Presenting and preserving the change in taxonomic knowledge for linked data
Abstract
Taxonomic knowledge provides a scientific name to each organismal group and is thus indispensable information for understanding biodiversity. However, the various perspectives of classifying organisms and changes in taxonomic knowledge have led to inconsistent classification information among different databases and repositories. To have a precise understanding of taxonomy, one needs to integrate relevant data across taxonomic databases. This is difficult to establish due to the ambiguity in taxon interpretation. Most researchers in earlier stages employed the Linked Open Data (LOD) technique to establish links in taxonomy transition. However, they overlooked the temporal representation of taxa and underlying knowledge of the change in taxonomy, so it is difficult for learners to gain perspective on how some identifiers of taxa are linked. To this end, this research is aimed at developing a model for presenting and preserving the change in taxonomic knowledge in the Resource Description Framework (RDF). Specifically, the proposed model takes advantage of linking Internet resources representing taxa, presenting historical information of taxa, and preserving the background knowledge of the change in taxonomic knowledge in order to enable a better understanding of organisms. We implement a prototype to demonstrate the feasibility and the performance of our approach. The results show that the proposed model is able to handle various practical cases of changes in taxonomic works and provides open and accurate access to linked data for biodiversity.
1.Introduction
Knowledge about biodiversity has been written about everywhere throughout the world. Researchers need to exchange knowledge about biodiversity across communities, so the link between communities’ knowledge becomes a challenging issue, and an intermediary is required. Nomenclature or a system of names was originally introduced to give a unique and stable name, also known as a scientific name, as an identity for every organism on Earth [28,48]. Ideally, a scientific name should be unique and be a medium of linked data; however, in fact, there is confusion due to ambiguous taxonomic notations. This leads to a change in taxonomic knowledge that becomes a serious problem, such as a change in taxa or nomenclatures [28,33,48,49]. This therefore results in imprecise linked knowledge, causing a single taxon to be misunderstood.
Fortunately, in the present age, the Internet and Semantic Web technologies provide a rich platform for linking data [18]. The idea of data interoperability enables a way to exchange data among different information systems. Information systems can be developed on the basis of their own requirements and own data structures. When researchers publish their own datasets, they should consider the ability to link, to be recognized by humans, and to be consistent with standards. Moreover, local vocabularies used by an individual system should be reused from or associated to existing ones in order to build effective linked data [18].
Therefore, developing a taxonomic information system that places importance on both knowledge management and linked data would be conducive to the better understanding of taxonomic knowledge. For this reason, our research is aimed at introducing a logical model for linking taxonomic knowledge on the basis of the following objectives.
– To preserve the change in taxonomic knowledge.
– To present and publish taxonomic knowledge as linked data.
To accomplish these objectives, we considered utilizing the idea of the Contextual Knowledge for Archives (CKA) approach [6] and the Meta-Ontology of Biological Name (TaxMeOn) [46] to capture the changes in taxonomic data and their context. We have, moreover, reused some taxonomic terms from Linked Open Data for ACademia (LODAC) [30], employed Simple Knowledge Organization System (SKOS) [41] vocabulary to manage the relationships between concepts, and publicized data to the Linked Open Data (LOD) Cloud [27]. In addition, we implemented a prototype to prove the feasibility of our proposed model. Finally, we evaluated this work against the real cases of changes in the taxonomy of moths under the family Saturniidae [19,21,24].
We give the background for our research in Section 2. We introduce our approach and fundamental concepts in Section 3. The prototype is presented in Section 4. The approach is evaluated in Section 5. Then, we discuss the outcome of our work in Section 6. Last, we draw conclusions and suggest some future improvements in Section 7.
2.Background
To analyze the change in taxonomic knowledge and provide the basis of our model, we here review relevant research and then give details on the change in taxonomic knowledge, online databases, and linked data.
2.1.Change in taxonomic knowledge and consequent impact
A large number of species throughout the world have been described and classified with appropriate naming according to their characteristics such as morphological characters, living behaviors, and DNA sequences [28,48]. Many taxonomists have dedicated themselves to studying organismal groups, and their knowledge has been published for more than hundred years. However, this knowledge has not always been shared among all researchers around the world. In addition, there is no consensus on classification systems among all taxonomists. In other words, taxonomists might have different perspectives when it comes to classifying and naming organismal groups. As a consequence, a single species is often classified and named differently [48]. To describe this situation more clearly, in this part, we demonstrate cases of change in taxonomic knowledge.
The first example shows that one organism may have different names. If we take the Chinese yellow swallowtail, named Papilio xuthus Linnaeus, 1767 as an example, we see that taxonomists at different research institutes have given this species different names, such as xuthulus Bremer, 1861, chinensis Neuburger, 1900, koxinga Fruhstorfer, 1908, and neoxuthus Fruhstorfer, 1908 [48].
Second, when two or more taxa were recognized as the same thing, only one name became accepted [20]. Thus, some species have to be reclassified and renamed due to the naming system [28,48]. For example, in 2008, Hoare established the genus Kendrickia (ostracods). Then, in 2010, Kempf found that this genus was a primary junior homonym for Kendrickia Solem, 1985 (gastropods) and proposed the name Dickhoarea as a replacement name for the Kendrickia Hoare, 2008. This led to the subsequent change in species names; for instance, Kendrickia asketos has subsequently been renamed Dickhoarea asketos since Kempf announced the name in 2010 [48].
Next, the progress of taxonomic studies frequently causes the redefinition of taxon concepts, i.e., the circumscription of the taxon [48]. Sometimes, it results in the change in species name. For example, the genus Columba (pigeons) has been split into five genera, Patagioenas, Chloroenas, Lepidoenas, Oenoenas, and Columba, where the latter Columba is narrower than the former one. Some species of the genus Columba have been assigned to one of these newly separated genera, for instance, Columba speciosa was changed to Patagioenas speciosa [2].
Another situation is to merge taxa such as on the genus level. When some genera were decided to be merged into a single taxon, their lower taxa such as species had to be transferred to the newly accepted genus [20]. According to nomenclature, these species had to be renamed to be consistent with the new genus name [28,48]. For instance, two genera of owls, Bubo and Nyctea, were merged into the prior genus Bubo. Following the change in these genera, the scientific name of the snowy owl Nyctea scandiaca has been subsequently changed to Bubo scandiacus in order to satisfy the zoological nomenclature [47].
Moreover, some researchers may have an incorrect understanding of some taxon concepts as a result of them having been reclassified frequently, for example, a reclassification of the Baltimore oriole (Icterus galbula Linnaeus, 1758) and the Bullock’s oriole (I. bullockii Swainson, 1827). In 1964, Sibley and Short argued that these two species should be merged into a single species [40]. As a result, the former name, I. galbula, became the accepted name, whereas I. bullockii was a junior synonym of I. galbula. In contrast, in 1995, research results regarding the DNA sequences of the two species led to the splitting of I. galbula into I. galbula and I. bullockii again [15]. Although these two species are currently separate, some information on I. galbula, especially which recorded between 1964 and 1995, might include important details on I. bullockii. Researchers sometimes obtain imprecise information when they simply search for information by using the name I. galbula only.
In these studies, we regard change in taxonomic knowledge to possibly be change in name and change in classification [14,28,46,48]. The example cases demonstrated the problems that occur when each name reflects particular details observed by each researcher. Due to such a change in taxonomic names, when one who studies species data accesses only information containing only the present scientific name, she or he sometimes misses important information that was recorded with its former scientific names. This means that the scientific names and taxonomy lack a single interpretation in biology [29,49]. Thus, to understand taxonomy thoroughly, we therefore need to know all synonyms across multiple datasets and then link their associated information together via the Internet [18]. Learning taxonomy with a single name may not be enough. To learn the precise knowledge of taxonomy, researchers have to pay attention to the significance of the change in taxa over time. Finding associations among the background knowledge of changes is also needed to be studied in order to understand the taxonomic knowledge more correctly.
2.2.Informatics on taxonomic databases
In light of the issue previously mentioned, this study is an attempt to address the problem of incorrect interpretation of taxonomic data. An approach to linking taxonomic data along with the precise context and preservation of their background information is clearly needed.
Therefore, in this section, we review several pieces of research that deal with solving this issue. A poor data model leads to the lack of linkability among different datasets [37]. A scientific name alone is not enough for introducing a precise link [3,22,23,25,32,33,37,38,49]. The International Organization for Plant Information (IOPI) model [3] used taxonomic names together with circumscription references as potential taxa for linking data among multiple taxonomic views. The Biodiversity Information Standard (TDWG) [5,43] developed a standard for taxonomic data sharing among different datasets, adopted Life Science Identifiers (LSIDs) as Globally Unique Identifiers (GUIDs) for indexing taxa, and allowed having versions of taxon concepts. It also provided Darwin Core schema [9] containing vocabularies for describing taxonomic data. Page [32] and Jones et al. [22] employed LSIDs for taxonomic databases, and the links of LSIDs can associate information among various data sources. The Universal Biological Indexer and Organizer (uBio) also gave LSIDs to taxa for enhancing the power of federated search engines [37]. As every taxon has been indexed with an ID, relations between taxa can be given by using links between IDs [23]. Schulz et al. [38] embedded the taxonomy of living things into an ontology by using semantic technology. The hierarchy of taxon concepts was represented in the Resource Description Framework (RDF) [38,43].
However, these researches have not yet mentioned about the preservation of changes in taxonomic knowledge. For this reason, TaxMeOn [46] developed a Semantic Web-based meta-ontology of biological names that managed and presented the changes in the scientific preposition of biological names and taxonomies such as splitting and lumping, and it emphasized how the biological names were published by referring to related publications. However, to the best of our knowledge, there is less discussion about the information structure of associations between any reasons behind changes or background knowledge, which is needed to make a clear understanding of taxonomy.
This challenge puts forward the view that an underlying knowledge of the changes in taxonomic knowledge is required for the correct interpretation of taxonomic data. The study of biodiversity informatics should focus on the inclusion of the historical changes in taxa and the context information that is essential for understanding the situation regarding their changes and how names are related as well.
2.3.Taxonomic knowledge and linked data
To materialize the conception of linked data, in this part, we studied how to utilize an Internet resource as an identifier for representing a taxon. There are several views on using identifiers such as LSIDs or URIs and human-readable or non-human-readable identifiers, which are reviewed as follows.
The use of LSIDs as GUIDs promoted by TDWG [5,43] resulted in taxonomic data becoming globally available and linked. Several information models adopted the LSID as a unique key representing a taxon in their databases [3,22,32]. Jones et al. [22] resolved the multiple names by assigning separated LSIDs for a name (NAMELSID) and for a taxon (TAXONLSID) and integrated the LSID into the uniform resource identifier (URI). In addition, the authors of [25] compared the differences between the LSID and the URI and recommended using a URI as a resource of taxonomic data in order to gain benefit from the Linked Data approach. TaxMeOn [46] also put forward the view that taxon concepts are always changed, so a fixed identifier might not proper for every concept. Therefore, when a taxon’s circumscription was changed, that concept needed to be recognized as a new identifier. For instance, the genus Bubo, before merging with the genus Nyctea, must not have the same Semantic Web-based identifier as the Bubo after merging because the latter Bubo is broader than the former one [47,48]. The model also allowed having a URI for a taxon concept and a URI for its name. It therefore had minimal redundancy and was flexible for updating either names or concepts. Nevertheless, TaxMeOn propounded the view that a taxon concept and its name were treated as one unit in a name collection. The domain or the range of properties is allowed to be a union of the scientific name and taxon concept.
Patterson et al. [33] additionally introduced the Global Names Architecture (GNA) and supported the view that names were keys to access biological information. GNA, which mainly treat names with implicit taxon concepts, has three layers, but two layers are related to this topic. One is the Global Names Index (GNI), which is aimed at collecting name strings used in various information sources and normalized spellings. Another one is the Global Names Usage Bank (GNUB). It is aimed at describing name uses, which is a combination of a name and a reference, and nomenclatural issues. This name-centric model also provided features for identifying relationships between names, and it was integrated into online official repositories of names such as ZooBank [35] and MycoBank [7]. The authors of [25] argued that it was very challenging to combine a name and a taxon concept into a single unit because doing so decreased the granularity of information but gave high simplicity. In addition, naming conventions for identifiers are different among different systems. The Global Biodiversity Information Facility (GBIF), which is an international organization aiming to construct an information infrastructure for sharing information on biodiversity globally, gave a reference guide for GNA. It is a guide for an information system to select valid, accepted names among all names used for living beings, and it recommends using an unfriendly label for a persistent identifier because a taxonomic name is not stable [8,34,36,44]. The authors of [25] used non-human-readable local names in URIs. TaxMeOn [46] does not specify the format of the URIs for data instances, so it is possible to use either human-readable or non-human-readable URIs. Furthermore, LODAC [30], which provided a linked data hub for biodiversity, denoted a URI as an Internet resource for representing a piece of taxonomic data. LODAC also considered including a human-readable label in URI in order to make the model be lightweight and human-friendly such as lodac:Bubo. It is consistent with the URIs of Internet resources used by DBpedia [26]. In this case, the human-readable URI is sometimes viewed as either a name or a taxon concept depending on the context. It also gives an advantage to humans, especially biologists, who are involved with linked data because the human-readable URI reduces the gap between machines and normal users.
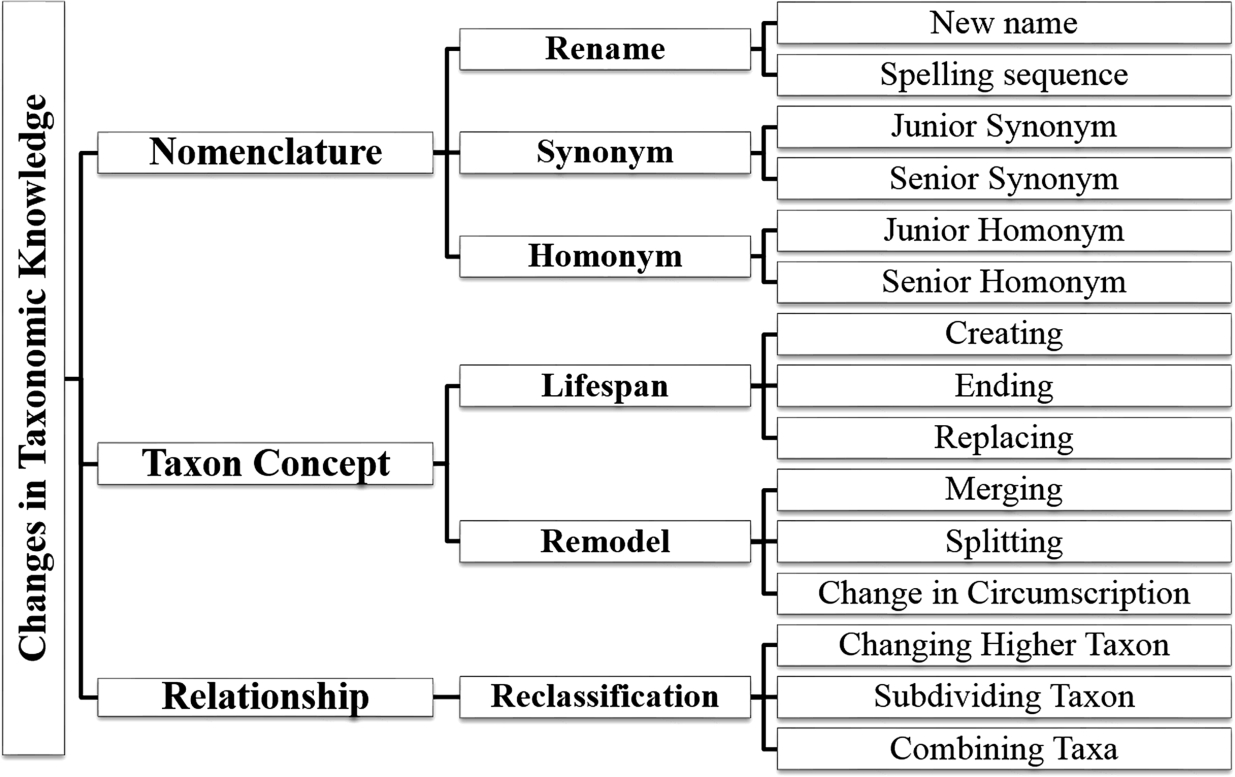
Fig. 1.
Analysis of changes in taxonomic knowledge.
3.Logical model for linking taxonomic knowledge
Here, we present a logical model named “Linked Taxonomic Knowledge” (LTK) for preserving and presenting the change in taxonomic knowledge for linked data. To achieve the goals and issues addressed in the previous sections, our logical model was developed on the basis of the following points.
– The model can manage the changes in taxonomic knowledge.
* The model preserves the changes as an event along with aspects of time and provenance.
* The model supports the changes in either taxa or association between taxa.
* The model allows tracing the background knowledge of the changes by linking the cause and effect between them.
– The model can be used to publish a suitable format for a dataset for linked open data.
* The linked data model deals with simple identifiers of Semantic Web resources in order to make the linked data be easily recognized by both humans and machines.
* The model provides a sequence of changes in taxa.
* The model presents temporal data on the basis of a given time point.
In this section, we illustrate the types of changes in taxonomic knowledge, terms and descriptions, a formal definition of LTK, a use of the data model, a description of the rules, and a method for utilizing our approach in the Resource Description Framework (RDF). This section usually uses shorthand aliases for URIs, so their namespaces can be referenced in Appendix A.
3.1.Structure to represent change in taxonomic knowledge
In this part, we studied how to classify a change in taxonomic knowledge. On the basis of these changes analyzed from actual use cases [14,15,19,21,24,40,46–48], we summarized the practical cases in Fig. 1. The figure shows that there are three main categories: changes in nomenclature, taxon concept, and relationship.
First, the category nomenclature refers to the change in name including rename, synonym, and homonym. A synonym is used when different names are assigned for the same taxon, whereas a homonym is used when the same name is assigned for different taxa.
Second, the change in taxon concept denotes the change in the description of a taxon. It includes the life span of the use of taxa that are initially stated (creating) and made obsolete (ending) and the replacement of taxa in checklists. It also includes the change in the scopes of taxa, which are merging, splitting, and change in circumscription. Merging means to lump taxa into a single taxon, splitting is to separate a taxon into several taxa, and change in circumscription means to modify the scope of a single taxon. In this case, the taxa before the change are assumed to be made obsolete from the dataset, and then, the other taxa after the change become newly created.
Last, the change in relationship means a modification made to a link between concepts. In terms of the Semantic Web, it is a change in a triple. In this figure, three changes are mentioned. The change in a higher taxon moves a lower taxon from a higher taxon to another one. The subdividing taxon is to create new sub-taxa under the given taxon. This differs from splitting because the given taxon remains accepted, and its description does not change. For example, a species Aus aus was subdivided into subspecies A. aus aus and A. aus bus. Conversely, in case of combining sub-taxa, the sub-taxa of a given taxon are no longer used when all sub-taxa are combined into one concept and no subdivision is applicable. For example, when the two subspecies A. aus aus and A. aus bus are combined into one subspecies and there are no other valid subspecies, all subspecies names are no longer used.
3.2.Preliminary definitions
How to describe the changes in taxonomy along with context knowledge is a challenging task. In this research, we primarily employed the CKA approach, which offers a logical model for presenting the change in the underlying community knowledge based on the theory of Flouris and Meghini [13]. CKA offers a data model for an event that assures entities of changes and binds a time interval and some references. The entity of change or the operation of change captures the change in conception such as splitting and merging and the change in association between concepts such as changing membership. CKA also provides ideas for transforming the event of change into timeline and temporal data, which basically respond to the requirements of digital archives. However, we have to enhance the CKA approach for satisfying the specific requirements of biodiversity informatics and also introduce some of the terms used by our research.
3.2.1.Entities for LTK
An entity in LTK is a URI for responding to specific positions, for example, entities for representing taxa, operations of changes, and events describing the changes. In this case, some terms are needed to be defined and clarified.
Nominal entity Semantic technology encourages that everything should be represented as an Internet resource identified by a URI [18]. In this research, a nominal entity is a concept and an Internet resource used for taxonomic knowledge, and it includes taxon concepts and names.
Simple nominal entity This research moreover introduces a simple nominal entity as a subset of the nominal entity, and each of these entities corresponds to a single scientific name. Due to the change in knowledge, the role of a taxon has a lifespan. The simple nominal entity, which is an Internet resource, can act as either a taxon concept or a name according to the following situations. If a scientific name of any entity had been accepted for a certain period, that entity could be viewed as a taxon concept at any time in that period. In contrast, it becomes viewed as a taxonomic name when it is mentioned in other times. Thanks to an advantage from DBpedia [26] and LODAC [30], a human-readable URI makes RDF statements be human friendly in linked data graphs, for example, dbpedia:Bubo and lodac:Bubo. We recommend using simple nominal entities for several reasons. A model is simple and lightweight, presented data are easily recognized by normal users, and a triple in linked data is more understandable. In addition, the issue of homonyms can be solved by using a different namespace.
Contextual nominal entity Change in knowledge sometimes has an impact on some representative taxa, and their circumscription or their name may be changed. Our work deals with this problem by applying the idea of TaxMeOn [46], which creates different URIs for the same taxon when its description is changed. We additionally define that every representation of taxonomy used in LTK is viewed as a version of a nominal entity. In the case of supporting a simple nominal entity, this research provides the following recommendations.
1) A URI should include a scientific name and a version. We recommend using a year of the change as a version number such as genus:Bubo_1999.
2) If a change affects the change in nomenclature, a new URI should be created, and a link between the former and the latter URIs is developed to show the relationship between them.
3) In case that a new URI of a taxon concept is recreated for some purpose without a change in scientific name, the version number in the URI string should be updated.
The created contextual nominal entity can link to nominal entities from external datasets in order to make data be globally linkable. According to the standard of TDWG [5], our research uses the property dct:isVersionOf for linking between a contextual nominal entity and a nominal entity.
In practice, we make a simple nominal entity a representative of an external URI for maintaining links between the LTK dataset and external datasets. It is possible to link a contextual nominal entity with other taxonomic data such as the URIs or LSIDs from TDWG [5], GBIF [8,36,44], Catalog of Life (CoL) [22], LODAC [30], and DBpedia [26] via those representatives. For example, the following statement addresses an association among the contextual nominal entity (genus:Bubo_1999), the simple nominal entity as the representative of any external URIs (genus:Bubo), and the external URIs and LSIDs viewed as the nominal entity (gbif:5959091, lodac:Bubo, and urn:lsid:ubio.org:namebank:2473659).
genus:Bubo_1999 dct:isVersionOf genus:Bubo . genus:Bubo owl:sameAs gbif:5959091 , lodac:Bubo , <urn:lsid:ubio.org:namebank:2473659>.
Change entity (operation) A change entity or operation of change is a type of change in taxonomic knowledge, which was previously described, for example, replacing, merging, splitting, reclassifying, etc. In practice, these operations are subclasses of either cka:ConceptEvolution or cka:RelationshipEvolution. Our research generally uses instances of operations for managing changes in contextual nominal entities. Moreover, a link between operations can viewed as a link between background knowledge.
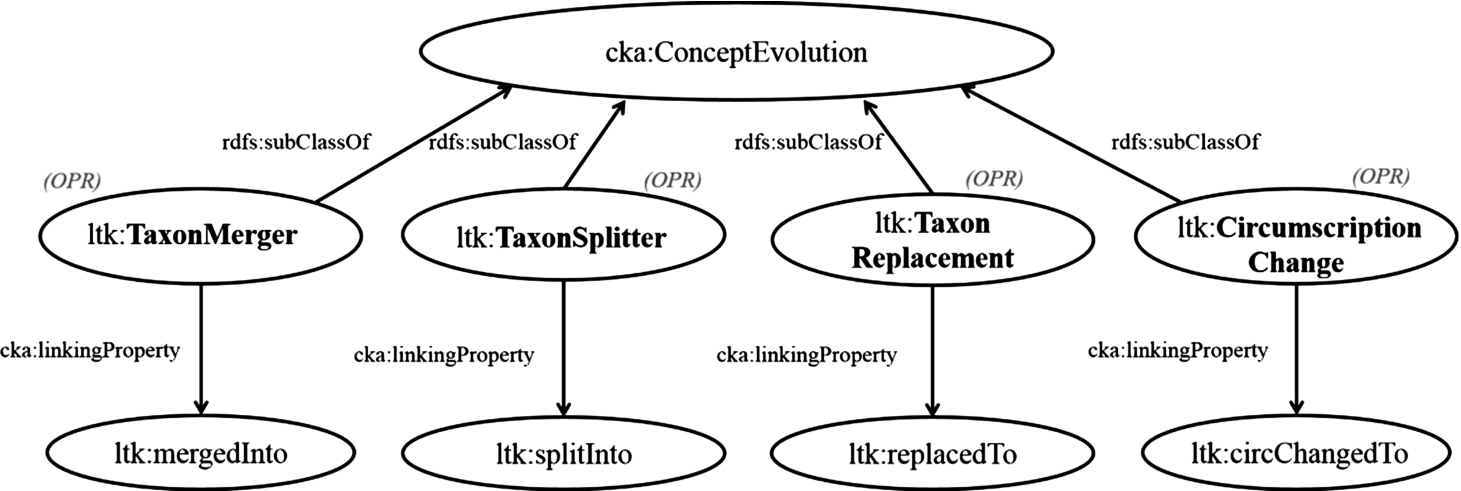
Fig. 2.
Model: Declaration of operations for changes in conception.
Event entity To reduce data redundancy, an event entity is created to group some operations that share the same aspects of time and provenance. Thus, the time interval and references are assigned to the event entity. In practice, the event entity is an instance of cka:CommunityKnowledge.
For the use of each entity, it is noted that our work does not restrict the representation of URIs. A simple nominal entity, unfriendly identifier, or separation of the scientific name and taxon concept are possible to use in our model.
In addition, in this research, we view the nominal entity, simple nominal entity, and contextual nominal entity as concepts, which are a subclass of skos:Concept. Because a change usually performs an action with concepts, from now on, when we mention the term “concept” in the context of change or with an operation of change, we mostly refer to a contextual nominal entity.
Last, since each entity is a Semantic Web resource, we added symbols to the figures in order to distinguish the types of entities:
– (nom) is an instance of a nominal entity,
– (sim) is an instance of a simple nominal entity,
– (con) is an instance of a contextual nominal entity,
– (OPR) is a class of a change entity (operation),
– (opr) is an instance of an operation, and
– (event) is an instance of an event entity.
3.2.2.Data models for LTK
In addition, to have researchers interpret data precisely, our knowledge management introduces various models of knowledge representations.
Event-centric model The event-centric model is a data structure that is used to preserve the change in taxonomic knowledge in RDF. It is based on the idea of CKA [6] that uses the n-ary relation for creating context-dependent RDF statements including operations, time intervals, and references [6,13,17]. Thus, the RDF presentation of this model is quite complicated by design. Although the model is expensive due to a lot of triples required, it is advantageous to various applications, especially in knowledge management systems.
Transition model The transition model is a model for presenting the chain of changes in contextual nominal entities. This model is transformed from the event-centric model by using Semantic Web rules. This model is presented as a general graph including only contextual nominal entities and their links, so it is simpler than the event-centric model and it works easily with linked data, but it is not good for representing background knowledge in detail.
Snapshot model The snapshot model is a set of simple RDF statements like the transition model, but it is generated according to a given time point. This model demonstrates how the information of a taxon changes over time.
3.3.Formal model for change in taxonomic knowledge
As mentioned in the previous section, the change in contextual nominal entities and the change in the relationship between them are key players in linking taxonomic knowledge. To present general definitions for the change in taxonomic knowledge, we propose a formal model for preserving and presenting the change in taxa for linked data. Our formal model is basically derived from the CKA approach [6]. The approach introduces a basic idea for how to reuse super classes from CKA to create an operation of the change in concepts and an operation of the change in relationship between two concepts and how to map an operation with a Semantic Web property.
3.3.1.Change in conception
In this part, we review the function of the change in concepts, cka:ConceptEvolution, which deals a set of concepts before the change and a set of concepts after the change. By reusing this function, we categorize the change in contextual nominal entities into four functions: ltk:TaxonMerger, ltk:TaxonSplitter, ltk:TaxonReplacement, and ltk:CircumscriptionChange. These operations are associated with four properties: ltk:mergedInto (merged into), ltk:splitInto (split into), ltk:replacedTo (replaced to), and ltk:cirChangedTo (circumscription changed to), respectively. Figure 2 shows the derivation of these operations, which are subclasses of cka:ConceptEvolution, and links to their associating properties. In special cases, basic changes such as merging and splitting occurring at once. This complex case is solved by the operation named ltk:TaxonComplexChange, which is a subclass of cka:ConceptEvolution. This operation allows multiple concepts before and after a change to be had, and the linking property of this operation is ltk:cpxChangedTo (complexly changed to). However, if it is possible to do, we recommend categorizing complex changes into simple operations, that is, merging, replacing, and splitting, for better understanding.
3.3.2.Change in relationship between taxa
In addition to the change in conception, we considered the operation of the change in the relationship between two things. The term relationship is used as a link not only for the same rank of taxon such as synonym but also for different ranks such as the hierarchical relationship. CKA [6] provides a superclass cka:RelationshipEvolution for capturing the change in association between two concepts. This operation generally records the transition of a triple by changing the object of the triple, but the subject and the predicate of the triple remain unchanged.
In LTK, we introduce operations to give evidence of the change in the relationship between two contextual nominal entities. For example, the change in the classification of a taxon, ltk:ChangeHigherTaxon, contributed a procedure for recording the change in the higher taxon rank of a taxon by switching the object of a predicate named ltk:higherTaxon to another one. Therefore, the operation ltk:ChangeHigherTaxon, which is a subclass of cka:RelationshipEvolution, is in charge of alternating a triple containing the relationship property named ltk:higherTaxon, as shown in Fig. 3.
Fig. 3.
Model: Example declaration of change in relationship between taxa.
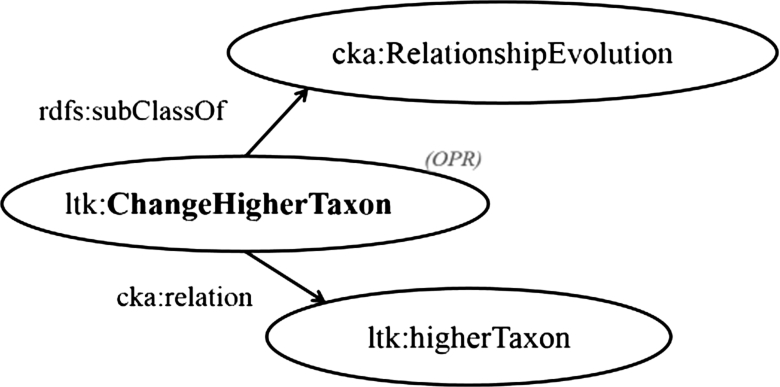
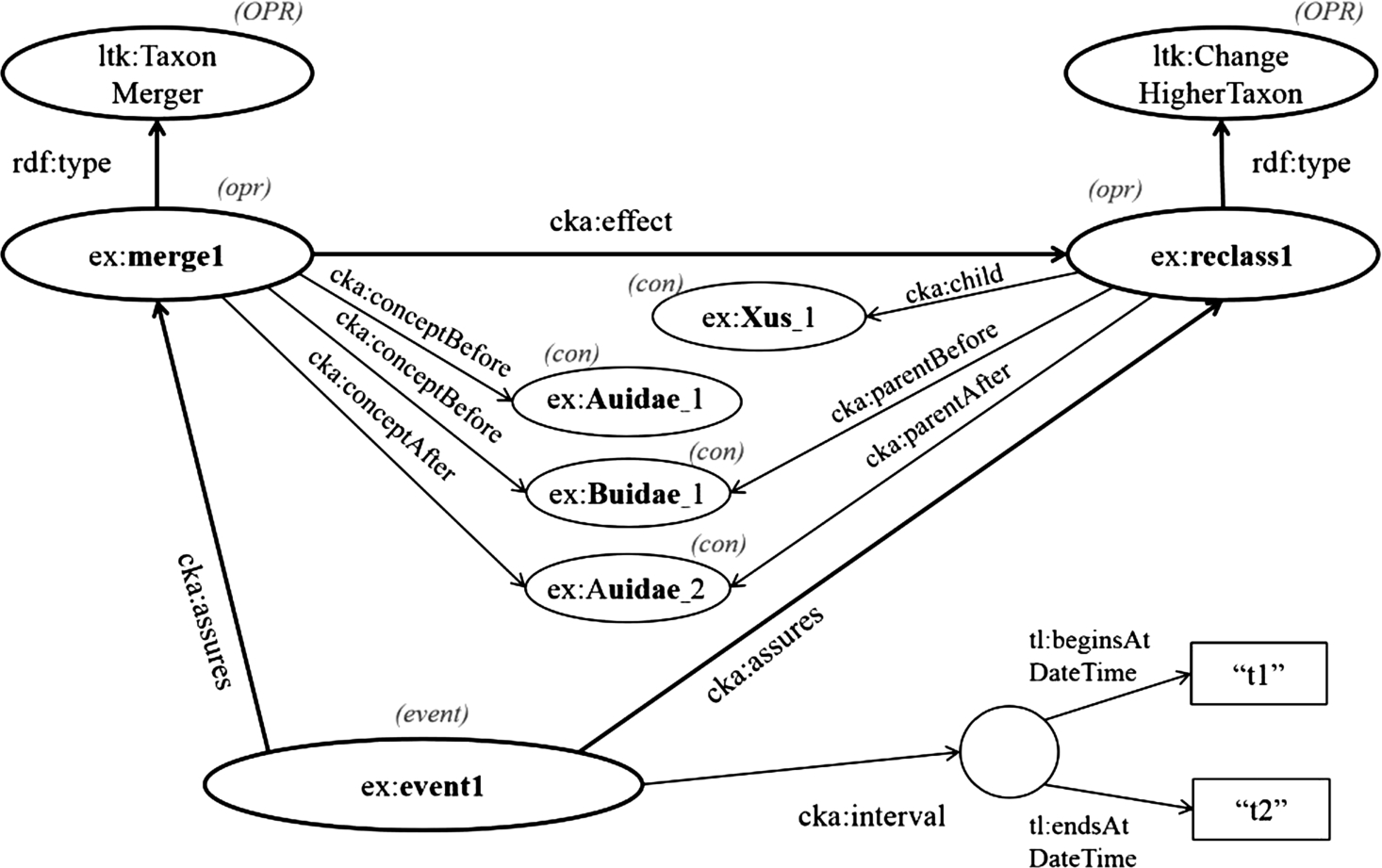
Fig. 4.
Model: Example event-centric model for representing changes in taxonomic knowledge.
3.4.Working with event-centric model
In this part, we present how to work with the even-centric model in order to capture the change in taxonomic knowledge. Here, we suppose the following simple test case. There are two families, Audae and Buidae, and Buidae includes one genus, Xus; then, at time t1, Buidae is merged into Audiae, and subsequently, the genus Xus is regarded as a member of a new URI of Auidae. This scenario is assumed to end at time t2; however, the end time point can be ignored if this event is still valid. In this case, we first assign URIs of contextual nominal entities for Auidae, Buidae, and Xus, which are ex:Auidae_1, ex:Buidae_1, and ex:Xus_1, respectively. When two families are merged into Audae at time t1, according the use of the contextual nominal entity, the model has to create a new URI of Auidae to be ex:Auidae_2. Then, the genus ex:Xus_1 is transferred to the newer accepted family. In nomenclature, a taxon higher than the genus level does not need its scientific name changed when it is transferred to another higher taxon [20,28]. Thus, the current URI of the genus ex:Xus1 is retained. However, if the change affects the scientific name of a taxon, a new contextual nominal entity has to be created, and a link between an old concept and a new concept has to be identified. Figure 4 demonstrates the changes in taxa, the change in relationship between them, and the event entity. First, the operation, ex:merge1, is the merging of ex:Auidae_1 and ex:Buidae_1 into ex:Auidae_2. Thus, the given values of cka:conceptBefore are ex:Auidae_1 and ex:Buidae_1, while the given value of cka:conceptAfter is ex:Auidae_2. Second, the change in relationship between contextual nominal entities, ex:reclass1, is the reclassification of ex:Xus_1 from ex:Buidae_1 to ex:Auidae_2. Hence, ex:Xus_1, ex:Buidae_1, and ex:Auidae_2 are assigned to the properties cka:child (≡cka:subject), cka:parentBefore (≡cka:objectBefore), and cka:parentAfter (≡cka:objectAfter), respectively. Moreover, according to this scenario, ex:merge1 results in ex:reclass1, so it can express that cka:effect maps ex:merge1 to ex:reclass1. Last, the event entity, named ex:event1, which is an instance of cka:CommunityKnowledge, confirms the two changes as mentioned above by using a property named cka:assures, and it identifies a temporal identity by using a property named cka:interval. The temporal identity mentions the begin time point “t1” by using the property tl:beginsAtDateTime and the end time point “t2” by using a property named tl:endsAtDateTime.
3.5.Working with Semantic Web rules
The examples mentioned in the previous section, which introduced context-dependent RDF statements, are general patterns for representing the change in taxonomic knowledge. The event-centric model is complex by design because it is used to preserve the change with context information. For the use of linked data, the complex expression detailed by the event-centric model is not suitable because it is difficult to use for making implicit links with existing semantic reasoners. Therefore, it has to transform the event-centric model into two easily-linkable models: the transition model and the snapshot model.
Fig. 5.
Rule: Transforming event-centric model into transition model.
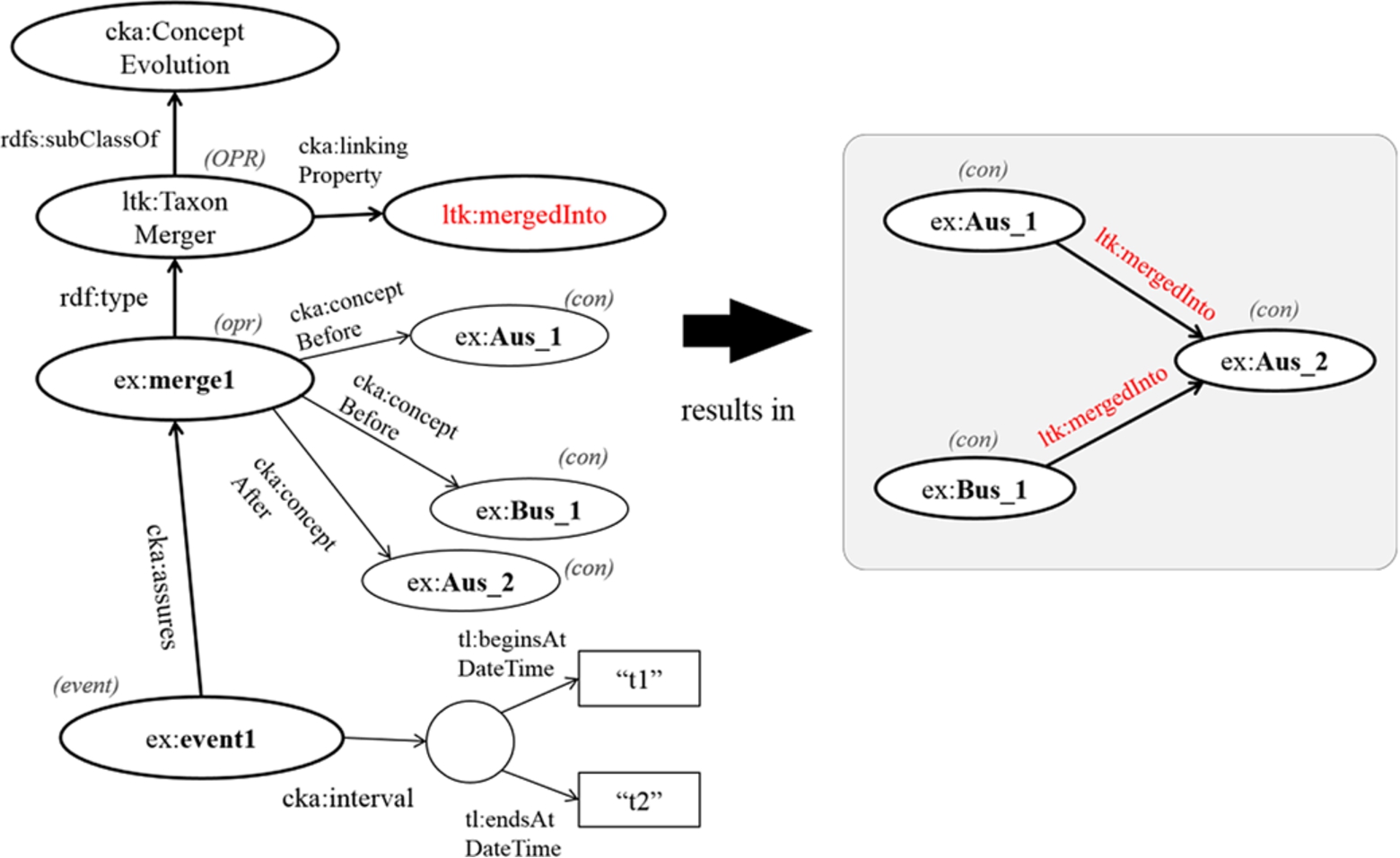
3.5.1.Generating transition model
First, we transform the event-centric model into the transition model. The following example Semantic Web rule gives a link between contextual nominal entities before and after merging.
TaxonMerger(?opr) ∧ conceptBefore(?opr,?c1) ∧ conceptAfter(?opr,?c2) ⇒ mergedInto(?c1,?c2)
The rule contains symbols named ?opr, ?c1, and ?c2, which are the variables of an operation of change, a contextual nominal entity before the change, and a contextual nominal entity after change, respectively. In this case, this is a merging operation, so the variable ?opr must be defined as an instance of the operation TaxonMerger. Executing this rule results in the linked data of taxa, that is, ex:Aus_1, ex:Bus_1, and ex:Aus_2, as shown in Fig. 5. In this research, the change is usually transformed into the transition model without any time references in order to demonstrate a timeline graph, but the time interval is employed by the snapshot model for displaying temporal changes in a concept. Moreover, in practice, we define a generic rule for each case, so the class named TaxonMerger and the property named mergedInto have to be represented by variables instead. The following statement expresses the common rule for linking concepts before the change (?c1) and concepts after the change (?c2), where the link is represented by a property (?p) bound with the operation of change (?OPR). Then, a triple containing ?c1, ?p, and ?c2 is produced.
subClassOf(?OPR, ConceptEvolution) ∧ linkinProperty(?OPR,?p) ∧ type(?opr,?OPR) ∧ conceptBefore(?opr,?c1) ∧ conceptAfter(?opr,?c2) ⇒?p(?c1,?c2)
3.5.2.Generating snapshot model
Second, we introduce a rule to transform the event-centric model into the snapshot model. Before executing the following rule, it is necessary to use a query statement to find only changes that contain a given concept and cover a given time point. After that, a property (?p), which is bound with an operation of the change in relationship (?opr), maps a subject (?s) and an object after the change (?oafter) to construct a triple.
subClassOf(?OPR, RelationshipEvolution) ∧ relation(?OPR,?p) ∧ type(?opr,?OPR) ∧ subject(?opr,?s) ∧ objectAfter(?opr,?oafter) ⇒?p(?s,?oafter)
Fig. 6.
Rule: Transforming event-centric model into different snapshot models according to different time points.
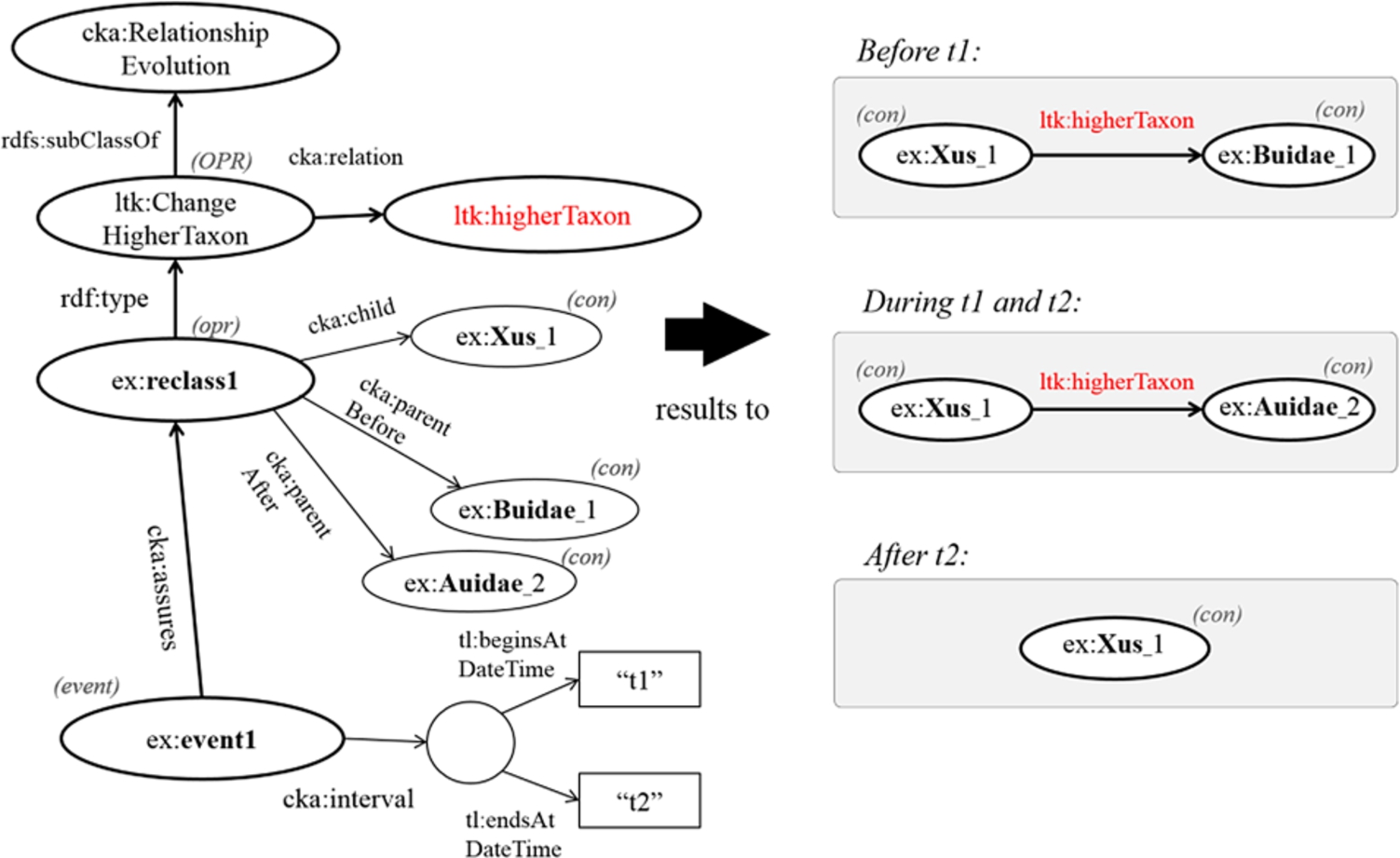
In addition, if the given time point is earlier than the begin time of a change, an object before the change (?obefore) becomes an object of a triple formed by the following rule. However, any changes ending before the given time point are not considered in this process.
subClassOf(?OPR, RelationshipEvolution) ∧ relation(?OPR,?p) ∧ type(?opr,?OPR) ∧ subject(?opr,?s) ∧ objectBefore(?opr,?obefore) ⇒?p(?s,?obefore)
Consequently, Fig. 6 shows that the classification of the genus ex:Xus_1 is interpreted variously according to different time points. The result after performing the rules is that the concept ex:Xus_1 is under the family ex:Buidae_1 before time t1, while ex:Xus_1 becomes under the family ex:Auidae_2 during the time between t1 and t2.
3.6.Representing LTK approach in RDF
Having proposed the formal description and rules, we now demonstrate how to utilize the RDF model to present and execute the change in taxonomy described in the previous sections. According to the change in the genus Columba, the following statements give the data of Columba in the RDF format. Initially, our work presents the relationship between a species and a genus by using the property ltk:higherTaxon and uses the notation species: and genus: as namespaces of species and genera, respectively.
species:Columba_speciosa_1789 ltk:higherTaxon genus:Columba_1758 .
Then, the following RDF statements express the event entity and operation for splitting the genus Columba together with a reference time point.
ex:event2003 cka:interval [tl:beginsAtDateTime"2003"] ; cka:assures ex:split1 . ex:split1 rdf:type ltk:TaxonSplitter ; cka:conceptBefore genus:Columba_1758 ; cka:conceptAfter genus:Patagioenas_2003 , genus:Chloroenas_2003 , genus:Lepidoenas_2003 , genus:Oenoenas_2003 , genus:Columba_2003 .
Furthermore, the framework provides a technique for transforming the event-centric model into the transition model along with a given concept. For example, links between the genus Columba and the new concepts after splitting can be shown as:
genus:Columba_1758 ltk:splitInto genus:Patagioenas_2003 , genus:Chloroenas_2003 , genus:Lepidoenas_2003 , genus:Oenoenas_2003 , genus:Columba_2003 .
3.6.1.Linking background knowledge
Fig. 7.
Cause and effect between two changes.
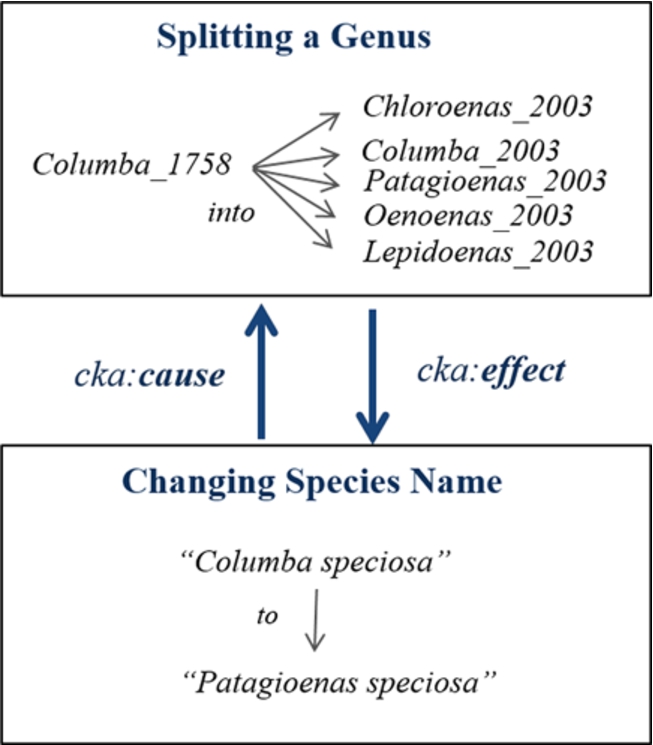
In addition, this model offers an association between related operations of changes by having two properties, cka:cause and cka:effect, to express the reason and the outcome of a change, respectively. For example, Fig. 7 shows the previous information of the newly registered name Patagioenas speciosa. Moreover, the property cka:detail is sometimes used for linking details of a concept after a change such as adding metadata. Consequently, we can find the history of the name Patagioenas speciosa and then use its background concepts, such as the old name Columba speciosa, to explore more information published in LOD.
3.6.2.LTK model in practice
To link data with the LOD Cloud, we proposed useful operations that specify the change in concepts, the changes in the details of a concept, the changes in relationships between concepts, and the background information of the change. All operations are defined by extending vocabularies from the well-known ontology named “SKOS” and properties from LODAC and CKA. The namespaces and example properties used by our model are described in Appendixes A and B. As a result, the data from our approach can be linked to data from other repositories.
For instance, the old concepts genus:Nyctea_1826 and genus:Bubo_1805 have been merged into a new concept named Bubo. As stated previously, the new identifier of the genus Bubo has to be initiated because its new scope is larger than the former one. According to the recommendation, the identifier should be ended with a string representing the year in which the new URI was created, so the new identifier of genus:Bubo_1805 becomes genus:Bubo_1999. To link between concepts before and after the change, LTK provides the property named ltk:mergedInto to represent the relationship between a concept before and a concept after merging. As a result, the relationship between genus:Nyctea_1826 and genus:Bubo_1999 remains to be represented by the property ltk:mergedInto. Moreover, in the case where a former concept and latter concept have the same name or their circumscriptions are very close, the property ltk:majorMergedInto is recommended for demonstrating the very close relationship between them, such as genus:Bubo_1805 and genus:Bubo_1999. To handle this situation, the model allows the use of the property cka:majorConceptBefore for the operation of merging and the property cka:majorConceptAfter for the operation of splitting. As the genus Nyctea was merged into the genus Bubo, all species under the genus Nyctea, such as N. scandiaca, have to be transferred to the genus Bubo; in this case, the name of this species has to be changed to B. scandiacus according to the nomenclature [20,28,47,48]. The following RDF statements describe the merging of two genera, the renaming of a species under the genus Nyctea, and the change in a species under the genus Bubo. In this case, the species:Bubo_scandiacus_1999 is newly generated without any higher taxa, so this event has to give it a higher taxon by using the operation ltk:HigherTaxonAddition to originate a higher taxon of a newly generated URI. In addition, references can be assigned to the event entity. They are researchers who discovered the changes (bibo:performer), researchers who published the changes (bibo:issuer), and publications (dct:source).
ex:event1999 bibo:performer pp:Wing, pp:Heidrich ; bibo:issuer pp:Richard ; dct:source pub:5224773 ; cka:interval [tl:beginsAtDateTime "1999"] ; cka:assures ex:mg1, ex:rp1, ex:ac1 . ex:mg1 rdf:type ltk:TaxonMerger ; cka:majorConceptBefore genus:Bubo_1805 ; cka:conceptBefore genus:Nyctea_1826 ; cka:conceptAfter genus:Bubo_1999 . ex:rp1 rdf:type ltk:TaxonReplacement ; cka:conceptBefore species:Nyctea_scandiaca_1826 ; cka:conceptAfter species:Bubo_scandiacus_1999 . ex:ac1 rdf:type ltk:HigherTaxonAddition ; cka:child species:Bubo_scandiacus_1999 ; cka:parent genus:Bubo_1999 . ex:mg1 cka:effect ex:rp1 . ex:rp1 cka:detail ex:ac1 .
3.6.3.Semantic Web rules
After that, Semantic Web rules are implemented in order to transform the event-centric model into the transition model in RDF. For example, a Jena rule [1] that infers the merging operation that uses the cka:conceptBefore of taxon concepts is
[rule_merge: (?opr rdf:type ltk:TaxonMerger), (?opr cka:conceptBefore ?before), (?opr cka:conceptAfter ?after) ->(?before ltk:mergedInto ?after) ]
In addition, the rule for cka:majorConceptBefore is the modification of the rule rule_merge made by changing cka:conceptBefore to cka:majorConceptBefore and changing ltk:mergedInto to ltk:majorMergedInto.
Moreover, the entered (cka:entered) and expired (cka:expired) time points of a concept are also generated by using the following example rule. However, in practice, this rule should be split into several ones in order to handle all possible cases that contain only some of the properties such as tl:beginsAtDateTime, tl:endsAtDateTime, cka:conceptBefore, and cka:conceptAfter.
[rule_time_span: (?event cka:interval ?inv), (?inv tl:beginsAtDateTime ?begin), (?inv tl:endsAtDateTime ?end), (?event cka:assures ?opr), (?opr rdf:type cka:ConceptEvolution), (?opr cka:conceptBefore ?before), (?opr cka:conceptAfter ?after) ->(?before cka:expired ?begin), (?after cka:entered ?begin), (?after cka:expired ?end) ]
In practice, the rules for the transition model are also performed, so the change in a given concept itself at a given time point is also presented. When all rules are executed, the following inferred RDF statements are produced to present the associations between changed taxa.
genus:Nyctea_1826 ltk:mergedInto genus:Bubo_1999 . genus:Bubo_1805 ltk:majorMergedInto genus:Bubo_1999 . species:Bubo_scandiacus_1999 ltk:higherTaxon genus:Bubo_1999 . species:Bubo_scandiacus_1999 ltk:synonym species:Nyctea_scandiaca_1826 . genus:Nyctea_1826 cka:expired "1999" . genus:Bubo_1805 cka:expired "1999" . genus:Bubo_1999 cka:entered "1999". species:Nyctea_scandiaca_1805 cka:expired "1999" . species:Bubo_scandiacus_1999 cka:entered "1999" .
A transfer into a simple RDF statement containing a subject, a predicate, and an object is useful for a client. This simple format is easier for working with well-known ontologies in order to query by well-known properties as defined in Appendix B. For example, the properties skos:exactMatch and lodac:hasSuperTaxon in query statements can produce the same results as the ones from ltk:synonym and ltk:higherTaxon, respectively. This approach also allows users to check the existence of a concept by inquiring about either the property cka:entered or the property cka:expired.
3.6.4.Working with other operations
Technically, the CKA framework allows other ontologies to customize their own operations of changes for particular purposes. This is done by extending either the class cka:ConceptEvolution for the change in a concept’s scope or the class cka:RelationEvolution for the change in the binary relationship between two concepts. For example, the operations of the change in taxon concepts, such as ltk:TaxonMerger and ltk:TaxonSplitter, are descended from cka:ConceptEvolution. Thus, when there are new properties that are not a part of either CKA or LTK, such as morphological, molecular, or ecological traits, new operations need to be initiated by extending one of the mentioned classes from CKA and then binding the new operations with related properties.
Fig. 8.
Prototype: System architecture.
In addition, although this research focuses on the change in taxonomic data, some triples that are not changed over time are recommended to be preserved by the even-centric model because it can present essential metadata such as a date added and references. Moreover, if some domains require more operations of changes, the operations can be created by extending cka:RelationshipEvolution. This method is also compatible with systems that separate a taxon concept and a name. Our model also allows having operations for either the object property or datatype property. Example properties or attributes are those such as skos:prefLabel [41], foaf:depiction [16], dwc:identificationID [9], dwc:taxonID [9], dwc:scientificNameID [9], dwc:scientificName [9], and lodac:hasCommonName [30]. Some details of them are described in Appendix C.
In conclusion, the introduced logical model includes the data model for the change in taxonomic knowledge and Semantic Web rules for transforming an event-centric model into a simple linked data model. It also presents how to use the model for real-world cases of the change in taxonomic knowledge in RDF. However, if more properties are needed for a specific purpose, developers can customize their operations by extending this framework.
4.Prototype
Our proposed logical model is intended for managing the change in taxonomic knowledge represented in RDF. To verify the possibility and feasibility of our work, a web application was developed. The main purpose of its implementation is to execute and present changes in taxonomic knowledge. The system architecture and a demonstration of this web application are also presented. Information on our prototype is available at the website “http://rc.lodac.nii.ac.jp/ltk/.”
4.1.Functionalities
The prototype is implemented on the basis of two key functions: defining and executing the change in taxonomic knowledge and presenting the temporal information of an Internet resource used in taxonomic knowledge.
The first function allows users to input changes in taxonomic knowledge by recording a list of operations, their parameters, and metadata. It also offers a bulk load feature for importing the event-centric model in RDF into the system directly. When the input data is submitted, rule-based reasoning produces the relationships between concepts that are the result of a change in taxonomic knowledge, and then, the system collects the RDF data in an RDF data store.
In addition to the execution of the event-centric model, the other function offers an interface for presenting temporal information and linked data of a given concept. The prototype lets users browse the URI of a given concept with a given time point in xsd:dateTime format, and it then displays the temporal information of the concept together with its related concepts that is a result of the change and any background information regarding changes.
4.2.Implementation
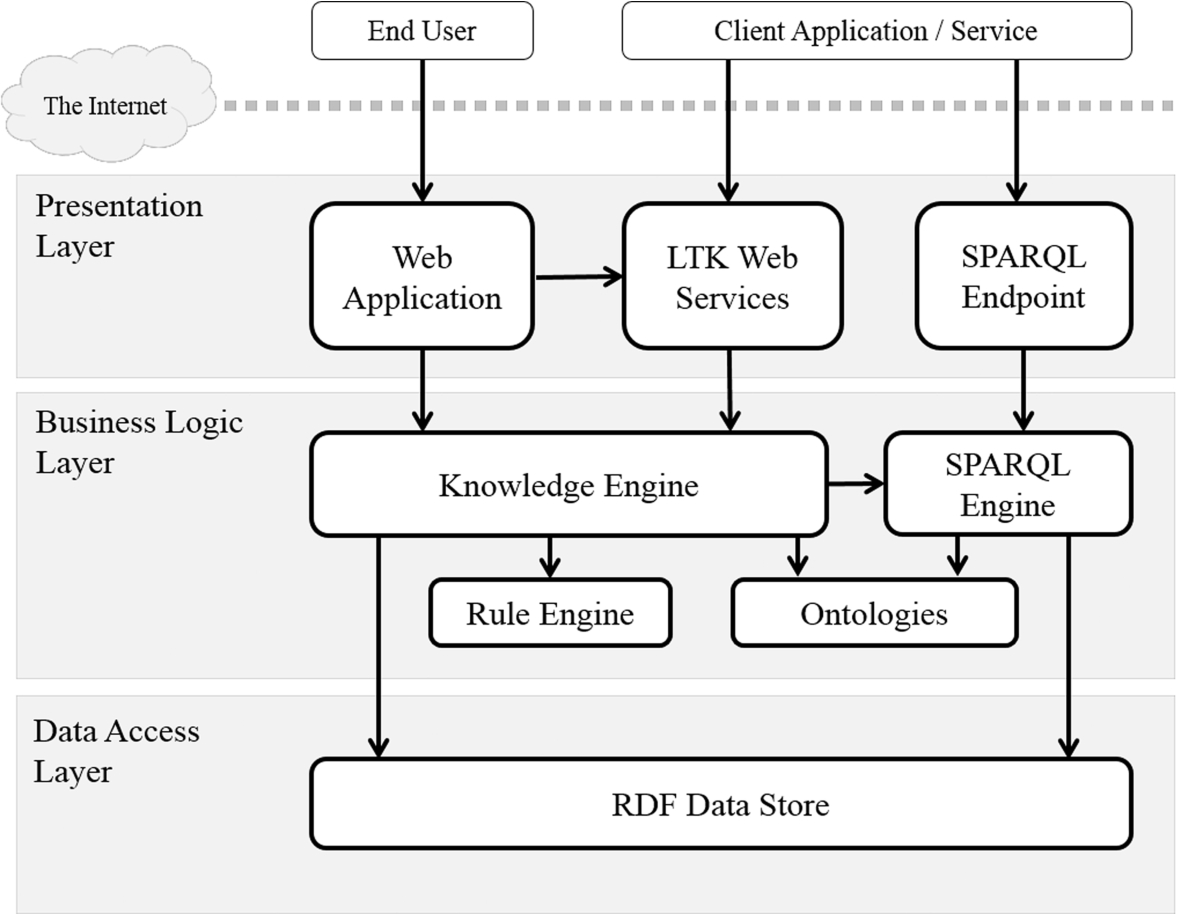
To accomplish these key activities, we analyzed the functions, designed the system architecture, employed well-known open source tools, and did the programming to implement the web application for end users and service interfaces for client applications. The architecture of the prototype is a web-based system, as shown in Fig. 8, comprising three layers: a presentation layer, business logic layer, and data access layer.
The presentation layer displays information related to such services as creating and executing the change in a given concept and presenting the taxonomic knowledge. It communicates with other service endpoints by outputting results to users or client applications. The user can browse the information by using a web application created by PHP, whereas the client applications can access the data by using LTK web services written in Java and SPARQL endpoint, which is provided by OpenRDF [31].
In addition to the presentation layer, the business logic layer controls an application’s functionality by performing data processing. Knowledge Engine, a Java-based component, is the main module that manages the RDF-based event-centric model together with Semantic Web rules and related ontologies in order to construct taxonomic knowledge and linked data of Internet resources for taxonomic data. Technically, this component normalizes and forwards RDF data to the data store directly. It also queries RDF data via the SPARQL engine with an API from OpenRDF. Moreover, a Semantic Web rule engine developed by using Apache Jena [1] transforms the event-centric model into the transition model and the snapshot model.
Last, the data access layer built for the storage and retrieval of triples collects subject-predicate-objects from components in the upper layers. Our experiment uses OpenRDF, which offers high capacity with great performance. It additionally offers an API that performs well with Jena.
All of these layers run on a server that is connected to the Internet, so the system is ready to provide LTK services to end users or client applications. Moreover, the system architecture is flexible to enable application to other domains. Developers can customize Semantic Web rules and ontologies to their own requirements and publish their data for open access.
4.3.LTK services
As a result of the services provided in the presentation layer, all interfaces are conveniently accessible over the Internet. In this section, we illustrate how to use services from this prototype by describing web application and web services.
4.3.1.Web application
Beginning with the web application, it contains two main parts, an administration interface and a user interface.
The administration interface provides a tool for importing a list of changes in concepts. Every change can be done by choosing an operation such as merging, replacing, and splitting, and then assigning a concept or a value to the required properties. After that, users can state the relationship between changes in the case where one change relates to another change by linking them with properties named “cause,” “effect,” or “detail.” Finally, the prototype allows users to prepare metadata of these changes, such as a begin time point, an end time point, performers, e.g., researchers, who discovered the change, reporters who announced the change, and references such as publications.
Fig. 9.
Prototype: View showing taxonomic knowledge of taxon.
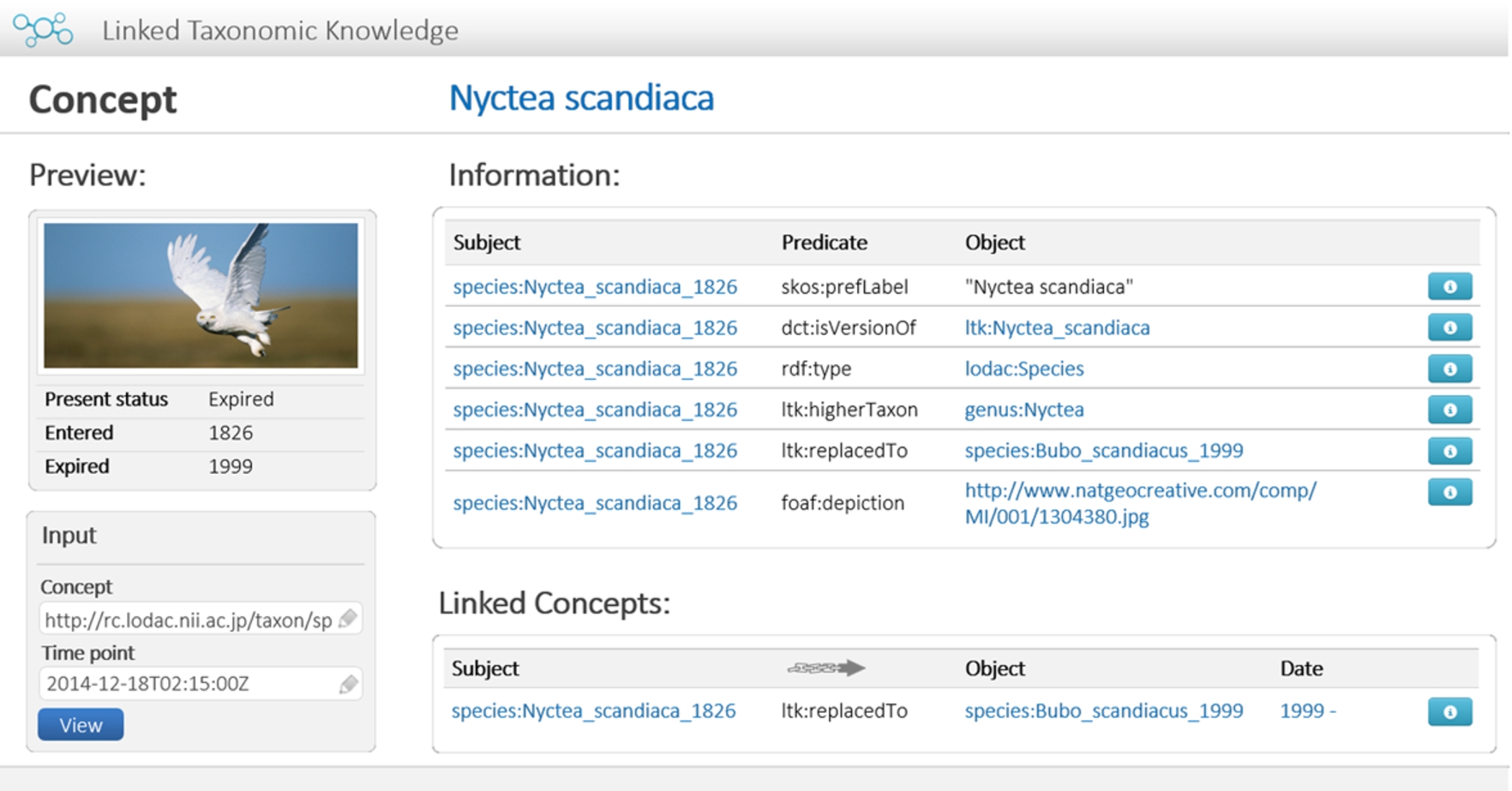
Apart from the administration interface, the user interface is implemented as a browser for presenting the information of a given concept. The web page shows historical information of a taxon concept including point temporal data, its related concepts that result from the change, and links of its related concepts. The user has to specify a URI of a concept together with a particular time. For this prototype, the URL pattern “http://[ltk_domain]/” denotes the domain name of our prototype, where the term “[ltk_domain]” is “rc.lodac.nii.ac.jp” in our experiment. The pattern of a request for displaying information of a given concept in a given time point is
http://[ltk_domain]/ltk/concept.php?concept=[concept]&date=[time_point],
http://[ltk-domain]/taxon/[rank]/[name]
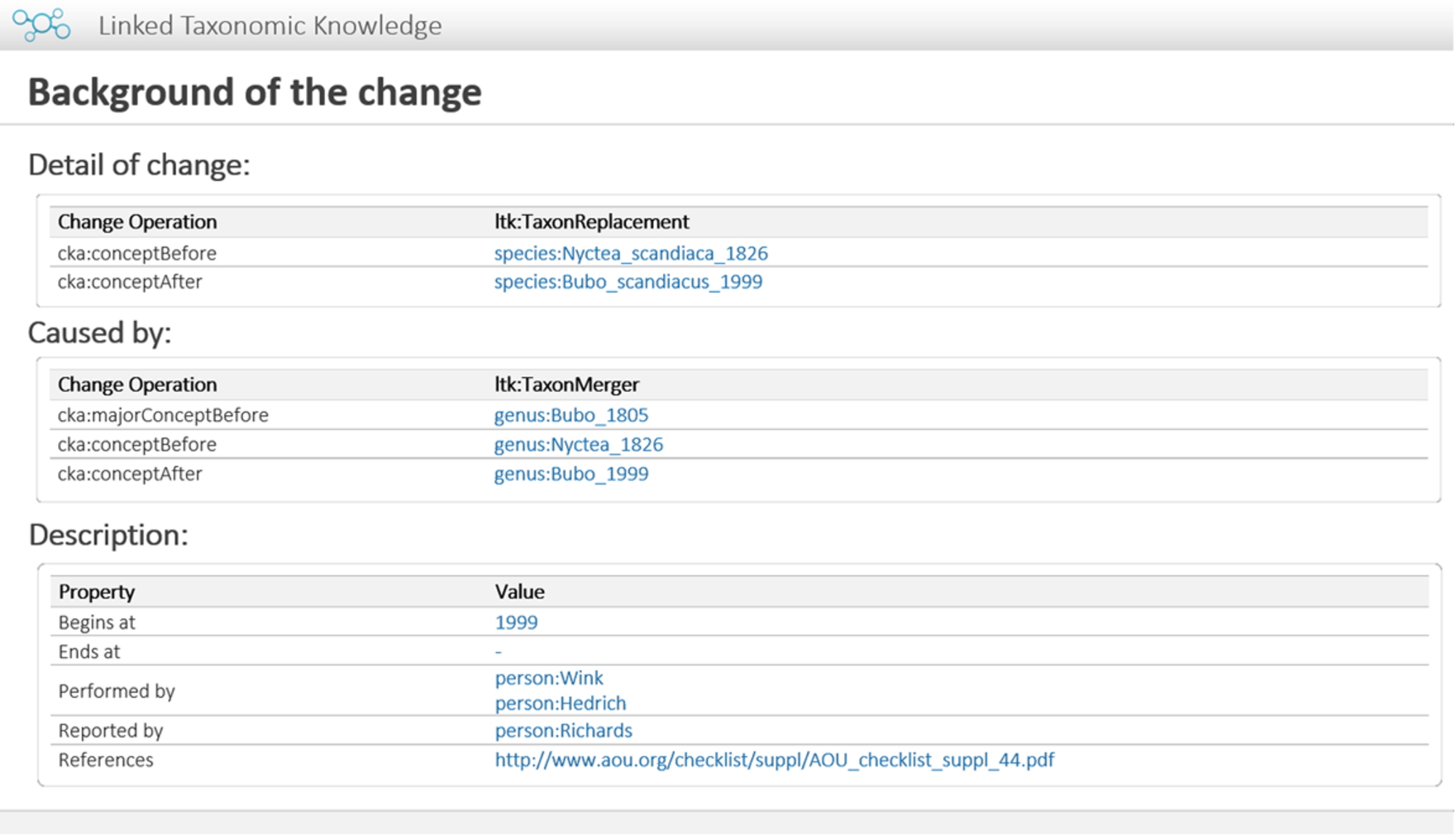
Fig. 10.
Prototype: Background information about change.
4.3.2.Web services
In addition to the web application, there are LTK web services and a SPARQL endpoint that provide data to client applications. Example datasets were loaded into OpenRDF [31] storage via LTK web service. The SPARQL endpoint for querying the links between concepts resulting from the changes can be accessed at the following URL.
http://[ltk_domain]/ltk-service/sparql/ltk
http://[ltk_domain]/ltk-service/context?concept=[concept]&date=[time_point]
http://[ltk_domain]/ltk-service/reason?subj=[subject_concept]&obj=[object_concept],
5.Evaluation
We proved the feasibility of our approach by experimenting on the prototype. We first evaluated our approach against use cases from domain experts and found that our research covers practical use cases. Second, we tested that the complexity of the event-centric model, which consumes many system resources, did not affect the overall performance of the prototype system.
5.1.Evaluation against use cases
We imported the example cases from Section 2 and some data on Japanese moths of the family Saturniidae published as three checklists (list of names): Inoue in 1982 [19], Jinbo in 2008 [21], and Kishida in 2011 [24]. One of the authors, Jinbo, analyzed the difference among these three checklists and finalized them into the changes in taxa among these checklists. The data cover operations of changes, which are creating a concept, making a concept obsolete, replacing a taxon, merging taxa, splitting a taxon, linking synonym, changing a higher taxon, subdividing a taxon, and combining taxa. This experiment contains 40 instances of operations together with 60 taxa from several taxonomic ranks: family, subfamily, genus, species, and subspecies. Here, we choose one example. In [19], the species Caligula boisduvalii has two subspecies, Caligula boisduvalii fallax and Caligula boisduvalii jonasii. In the subsequent study, this species was transferred from the genus Caligula to Saturnia, one of its subspecies jonasii was raised into a distinct species, and another subspecies, fallax, was regarded as a subspecies of jonasii. Hence, in that study, Caligula boisduvalii in [19] was redefined as two species, Saturnia boisduvalii and Saturnia jonasii. At the same time, the latter species was split into two subspecies, Saturnia jonasii jonasii and Saturnia jonasii fallax. These changes were adopted in the second checklist [21]. After a few years, both subspecies were combined into the species S. jonasii in [24]. These changes resulted in many links of synonyms. Even though these events are described in taxonomic papers, information on events is not included in each name and thus cannot be captured by the databases of scientific names. Some entities of background knowledge of the change in S. jonasii were linked so users could browse the accurate history of taxa, which is difficult to access for non-taxonomic experts. Therefore, the benefit of managing the change in concepts, such as presenting the links between concepts in the chain of the changes in taxonomic knowledge, temporal information about them, and the underlying knowledge of that change, made gathering correct data along with the precise context convenient. Therefore, it reduced confusion and helped avoid misunderstanding arising with respect to taxonomic data. This experiment proved that the LTK approach could deal with a real-world situation of changes in taxonomy.
5.2.Performance analysis
In addition to the usability evaluation, the performance of the prototype was tested. Our model essentially transforms a basic triple containing a subject, a predicate, and an object into a complex structure to express an event of a change in either a concept or a triple along with the reference time. As it consumes many more triples than the traditional form to present the same fact, the issue of performance becomes a key point in this research. We therefore verified the model with a great number of data and evaluated the query execution time by comparing our approach and a simple query as a baseline.
According to the data model, one event-centric model including 10 operations required about 100 triples. In this experiment, the number of test data in the repository was increased up to 1,000,000 triples. For every increase of 100,000 triples, we measured the performance and recorded all the results in a chart. All steps in this experiment were performed on Linux 3.11.0-12 (64 bit) installed on an Intel quad-core i5 3.40-GHz PC with 32 GB of memory. The changes in data were stored in OpenRDF SESAME Ver. 2.7.7. To optimize query performance, RDF schema and direct type hierarchy inferencing were enabled, so sequence triples were automatically generated from ones containing the properties rdf:type, rdfs:subClassOf, and rdfs:subPropertyOf. As a result, the dataset contains more than 5 million triples including inferred statements. The RDF repository additionally built two indexes: a subject-predicate-object-context (spoc) key pattern and a predicate-object-subject-context (posc) key pattern, where a context is generally viewed as a graph name [31].
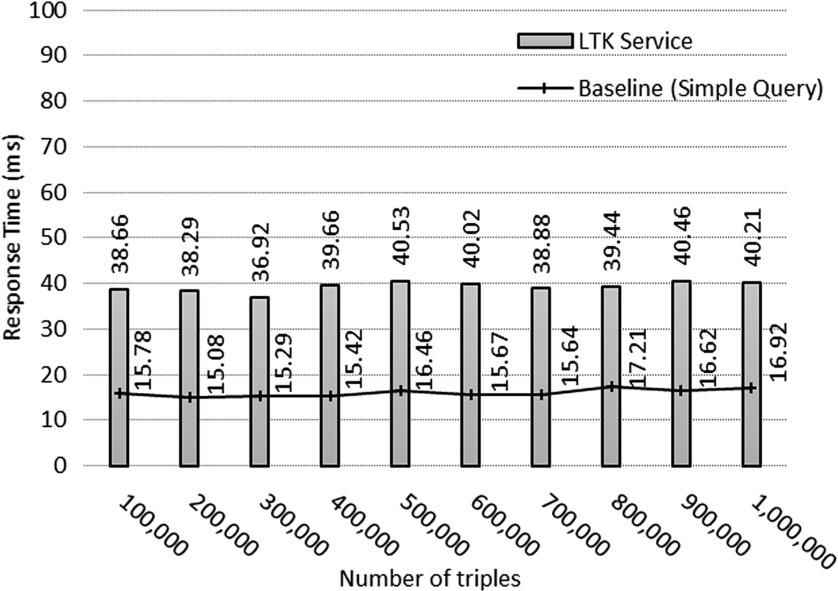
Fig. 11.
Query execution time in dataset.
Our verification step was performed by comparing the result from our approach with the baseline speed. To determine the basic speed of the SPARQL engine in our test, a baseline experiment was conducted by using the following simple SPARQL statement for searching information on a species.
SELECT ?p ?o WHERE
{ species:Nyctea_scandiaca ?p ?o .}Afterward, on the basis of our approach, we made an inquiry for the same information on the same species that is valid at a given time by using LTK web services. As the result is returned in accordance with a time input, the system has to produce the result on the fly depending on the defined time point. The service transforms data from the event-centric model into the snapshot model by using SPARQL statements together with Semantic Web rules, as mentioned in the previous sections. The performance was measured by recording the response time of the web method. For having more accuracy, data caching was disabled, and a given concept and a given time point were changed for every service request.
The results of the experiment are shown in Fig. 11, which shows that the execution time from our approach was almost constant at about 0.039 seconds for every 100,000 input triples added into the repository, while the value from the baseline was approximately 0.016 seconds. A closer look at the result indicates that our approach consumed slightly more execution time than did a simple query by a millisecond unit. The results of our experiment provide confirmatory evidence that our framework does not cause application performance problems in the current software development even if dealing with millions of pieces of data.
6.Discussion
Many approaches [3,22,32,37,38] usually focus on keeping up-to-date taxonomic data. In practice, the change in knowledge is necessary for comprehensively studying biodiversity; however, several previous pieces of work on taxonomic databases focused on the collection of name strings with proper identifiers at the first step of the integration of taxonomic information. Thus, the change in taxonomic knowledge is less discussed. Our work, LTK, provides a framework for preserving and presenting the change in taxonomic knowledge for linked data. We introduce operations for capturing the changes, such as merging, splitting, replacing, changing a higher taxon, etc., as shown in Appendix C. We discuss the values of our approach from four perspectives: knowledge representation, user engagement, system integration, and challenge.
6.1.Knowledge representation
In term of knowledge representation, our research responds to several requirements in order to have better understanding of biodiversity by giving different viewpoints of the change in taxonomic knowledge.
6.1.1.Historical change in taxa
Browsing chains of changes in concepts is a feature with which users can learn the historical changes in a given taxon. LTK provides properties indicating dynamic changes in taxa for this feature. Discussed in other pieces of work, the Taxonomic Concept Schema (TCS) [42] is one of the well-known approaches to describing a taxon concept in an informatics way. This approach was used to attempt to describe a concept expressed as RDF in a piece of work titled “Describing Taxon Concept as RDF” [10]. The TCS regarded each concept as more static and classified operations of change into proper categories, so most operations seem to be more static than LTK. In terms of using properties to represent dynamic changes in the conception of taxa, our work introduced the hierarchy and configuration of the properties in Appendix D. Properties such as ltk:mergedInto, ltk:splitInto, and ltk:replacedTo can be simply used in the query statement. These properties are asymmetric and non-transitive object properties, so the query result returns only directed-adjacent nodes of a given concept. LTK can also present the main concepts in the timeline by using the properties ltk:majorMergedInto and ltk:majorSplitInto, which are sub properties of ltk:mergedInto and ltk:splitInto, respectively. ltk:majorMergedInto and ltk:majorSplitInto show that their subject and object are dominant in the change, so the concepts connected by these properties have a stronger relationship than those linked by ltk:mergedInto and ltk:splitInto. In addition to getting the adjacent concepts, finding all concepts having the same history can be queried by using the properties cka:serialLinkTo and cka:semanticLink. The former, cka:serialLinkTo, is a transitive and asymmetric object property, so all concepts in only one direction in a timeline occurring before or after the change in the given concepts can be queried. In addition, if it needs to find out all concepts in the same history, the query expression should be mention the property cka:semanticLink, which is a transitive and symmetric property and also a super property of cka:serialLinkTo.
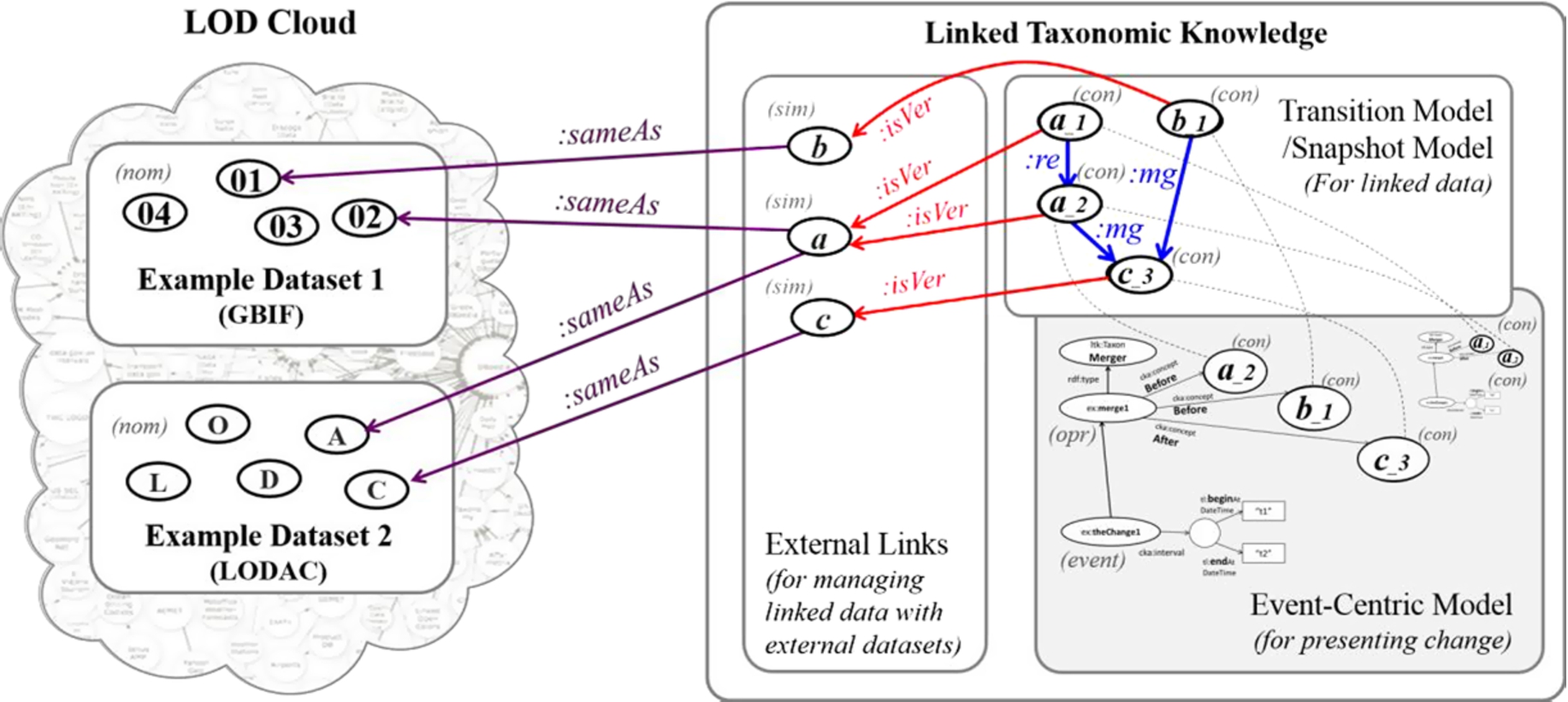
Fig. 12.
Role of LTK (right) in LOD Cloud (left) containing example datasets. Ovals with single alphabet or ID number are general concepts, ovals with version are versions of general concepts, dashed lines show same URIs, :same is owl:sameAs, :isVer is dct:isVersionOf, :re is ltk:replacedInto, and :mg is ltk:mergedInto.
6.1.2.Temporal information of taxa
The use of temporal data allows users to learn of the change in taxonomic knowledge in terms of the change in triples, for example the changes in classification, membership, metadata, etc. Operations of changes that are found in the same publication or event are grouped into one event-centric model, and aspects about time and provenance are assigned. Each operation assured by the event entity can be transformed into two triples, one happening before the begin time point and the other one happening during the begin and end time points, but no triple generated after the end time point. However, these two triples are not directly stored in the database, so a client needs to use query expression with Semantic Web rules to produce a snapshot model of a given concept at a given time point. In the case a concept is given without a time point, the system assigns a current time by default. Although the event-centric model consumes many triples, the performance analysis from the previous section confirms that this is not an issue for current SPARQL engines. Thus, users do not only learn the association between data but also understand the precise context of the linked data by temporal information and references. They also recognize triples added or removed at different times, so they can learn the progress of taxonomic knowledge along with time.
6.1.3.Background knowledge of change
Our approach has similar objectives as TaxMeOn [46] in terms of managing the changes and linked data, but both pieces of work are technically different due to specific purposes. TaxMeOn regularly presents a change by using one triple containing an old taxon concept, a property indicating taxonomic change, and a new taxon concept, and it sometimes uses instances of operations of changes such as lumping and splitting. Thus, data model gives a simple and easily understandable timeline of the changes in taxon concepts. However, in the case of using only one triple for representing a change, it is limited to giving a link between changes, so associations between background knowledge cannot be implemented directly. In this case, the event-centric model becomes more advantageous for meeting this requirement because an instance of an operation can also be regarded as background knowledge, so the link between operations allows users to trace back to the information behind the change. The properties cka:cause and cka:effect are used in a query string to find the reason and the result of a particular change, respectively. Our prototype demonstrates how two concepts are related by finding operations that are the background knowledge of a link between the given subject and object.
6.1.4.Ability to publish linked data
The LTK approach was developed on the basis of the Semantic Web and the underlying community knowledge [6,13], so it can act as a medium that collects links among taxonomic data from different datasets and provides background knowledge about how concepts are changed or linked. We encourage linking contextual nominal entities with external nominal entities from known datasets that are commonly referred to by many applications and publications such as GBIF [44], CoL [22], uBio [37], and LODAC [30] by using the property dct:isVersionOf in order to enable global access on data. The role of LTK in terms of linked data is demonstrated in Fig. 12. In the figure, LTK becomes the medium of linked data having three parts. The first part consists of external links for representative concepts and links to external datasets. The second part includes the transition model and snapshot model. The third part contains the event-centric model that acts as the background knowledge of change. Our approach can publish data to the LOD Cloud by using open access data via SPARQL, making URIs be dereferenceable, and linking data to known datasets [18].
6.2.User engagement
Another important task of building a taxonomic information system is to encourage users such as taxonomists, ecologists, and molecular biologists to participate in providing and utilizing data. However, many of them are non-computer-expert users. Since linked data, the Semantic Web, and RDF syntax are relevant to each other, which is, as far as we know, the current situation of the Semantic Web, we recommend users understand basic RDF syntax in order to benefit from linked data. In this research, we intend to keep taxonomic knowledge representation as simple as possible under the boundary of the RDF framework.
6.2.1.Human readability
Since the event-centric model is considered to represent data in various dimensions, RDF representation is complicated by designed. However, the simplicity of the model can be improved by the simple use of identifiers, making the transition model and snapshot model become consequently simpler. In terms of human readability, the uses of the contextual nominal entity and simple nominal entity are consistent with the idea of GNUB, which describes the usage of a name, and GNI, which collects name strings, respectively [33]. Thus, normalized and valid readable names are tied to a checklist such as CoL [22]. In another viewpoint, GBIF [8,36] suggested that the persistent identifiers of taxa should be unfriendly to read, and a taxon concept and name should be presented separately so that the identifiers still endure, while the names change. This idea is basically consistent with the normalized database design that eliminates the difficulty of updating data, but the data model is much more complex for accessing. In this research, we more focus on accessing linked data, but updating is less emphasized because the change in knowledge is recorded by appending a new revision. Working with a revision of knowledge, an identifier is does not necessarily have to be viewed as a persistent thing. This viewpoint leads to the idea that designing a data model is more relaxed than the use of persistent identifiers. Thus, it is possible to encapsulate a taxon concept and a taxonomic name within a single identifier, and using a human-readable string in a URI is also possible. This simple representation comes with several advantages: lightweight data, recognizable URIs, and understandable linked data. Although it results in a slight decrease of information granularity, it improves user satisfaction in contributing and consuming data. However, this model does not restrict the use of URIs; either separating a taxon concept and name or using unreadable URIs is possible to implement.
6.2.2.Data preparation
In this field, data are usually provided by domain experts, especially taxonomists. The prototype provides a form-based web application with text fields for user input. It is good for a small number of data in practice. However, when dealing with a large number of data, we recommend users upload a text file containing the event-centric model. Since this research is not aimed at user experience design, in this phase, we encourage users to understand the basic syntax of RDF N-Triples. The data preparation steps are simply demonstrated as the following steps.
1) Giving contextual nominal entities for every taxon with every change.
2) Creating an event entity with a time interval and references.
3) Creating instances of proper operations for every change.
4) Assigning contextual nominal entities before and after a change.
5) Giving links for causes and effects between operations.
6) Creating representatives of external nominal entities for all taxa.
7) Giving links between contextual nominal entities and representatives of external nominal entities.
8) Searching taxa from the Internet.
9) Giving links between representatives and external URIs.
Since all operations are employed in similar ways and URIs are human-readable, non-computer-expert users can create data and import them into the system. However, we learned that finding available URIs from known online datasets requires a lot of effort. In the future, we will find proper solutions to support this task and create a spreadsheet template for bulk upload.
6.3.System integration
For the design of the data model, apart from satisfying the present requirements, the viewpoints of framework enhancement and data exchange are discussed.
6.3.1.Extensibility
There are a lot of kinds of relationships available in taxonomic documents such as comprehensive relationships documented by TCS [42]. There are many minor relationships between names and concepts, but usually, these relationships are summarized as valid (accepted), invalid (not valid but correctly proposed), and unavailable (neither valid nor correctly proposed). Some of the properties collected by TCS are is-homotypic-synonym-of, is-later-homonym-of, is-validation-of, is-vernacular-for, has-conserved-name, is-second-parent-for, and is-hybrid-parent-of. However, our present work is focused mainly on the changes in taxonomic knowledge with simple situations, and the introduction of more terms is a future challenge. In this case, our framework allows increasing the capability of a system with other vocabularies by creating operations under either the classes of the change in conception (cka:ConceptEvolution) or the change in triple (cka:RelationshipEvolution) and reusing or adapting the Semantic Web rules.
6.3.2.Interoperability
Thanks to the progress of Semantic Web technology, current RDF repositories can maintain billions of pieces of data. However, in reality, the technology does not rely on a single data source. The integration among taxonomic information systems is able to be done via the Internet by using either web services or SPARQL endpoints together with commonly accepted data models.
6.4.Challenge
In this research, we assume that every change in taxonomy is clearly described. The representations of any changes are based on explicit evidence such as publication. In our experiment, before creating RDF data presenting the changes, a domain expert has to analyze the difference between several checklists, finding how names are different, and summarize them into operations of changes. For this reason, the precision of the model relies on the completeness and the correctness of collected data. However, even existing references such as books and publications contain only insufficient information. For example, a synonymic catalogue, also called a “synonym list,” is a standard way in taxonomy to present a historical summary of taxonomic studies on each species, including unaccepted names, misidentifications, references, etc. Here, a statement from the synonymic catalogue [19],
Adela Latreille, 1796
35. reaumurella (Linnaeus, 1958),
Syst. Nat. (Edn 10) 1:540 (Phalaena).
viridella (Scopoli, 1763), Ent. Carniolica: 250 (Phalaena).
In practice, a publication sometimes does not describe an exact date of a particular change clearly, so a published date of the earliest publication that announced the change can be used to assign in the knowledge base as a workaround. A published date is generally written only with a year, but due to the constraint of the datatype xsd:dateTime, which is the range of the property tl:interval of the Timeline ontology [45], other components such as a day and a month are also required. In this manner, regarding the determination of date recommended by International Code of Zoological Nomenclature (ICZN) [11], if a date is not completely specified but either a month-year or a year is known, the last day of the known period should be entered in a knowledge base. In case a developer considers that this format shows too much detail to users, an application can select a suitable part of the date and time string such as a month-year or a year number for interacting with users.
For the other important issue, having no single globally-accepted taxonomy is also a great challenge at the moment. There are multiple branches of taxonomies and each of them is agreed by different communities of taxonomists. Since the change in taxonomic knowledge across multiple accepted taxonomies is not generally found, this research focuses on a scope of the management on historical changes within a single accepted taxonomy. For this issue, it is recommended that the administration of multiple accepted taxonomies is possible to be performed by using some separated installations of taxonomic information systems and linking some Internet resources of the same taxa across all data repositories.
7.Conclusion
We presented a logical model and ontology for linking concepts that comprise a series of changes, a diversity of taxonomic classifications, and a variety of scientific names. For the purpose of linking data, we developed our model by employing an ontology of contextual knowledge evolution together with widely accepted ontologies such as SKOS. A single and readable Internet resource for representing a version of concepts used in taxonomic knowledge can be viewed as either a name or a taxon concept. The result is that triples become lightweight, simple, and easy to understand by both machines and non-computer-expert users. Our model can deal with both the complex format of the event-centric model and easily-linkable triples from the transition model and snapshot model in RDF, and hence, can trace the background knowledge of given associated concepts. In addition, we implemented a prototype that utilizes the proposed model for managing the change in taxonomic knowledge and offering open access in order to give an opportunity to link our data to the LOD Cloud. As a consequence, other applications that need linked concepts can readily connect to these data. By giving links to and reusing existing URIs from well-known taxonomic databases, it is possible to associate our dataset with the large amount of taxonomic data across repositories in order to discover a broader knowledge of biology.
Our approach was designed mainly on the basis of test cases in zoology. Some requirements from these domains, such as botany and mycology, sometimes differ from zoology. Thus, some operations of changes, some configurations of property, and some Semantic Web rules have to be improved in order to satisfy the needs of those domains. Moreover, this approach provides features for managing changes in taxonomic knowledge in RDF; however, building a practical taxonomic information system requires additional functionalities. To have a fully functioning system, developers have to consider further points. First, RDF data can be contributed by many providers. To encourage non-computer-expert users to get involved with the system, an application should have a good user experience design. Second, to have proper data management, the systems needs high quality functions for authentication, authorization, and administration that can manage user privileges and access controls at the data level. The licensing of data must also be properly declared. In addition, automated data matching would need to be provided; otherwise, data providers would have to collect external URIs and link their own contextual nominal entities with them in order to link to the LOD Cloud. Next, a data converter that can migrate other legacy datasets into the LTK model should be developed. Last, the management on multiple accepted taxonomies can be the next step towards enhancements.
Acknowledgements
We thank Dr. Isao Nishiumi of the National Museum of Nature and Science and Dr. Takeshi Yamasaki of the Yamashina Institute for Ornithology, who provided us with valuable comments on the taxonomy of birds. We also thank the researchers from the Asian Institute of Technology, Chulalongkorn University, Kasetsart University, Thammasat University, the Consortium of International Agricultural Research Centers, and the National Institute for Agro-Environmental Science for the many valuable ideas that helped us improve our research. We appreciate the financial support from the National Institute of Informatics that fully funded our research activities.
Appendices
Appendix A.
Appendix A.Example namespaces used by LTK
This section gives information about prefixes and namespaces used in this paper.
| Prefix | Namespace |
| bibo: | Bibliographic Ontology [4] http://purl.org/ontology/bibo/ |
| cka: | Contextual Knowledge for Archives [6] http://www.cka.org/2012/01/cka-onto# |
| dct: | Dublin Core Terms Namespace [12] http://purl.org/dc/terms/ |
| dbpedia: | DBpedia Namespace [26] http://live.dbpedia.org/resource/ |
| dwc: | Darwin Core [9] http://rs.tdwg.org/dwc/terms/ |
| foaf: | Friend of a Friend Ontology [16] http://xmlns.com/foaf/0.1/ |
| gbif: | Global Biodiversity Information Facility [44] http://www.gbif.org/species/ |
| genus: | Namespace for genera used in LTK http://rc.lodac.nii.ac.jp/taxon/genus/ |
| lodac: | LODAC Species [30] http://lod.ac/species/ |
| ltk: | Linked Taxonomic Knowledge Ontology http://rc.lodac.nii.ac.jp/ns/ltk# |
| skos: | Simple Knowledge Organization System Namespace [41] http://www.w3.org/2004/02/skos/core# |
| species: | Namespace for species used in LTK http://rc.lodac.nii.ac.jp/taxon/species/ |
| soic: | Semantically-Interlinked Online Communities Core Ontology [39] http://rdfs.org/sioc/ns# |
| tl: | Timeline Ontology [45] http://purl.org/NET/c4dm/timeline.owl# |
| tmo: | Meta-Ontology of Biological Name [46] http://www.yso.fi/onto/taxmeon/ |
Appendix B.
Appendix B.Example LTK properties
This section gathers some properties provided by LTK.
| Properties | rdfs:subPropertyOf |
| ltk:higherTaxon | – cka:higherClass – skos:broaderTransitive – tmo:isPartOfHigherTaxon – lodac:hasSuperTaxon |
| ltk:replacedTo | – cka:serialLinkTo – tmo:congruentWithTaxon – skos:exactMatch |
| ltk:mergedInto | – cka:serialLinkTo – skos:broadMatch |
| ltk:majorMergedInto | – cka:serialLinkTo – skos:closeMatch |
| ltk:splitInto | – cka:serialLinkTo – skos:narrowMatch |
| ltk:majorSplitInto | – cka:serialLinkTo – skos:closeMatch |
| ltk:dsynonym | – skos:exactMatch – lodac:hasSynonym |
| ltk:synonym | – skos:exactMatch – ltk:dsynonym – lodac:hasSynonym |
| ltk:cpxChangedTo | – skos:relatedMatch |
| ltk:circChangedTo | – skos:closeMatch |
| ltk:subdividedInto | – skos:narrowMatch |
| ltk:combinedInto | – skos:broadMatch |
Appendix C.
Appendix C.Example LTK operations
The following list declared operations and their parameters, which are provided by LTK ontology. An italic symbol in the parentheses of each parameter indicates its cardinality for every operation. The symbol “(1)” allows only one value, the symbol “(2..*)” expects at least two values required, and the symbol “(0..1)” presents one optional value.
ltk:TaxonMerger
Description For merging some concepts (before) into one concept (after). Parameters – cka:conceptBefore (2..*)
– cka:majorConceptBefore (0..1)
– cka:conceptAfter (1)Example input RDF ex:opr rdf:type ltk:TaxonMerger .
ex:opr cka:conceptBefore
ex:be1, ex:be2 ;
cka:majorConceptBefore
ex:mb0 ;
cka:conceptAfter ex:af1 .Example result ex:be1 ltk:mergedInto ex:af1 .
ex:be2 ltk:mergedInto ex:af1 .
ex:mb0 ltk:majorMergedInto ex:af1 .Example entailment ex:be1 skos:broadMatch ex:af1 .
ex:be2 skos:broadMatch ex:af1 .
ex:mb0 skos:closeMatch ex:af1 .ltk:TaxonSplitter
Description For splitting a concept (before) into new concepts (after). Parameters – cka:conceptBefore (1)
– cka:conceptAfter (2..*)
– cka:majorConceptAfter (0..1)Example input RDF ex:opr rdf:type ltk:TaxonSplitter .
ex:opr cka:conceptBefore ex:be1 ;
cka:conceptAfter
ex:af1, ex:af2 ;
cka:majorConceptAfter
ex:ma0 .Example result ex:be1 ltk:splitInto ex:af1 .
ex:be1 ltk:splitInto ex:af2 .
ex:be1 ltk:majorSplitInto ex:ma0 .Example entailment ex:be1 skos:narrowMatch ex:af1 .
ex:be1 skos:narrowMatch ex:af2 .
ex:be1 skos:closeMatch ex:ma0 .ltk:TaxonReplacement
Description For replacing one concept (before) to another one (after). Parameters – cka:conceptBefore (1)
– cka:conceptAfter (1)Example input RDF ex:opr rdf:type
ltk:TaxonReplacement .
ex:opr cka:conceptBefore ex:be1 ;
cka:conceptAfter ex:af1 .Example result ex:be1 ltk:replacedTo ex:af1 . Example entailment ex:be1 skos:exactMatch ex:af1 .
ex:be1 tmo:congruentWithTaxon ex:af1 .ltk:TaxonComplexChange
Description For a complex case that many concepts (before) are merged and split into many other concepts (after). Parameters – cka:conceptBefore (2..*)
– cka:conceptAfter (2..*)Example input RDF ex:opr rdf:type
ltk:TaxonComplexChange .
ex:opr cka:conceptBefore
ex:be1, ex:be2 ;
cka:conceptAfter
ex:af1, ex:af2 .Example result ex:be1 ltk:cpxChangedTo ex:af1 .
ex:be1 ltk:cpxChangedTo ex:af2 .
ex:be2 ltk:cpxChangedTo ex:af1 .
ex:be2 ltk:cpxChangedTo ex:af2 .Example entailment ex:be1 skos:relatedMatch ex:af1 .
ex:be1 skos:relatedMatch ex:af2 .
ex:be2 skos:relatedMatch ex:af1 .
ex:be2 skos:relatedMatch ex:af2 .ltk:CircumscriptionChange
Description For changing circumscription of one concept (before) to another one (after). Parameters – cka:conceptBefore (1)
– cka:conceptAfter (1)Example input RDF ex:opr rdf:type
ltk:CircumscriptionChange .
ex:opr cka:conceptBefore ex:be1 ;
cka:conceptAfter ex:af1 .Example result ex:be1 ltk:circChangedTo ex:af1 . Example entailment ex:be1 skos:closeMatch ex:af1 . ltk:ChangeHigherTaxon
Description For reclassifying a lower concept (child) by moving from a higher concept (before) to another one (after). Parameters – cka:child (1)
– cka:parentBefore (1)
– cka:parentAfter (1)Example input RDF ex:opr rdf:type
ltk:ChangeHigherTaxon .
ex:opr cka:child ex:c1;
cka:parentBefore ex:p1 ;
cka:parentAfter ex:p2 .Example result ex:c1 ltk:higherTaxon ex:p2 .
ex:p2 ltk:lowerTaxon ex:c1 .Example entailment ex:c1 skos:broaderTransitive ex:p2 .
ex:p2 skos:narrowerTransitive ex:c1 .
ex:c1 lodac:hasSuperTaxon ex:p2 .ltk:SubdivideTaxon
Description For subdividing a higher taxon (source) into some lower taxa (target). Parameters – cka:sourceConcept (1)
– cka:targetConcept (2..*)Example input RDF ex:opr rdf:type ltk:SubdivideTaxon .
ex:opr
cka:sourceConcept ex:h1 ;
cka:targetConcept ex:c1, ex:c2 .Example result ex:h1 ltk:subdividedInto ex:c1 .
ex:h1 ltk:subdividedInto ex:c2 .Example entailment ex:h1 skos:narrowMatch ex:c1 .
ex:h1 skos:narrowMatch ex:c2 .ltk:CombineTaxa
Description For combining lower taxa (source) into a higher taxon (target). Parameters – cka:sourceConcept (2..*)
– cka:targetConcept (1)Example input RDF ex:opr rdf:type ltk:CombineTaxa .
ex:opr
cka:sourceConcept ex:c1, ex:c2 ;
cka:targetConcept ex:h1 .Example result ex:c1 ltk:combindedInto ex:h1 .
ex:c2 ltk:combindedInto ex:h1 .Example entailment ex:c1 skos:broadMatch ex:h1 .
ex:c2 skos:broadMatch ex:h1 .ltk:DirectSynonymLink
Description For identifying a synonym (target) of a concept (source). It is a directional synonym, which is always used in botany. Parameters – cka:sourceConcept (1)
– cka:targetConcept (1)Example input RDF ex:opr rdf:type
ltk:DirectSynonymLink .
ex:opr cka:sourceConcept ex:c1 ;
cka:targetConcept ex:c2 .Example result ex:c1 ltk:dsynonym ex:c2 . Example entailment ex:c1 skos:exactMatch ex:c2 .
ex:c2 skos:exactMatch ex:c1 .
ex:c1 lodac:hasSynonym ex:c2 .ltk:SynonymLink
Description For identifying a synonym (target) of a concept (source). It is a bidirectional synonym, which is generally used in many domains especially in zoology. Parameters – cka:sourceConcept (1)
– cka:targetConcept (1)Example input RDF ex:opr rdf:type ltk:SynonymLink .
ex:opr cka:sourceConcept ex:c1 ;
cka:targetConcept ex:c2 .Example result ex:c1 ltk:synonym ex:c2 . Example entailment ex:c2 ltk:synonym ex:c1 .
ex:c1 ltk:dsynonym ex:c2 .
ex:c2 ltk:dsynonym ex:c1 .
ex:c1 skos:exactMatch ex:c2 .
ex:c2 skos:exactMatch ex:c1 .
ex:c1 lodac:hasSynonym ex:c2 .
ex:c2 lodac:hasSynonym ex:c1 .ltk:SeniorSynonymLink
Description For identifying a senior synonym (target) of a concept (source). Parameters – cka:sourceConcept (1)
– cka:targetConcept (1)Example input RDF ex:opr rdf:type
ltk:SeniorSynonymLink .
ex:opr cka:sourceConcept ex:c1 ;
cka:targetConcept ex:c2 .Example result ex:c1 ltk:seniorSynonym ex:c2 .
ex:c2 ltk:juniorSynonym ex:c1 .Example entailment ex:c1 ltk:synonym ex:c2 .
ex:c2 ltk:synonym ex:c1 .
ex:c1 skos:exactMatch ex:c2 .
ex:c2 skos:exactMatch ex:c1 .
ex:c1 lodac:hasSynonym ex:c2 .
ex:c2 lodac:hasSynonym ex:c1 .ltk:HomonymLink
Description For identifying a homonym (target) of a concept (source). Parameters – cka:sourceConcept (1)
– cka:targetConcept (1)Example input RDF ex:opr rdf:type
ltk:HomonymLink .
ex:opr cka:sourceConcept ex:c1 ;
cka:targetConcept ex:c2 .Example result ex:c1 ltk:homonym ex:c2 .
ex:c2 ltk:homonym ex:c1 .
Appendix D.
Appendix D.Example part of LTK ontology
This section shows an example part of LTK ontology that deal with the transition model. The hierarchy and type of properties are defined as follows:
ltk:majorMergedInto rdfs:subPropertyOf ltk:mergedInto . ltk:majorSplitInto rdfs:subPropertyOf ltk:splitInto . ltk:mergedInto rdfs:subPropertyOf cka:serialLinkTo . ltk:splitInto rdfs:subPropertyOf cka:serialLinkTo . ltk:replacedTo rdfs:subPropertyOf cka:serialLinkTo . ltk:serialLinkTo rdf:type owl:TransitiveProperty; rdfs:subPropertyOf cka:semanticLink . ltk:semanticLink rdf:type owl:TransitiveProperty , owl:SymmetricProperty .
References
[1] | Apache Jena, http://jena.apache.org/. |
[2] | R.C. Banks, C. Cicero et al., Forty-fourth supplement to the American Ornithologists’ Union checklist of North American birds, The Auk 120: (3) ((2003) ), 923–931, The American Ornithologists’ Union. |
[3] | W.G. Berendsohn, A taxonomic information model for botanical databases: The IOPI model, Taxon ((1997) ) 283–309. |
[4] | Bibliographic Ontology, http://bibliontology.com. |
[5] | Biodiversity Information Standards (TDWG), http://www.tdwg.org. |
[6] | R. Chawuthai, V. Wuwongse and H. Takeda, A formal approach to the modelling of digital archives, in: The Outreach of Digital Libraries: A Globalized Resource Network, H.H. Chen and G. Chowdhury, eds, Springer, Heidelberg, (2012) , pp. 179–188. |
[7] | P.W. Crous, W. Gams et al., MycoBank: An online initiative to launch mycology into the 21st century, Studies in Mycology 50: (1) ((2004) ), 19–22. |
[8] | P. Cryer, R. Hyam, C. Miller et al., Adoption of persistent identifiers for biodiversity informatics: Recommendations of the GBIF LSID GUID task group, in: Tech Rep Global Biodiversity Information Facility (GBIF), 6 November 2009, Copenhagen, Denmark, (2010) . |
[9] | Darwin Core, http://rs.tdwg.org/dwc/terms/. |
[10] | Describing Taxon Concepts as RDF (draft), https://code.google.com/p/tdwg-rdf/wiki/TaxonInRDF. |
[11] | Article 21. Determination of date (online): In International Commission on Zoological Nomenclature (ICZN), http://www.nhm.ac.uk/hosted-sites/iczn/code/index.jsp?article=21. |
[12] | Dublin Core Metadata Initiative Terms, http://dublincore.org/documents/dcmiterms/. |
[13] | G. Flouris and C. Meghini, Terminology and wish list for a formal theory of preservation, in: Proc. of the PV2007 International Conference, 9–11 October 2007, (2007) . |
[14] | N. Franz and R. Peet, Towards a language for mapping relationships among taxonomic concepts, Systematics and Biodiversity 7: (1) ((2009) ), 5–20, Edited by B. Rosen, Taylor & Francis. |
[15] | S. Freeman and R.M. Zink, A phylogenetic study of the blackbirds based on variation in mitochondrial DNA restriction sites, Systematic Biology 44: (3) ((1995) ), 409–420. |
[16] | Friend of a Friend, http://xmlns.com/foaf/0.1/. |
[17] | C. Gutierrez, C. Hurtado and R. Vaisman, Temporal RDF, in: The Semantic Web: Research and Applications, L. Aroyo et al., eds, Springer, Heidelberg, (2005) , pp. 93–107. |
[18] | T. Heath and C. Bizer, Linked data: Evolving the web into a global data space, in: Synthesis Lectures on the Semantic Web: Theory and Technology, J. Hendler and F.V. Harmelen, eds, Morgan & Claypool Publishers, (2011) . |
[19] | H. Inoue, S. Sugi et al., Moths of Japan, Volume 2: Plates and Synonymic Catalogue, Kodensha Co. Ltd, Tokyo, (1982) . |
[20] | International Commission on Zoological Nomenclature, International Code of Zoological Nomenclature, The 4th Edition, The International Trust for Zoological Nomenclature, London, UK, 1999, p. 306. |
[21] | U. Jinbo, List-MJ: A checklist of Japanese moths 2004–2008, http://listmj.mothprog.com. |
[22] | A.C. Jones, R.J. White and E.R. Orme, Identifying and relating biological concepts in the catalogue of life, Journal of Biomedical Semantics 2: (7) ((2011) ). |
[23] | J. Kennedy, R. Kukla and T. Paterson, Scientific names are ambiguous as identifiers for biological taxa: Their context and definition are required for accurate data integration, in: The 2nd International Conference on Data Integration in the Life Sciences (DILS), 20–22 July 2005, San Diego, California, B. Ludascher and L. Raschid, eds, Springer-Verlag, (2005) , pp. 80–95. |
[24] | Y. Kishida, The Standard of Moths in Japan II, Gakken Education Publishing, (2011) . |
[25] | N. Laurenne, J. Tuominen, H. Saarenmaa and E. Hyvönen, Making species checklists understandable to machines – a shift from relational databases to ontologies, Journal of Biomedical Semantics 5: (1) ((2014) ). |
[26] | J. Lehmann, R. Isele et al., DBpedia – a large-scale, multilingual knowledge base extracted from Wikipedia, Semantic Web Journal (2014). |
[27] | Linked Open Data, http://linkeddata.org/. |
[28] | C. Linnaeus, Systema Naturae, 10th edn, Holmiae, (1758) . |
[29] | J. Mallet, Concept of species, in: Encyclopedia of Biodiversity, S.A. Levin, ed., Vol. 5: , Academic Press, (2001) , pp. 427–440. |
[30] | Y. Minami, H. Takeda, F. Kato et al., Towards a data hub for biodiversity with LOD, in: Proc. of the Second Joint International Conference, 2–4 December 2012, Japan, H. Takeda, Y. Qu, R. Mizoguchi et al., eds, Springer, (2013) , pp. 356–361. |
[31] | OpenRDF, http://www.openrdf.org. |
[32] | R.D.M. Page, Taxonomic names, metadata, and the Semantic Web, Biodiversity Informatics 3: ((2006) ). |
[33] | D.J. Patterson, J. Cooper et al., Names are key to the big new biology, Trends in Ecology & Evolution 25: (12) ((2004) ), 686–691. |
[34] | D.P. Remsen, M. Döring, R.T. Copenhagen, GBIF GNA Profile Reference Guide for Darwin Core Archives, version 1.2, 2011. |
[35] | L.P. Richard and M. Ellinor, ZooBank: Developing a nomenclatural tool for unifying 250 years of biological information, 2008. |
[36] | K. Richards, R. White, N. Nicolson and R. Pyle, A beginner’s guide to persistent identifiers, in: Tech Rep Global Biodiversity Information Facility (GBIF), Copenhagen, Denmark, (2011) . |
[37] | I.N. Sarkar, Biodiversity informatics: Organizing and linking information across the spectrum of life, Briefings in Bioinformatics 8: (5) ((2007) ), 347–357. |
[38] | S. Schulz, H. Stenzhorn and M. Boeker, The ontology of biological taxa, Bioinformatics 24: (13) ((2008) ), i313–i321. |
[39] | Semantically-Interlinked Online Communities Core Specification, http://www.w3.org/Submission/sioc-spec/. |
[40] | C.G. Sibley and L.L.J. Short, Hybridization in the orioles of the great plains in the condor, JSTOR 66: (2) ((1964) ), 130–150. |
[41] | Simple Knowledge Organization System, http://www.w3.org/TR/skos-primer/. |
[42] | Taxonomic Concept Schema Complementary Documentation for Draft Standard, http://tdwg.napier.ac.uk/doc/tdwg_tcs.doc. |
[43] | Taxonomic Names and Concepts Interest Group: Taxonomic concept transfer schema, 2005, http://www.tdwg.org/standards/117/. |
[44] | The Global Biodiversity Information Facility (GBIF), http://www.gbif.org. |
[45] | The Timeline Ontology, http://motools.sourceforge.net/timeline/. |
[46] | J. Tuominen, N. Laurenne and E. Hyvönen, Biological names and taxonomies on the semantic web: Managing the change in scientific conception, in: Proc. of the 8th Extended Semantic Web Conference, 29 May–2 June 2011, Greece, G. Antoniou et al., eds, (2011) , pp. 255–269. |
[47] | M. Wink and P. Heidrich, Molecular evolution and systematics of the owls (Strigiformes), in: A Guide to Owls of the World, C. Konig and J.H. Becking, eds, Yale University Press, Yale, (1999) , pp. 39–57. |
[48] | J.E. Winston, Describing Species: Practical Taxonomic Procedure for Biologists, Columbia University Press, New York, (1999) . |
[49] | N. Ytow, D. Morse and D. Roberts, Nomencurator: A nomenclatural history model to handle multiple taxonomic views, Biological journal of the Linnean Society 73: (1) ((2001) ), 81–98. |



