. Clin Cancer Res. (2006) ;12: (13):4043–54. |
[83] | Khanna C , Hunter K . Modeling metastasis in vivo . Carcinogenesis. (2005) ;26: (3):513–23. |
[84] | Overdevest JB , Thomas S , Kristiansen G , Hansel DE , Smith SC , Theodorescu D . CD24 offers a therapeutic target for control of bladder cancer metastasis based on a requirement for lung colonization. Cancer Research. (2011) ;71: (11):3802–11. |
[85] | Nicholson BE , Frierson HF , Conaway MR , Seraj JM , Harding MA , Hampton GM , et al. Profiling the evolution of human metastatic bladder cancer. Cancer Res. (2004) ;64: (21):7813–21. |
[86] | Smith SC , Nicholson B , Nitz M , Frierson HF , Smolkin M , Hampton G , et al. Profiling bladder cancer organ site-specific metastasis identifies LAMC2 as a novel biomarker of hematogenous dissemination. The American Journal of Pathology. (2009) ;174: (2):371–9. |
[87] | Klaunig JE , Barut BA . Role of the implantation site on metastatic ability of the murine MBT-2 transitional cell carcinoma. Urological Research. (1988) ;16: (1):19–21. |
[88] | Lodillinsky C , Rodriguez V , Vauthay L , Sandes E , Casabe A , Eijan AM . Novel invasive orthotopic bladder cancer model with highcathepsin B activity resembling human bladder cancer. The Journal of Urology. (2009) ;182: (2):749–55. |
[89] | Capes-Davis A , Reid YA , Kline MC , Storts DR , Strauss E , Dirks WG , et al. Match criteria for human cell line authentication: Where do we draw the line? International Journal of Cancer. (2013) ;132: (11):2510–9. |
[90] | Chiong E , Dadbin A , Harris LD , Sabichi AL , Grossman HB . The use of short tandem repeat profiling to characterize human bladder cancer cell lines. J Urol. (2009) ;181: (6):2737–48. |
[91] | Sibley K , Cuthbert-Heavens D , Knowles MA . Loss of heterozygosity at 4p16.3 and mutation of FGFR3 in transitional cell carcinoma. Oncogene. (2001) ;20: (6):686–91. |
[92] | Pinto-Leite R , Carreira I , Melo J , Ferreira SI , Ribeiro I , Ferreira J , et al. Genomic characterization of three urinary bladder cancer cell lines: Understanding genomic types of urinary bladder cancer. Tumour Biol. (2014) ;35: (5):4599–617. |
[93] | MacLeod RA , Dirks WG , Reid YA , Hay RJ , Drexler HG . Identity of original and late passage Dami megakaryocytes with HEL erythroleukemia cells shown by combined cytogenetics and DNA fingerprinting. Leukemia. (1997) ;11: (12):2032–8. |
[94] | Seager C , Puzio-Kuter AM , Cordon-Cardo C , McKiernan J , Abate-Shen C . Mouse Models of Human Bladder Cancer as a Tool for Drug Discovery. Current Protocols in Pharmacology: John Wiley & Sons, Inc; (2001) . |
[95] | Gartler SM . Apparent Hela cell contamination of human heteroploid cell lines. Nature. (1968) ;217: (5130):750–1. |
[96] | Nardone RM . Curbing rampant cross-contamination and misidentification of cell lines. Biotechniques. (2008) ;45: (3):221–7. |
[97] | Skloot R . The immortal life of Henrietta Lacks. New York: Crown Publishers; (2010) . |
[98] | Gey G , Coffman WD , Kubicek MT . Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. (1952) ;12: :264–5. |
[99] | Jager W , Horiguchi Y , Shah J , Hayashi T , Awrey S , Gust KM , et al. Hiding in plain view: Genetic profiling reveals decades old cross contamination of bladder cancer cell line KU7 with HeLa. J Urol. (2013) ;190: (4):1404–9. |
[100] | Johnen G , Rozynek P , von der Gathen Y , Bryk O , Zdrenka R , Johannes C , et al. Cross-contamination of a UROtsa stock with T24 cells–molecular comparison of different cell lines and stocks. PLoS One. (2013) ;8: (5):e64139. |
[101] | Tachibana M . Studies on cellular adhesiveness in five different culture cell lines derived from carcinoma of the urinary bladder. Keio J Med. (1982) ;31: (3):127–48. |
[102] | Hadaschik BA , ter Borg MG , Jackson J , Sowery RD , So AI , Burt HM , et al. Paclitaxel and cisplatin as intravesical agents against non-muscle-invasive bladder cancer. BJU Int. (2008) ;101: (11):1347–55. |
[103] | O’Toole CM , Povey S , Hepburn P , Franks LM . Identity of some human bladder cancer cell lines. Nature. (1983) ;301: (5899):429–30. |
[104] | Masters JR , Bedford P , Kearney A , Povey S , Franks LM . Bladder cancer cell line cross-contamination: Identification using a locus-specific minisatellite probe. Br J Cancer. (1988) ;57: (3):284–6. |
[105] | Dirks WG , MacLeod RA , Drexler HG . ECV304 (endothelial) is really T24 (bladder carcinoma): Cell line cross- contamination at source. In Vitro Cell Dev Biol Anim. (1999) ;35: (10):558–9. |
[106] | Phuchareon J , Ohta Y , Woo JM , Eisele DW , Tetsu O . Geneticprofiling reveals cross-contamination and misidentification of 6adenoid cystic carcinoma cell lines: ACC2, ACC3, ACCM, ACCNS, ACCSand CAC2. PLoS One. (2009) ;4: (6):e6040. |
[107] | Bian X , Yang Z , Feng H , Sun H , Liu Y . A combination of species identification and STR profiling identifies cross-contaminated cells from 482 human tumor cell lines. Sci Re. (2017) ;7: (1):9774. |
[108] | Huang Y , Liu Y , Zheng C , Shen C . Investigation of cross-contamination and misidentification of 278 widely used tumor cell lines. PLoS One. (2017) ;12: (1):e0170384. |
[109] | Almeida JL , Cole KD , Plant AL . Standards for cell line authentication and beyond. PLoS Biology. (2016) ;14: (6):e1002476. |
[110] | Reid Y , Storts D , Riss T , Minor L . Authentication of Human Cell Lines by STR DNA Profiling Analysis (2004) . |
[111] | Committee ICLA. Guide to Human Cell Line Authentication: ANSI/ATCC ASN-0002-2011. (2012) . |
[112] | Cooper JK , Sykes G , King S , Cottrill K , Ivanova NV , Hanner R , et al. Species identification in cell culture: A two-pronged molecular approach. In Vitro Cell Dev Biol Anim. (2007) ;43: (10):344–51. |
[113] | ICLAC. Register of Misidentified Cell Lines 2012 [Available from: http://iclac.org/ |
[114] | Fusenig NE , Capes-Davis A , Bianchini F , Sundell S , Lichter P . The need for a worldwide consensus for cell line authentication: Experience implementing a mandatory requirement at the International Journal of Cancer. PLoS Biology. (2017) ;15: (4):e2001438. |
[115] | Editorial. What’s Your Line? Clinical Cancer Research. (2009) ;15: (13):4251. |
[116] | Editorial. Identity crisis. Nature. (2009) ;457: (7232):935–6. |