Distinct Lysosomal Network Protein Profiles in Parkinsonian Syndrome Cerebrospinal Fluid
Abstract
Background: Clinical diagnosis of parkinsonian syndromes like Parkinson’s disease (PD), corticobasal degeneration (CBD) and progressive supranuclear palsy (PSP) is hampered by overlapping symptomatology and lack of diagnostic biomarkers, and definitive diagnosis is only possible post-mortem.
Objective: Since impaired protein degradation plays an important role in many neurodegenerative disorders, we hypothesized that profiles of select lysosomal network proteins in cerebrospinal fluid could be differentially expressed in these parkinsonian syndromes.
Methods: Cerebrospinal fluid samples were collected from PD patients (n = 18), clinically diagnosed 4-repeat tauopathy patients; corticobasal syndrome (CBS) (n = 3) and PSP (n = 8); and pathologically diagnosed PSP (n = 8) and CBD patients (n = 7). Each patient set was compared to its appropriate control group consisting of age and gender matched individuals. Select lysosomal network protein levels were detected via Western blotting. Factor analysis was used to test the diagnostic sensitivity, specificity and accuracy of the select lysosomal network protein expression profiles.
Results: PD, CBD and PSP were markedly different in their cerebrospinal fluid lysosomal network protein profiles. Lysosomal-associated membrane proteins 1 and 2 were significantly decreased in PD; early endosomal antigen 1 was decreased and lysozyme increased in PSP; and lysosomal-associated membrane proteins 1 and 2, microtubule-associated protein 1 light chain 3 and lysozyme were increased in CBD. A panel of lysosomal-associated membrane protein 2, lysozyme and microtubule-associated protein 1 light chain discriminated between controls, PD and 4-repeat tauopathies.
Conclusions: This study offers proof of concept that select lysosomal network proteins are differentially expressed in cerebrospinal fluid of Parkinson’s disease, corticobasal syndrome and progressive supranuclear palsy. Lysosomal network protein analysis could be further developed as a diagnostic fluid biomarker in parkinsonian syndromes.
INTRODUCTION
There is a compelling need to develop diagnostic biomarkers for Parkinson’s disease (PD) and atypical parkinsonian syndromes (APS), including corticobasal degeneration (CBD) and progressive supranuclear palsy (PSP). Due to their overlapping clinical manifestations, the diagnosis of these conditions can often be challenging, especially during their early clinical stages. Despite having similar clinical presentations, PD and the APS have different associated neuropathological features [1–4], which suggest that the underlying neurodegenerative mechanisms are also distinct, and their neurochemical footprints could be discerned through biofluid analysis for diagnostic purposes. Recent studies have explored the diagnostic and prognostic value of cerebrospinal fluid analysis of a number of proteins typically associated with neurodegenerative conditions, including amyloid-β1-42 (Aβ1-42), tau, alpha synuclein (α-synuclein) and neurofilament light chain (NFL) in Parkisonian syndromes [5]. Whereas studies for individual proteins in specific parkinsonian syndromes have sometimes been inconclusive [6], a panel approach, where a combination of several biomarkers is analyzed, has proven to reach higher accuracy and specificity [7]. Given that, protein misfolding, the key neuropathological changes in parkinsonian syndromes has been observed many years before the manifestation of clinical signs, some of the earliest pathogenic changes are hypothesized to involve mechanisms affecting protein autophagy, degradation and clearance [8–10]. Some of the earliest changes in Alzheimer’s disease (AD) have been described in connection to the lysosomal network, an interconnected vesicular network of endosomes, lysosomes and autophagosomes, that is crucial for protein homeostasis [11]. We recently observed select CSF lysosomal network protein increases in a large group of AD patients, including early endosomal antigen 1 (EEA1), lysosomal-associated membrane proteins 1 and 2 (LAMP-1, LAMP-2), lysozyme and microtubule-associated protein 1 light chain 3 (LC3) [12, 13]. The goal of this exploratory pilot study was to provide proof-of-principle that lysosomal network protein analysis reveals neurochemical differences in parkinsonian syndromes that could be further developed into diagnostic fluid biomarkers. We measured CSF levels of the select lysosomal network proteins EEA1, LAMP-1, LAMP-2, LC3 and lysozyme in PD, CBD and PSP patients, and determined whether their expression profiles are different compared to controls. A subset of analyses included CSF from pathologically confirmed CBD and PSP patients. Markedly different protein profiles were observed in PD and APS samples, compared to controls.
MATERIALS AND METHODS
Study population
CSF set 1: PD
CSF samples from PD patients (n = 18) who met the criteria for PD as defined by The United Kingdom PD Society Brain Bank [14], but not autopsy confirmed, were obtained from the Karolinska hospital. The patients were matched in age and gender, with controls suffering from benign neurological conditions, such as tension headache and unclassified sensory disturbances (n = 18). The characteristics of the patients included in CSF set 1 are presented in Table 1.
CSF set 2: Clinically diagnosed 4-repeat tauopathies
4-repeat tauopathy CSF samples, from clinically diagnosed CBS (n = 3) and PSP patients (n = 8), were obtained at the Karolinska hospital. Patients with PSP were clinically diagnosed with possible or probable PSP according to the National Institute of Neurological Disorders and Stroke-Society for Progressive Supranuclear Palsy (NINDS-SPSP) criteria [15]. Briefly, these include: 1) a gradually progressive disorder with onset at the age of 40 years or later and 2) vertical supranuclear gaze palsy and prominent postural instability within the first year of disease onset. Patients with CBS were diagnosed according to the Armstrong criteria [16]. The following features were required for a clinical diagnosis of CBS: 1) a slowly progressive course, 2) asymmetric limb or axial rigidity, present without reinforcement, 3) aphasia, visuospatial impairment or neglect, or apraxia, and 4) dystonia, myoclonus, cortical sensory loss, or alien limb phenomenon. The patients were matched in age and gender, with controls (n = 11) suffering from benign neurological conditions, such as tension headache and unclassified sensory disturbances. The characteristics of the patients included in CSF set 2 are presented inTable 1.
CSF set 3: Pathologically confirmed CBD and PSP
CSF samples from clinically diagnosed and pathologically confirmed PSP (n = 8) and CBD (n = 7) patients, were obtained from the University of California, San Francisco Memory and Aging Center. Patients with PSP were clinically diagnosed with possible or probable PSP according to the National Institute of Neurological Disorders and Stroke-Society for Progressive Supranuclear Palsy (NINDS-SPSP) criteria [15]. Patients with CBD were diagnosed according to the Armstrong criteria [16]. Pathological confirmation of the disease was performed on postmortem brains according to the previously described method [17]. The patients were matched in age and gender, with cognitively normal healthy elderly controls (n = 10) with no neurological complaints. The characteristics of the patients included in CSF set 3 are presented in Table 1.
CSF sampling
A standardized lumbar puncture (LP) procedure was performed fasting and sitting up, in accordance with the Alzheimer’s Disease Neuroimaging Initiative (ADNI) recommended protocol. Samples were collected into sterile polypropylene tubes, the first 2 mL were discarded and between 12 and 15 mL CSF from the first portion was collected and gently mixed in order to minimize the gradient influence and centrifuged in the original tube at 4000RPM for 10 min at +4°C. CSF samples were divided into aliquots put on dry ice before storing at –80°C until assayed. Blood-contaminated samples were excluded, and time between sample collection and freezing was maximum 1 hour. The clinically confirmed CBS and PSP CSF samples were collected during workup for parkinsonism. The pathologically confirmed CBD and PSP CSF samples were collected from live patients between 3.5 and 8 years after symptoms appeared, with an average disease duration of 6 and 10 years respectively.
Ethical statement
All the investigations of the patients and the analyses of their CSF were approved by the ethic committees of the respective institutions. Informed consent was obtained from the subjects.
Protein quantitation by Western immunoblotting
The CSF protein analysis via Western blotting has been described elsewhere [12], with the following change, blotting onto a 0.45 μm nitrocellulose membrane in transfer buffer (Tris-Base 25 mM, Glycine 192 mM, methanol 20%) for 1 h at 100 V and +4°C. The following antibodies were used: LAMP-1 and LAMP-2 (9835-01 and 9840-01, Southern Biotech, Burmingham, AL, USA), LC3 (NB600-1384, Novus Biologicals, Littleton, CO, USA), EEA1 (E4156, Sigma Aldrich, St. Louis, MO, USA), lysozyme and horseradish peroxidase-conjugated antibodies (A0099, P0448 and P0447, Dako, Glostrup, Denmark). The films were scanned and the immunoblots were quantified using the Image J program (available at http://rsbweb.nih.gov/ij/). All antibodies were confirmed to recognize their epitope via Western blot analysis of SH-SY5Y cell lysates, as well as overexpressing cell lines. Furthermore, most of the antibodies are highly specific and display only a single band of the CSF samples. Due to individual differences in protein concentrations in CSF, 20 μL of CSF were loaded per sample and equal sample loading was verified by Ponceau S (Sigma Aldrich) staining of total protein in each lane on the membranes (representative blot in Fig. 1C ). A standard CSF sample from one healthy control individual was loaded on each gel for normalization between the gels. Each gel was loaded with both control and patient samples and the samples were randomly distributed on the gels, and each sample set required multiple gels to fit all samples. Any difference in blot intensity was removed by subtracting the blot background for each sample separately.
Statistical analyses
Data were visually explored with histogram and box plots for normality and presence of outliers. Additional tests for normality were conducted with the Shapiro-Wilk test. All data were normally distributed and further data analysis proceeded with parametric tests. Mean densitometric Western blot values for each protein were compared with independent samples bidirectional Student’s t test. In set 2, data for CBS and PSP were combined into a single group of 4-repeat tauopathy. Factor analysis was used to test the sensitivity, specificity and accuracy of the select lysosomal network protein expression profiles to classify patients into the correct diagnostic group. Individual Western blot densitometric values for each lysosomal protein were used as independent variable predictors. Diagnoses (i.e. control, PD, CBD and PSP) were established as dependent grouping variables. Means and variance for control groups in the three different sets were similar. Thus, for factor analyses, data from sets 1, 2, and 3 were pooled, resulting in 39 controls, 18 PD, 10 CBD and 16 PSP cases. Statistical significance was defined for p-values of less than 0.05 (*), 0.01 (**) and 0.001 (***). Statistical analyses were performed using GraphPad Prism version 5.0 for Windows (GraphPad Software, San Diego, CA, USA) and SPSS version 23 (SPSS/IBM, Chicago, IL).
RESULTS
Decreased levels of the lysosomal network proteins LAMP-1 and LAMP-2 in PD CSF
We investigated the presence and levels of the lysosomal network proteins EEA1, LAMP-1, LAMP-2, LC3 and lysozyme in control and PD CSF (Table 1, CSF set 1) using Western blot analysis. Mature, functional LAMP-1 and LAMP-2 proteins are extensively glycosylated and run as spread out bands with a molecular weight between 100–150 kDa on Western blots. LAMP-1 and LAMP-2 levels were significantly decreased in the PD samples as compared with the control CSF samples (Fig. 1A and B). There was a 23% decrease in LAMP-1 levels, and a 30% decrease in LAMP-2 levels in PD CSF. Equal sample loading was confirmed via Ponceau S staining of total protein in each lane (Fig. 1C), and all proteins were detected in human SH-SY5Y neuroblastoma cell lysates, confirming the ability of the antibodies to recognize the epitopes (data not shown). EEA1, LC3 and lysozyme were present at detectable levels in the CSF, but no significant differences were observed between controls and PD patients. The complete western blots containing all CSF samples are demonstrated in Supplementary 1A.
Increased levels of lysosomal network protein LC3 in clinically diagnosed 4-repeat tauopathy CSF
Next, we investigated the levels of the lysosomal network proteins in CSF from clinically diagnosed 4-repeat tauopathy patients, consisting of CBS and PSP patients. Contrary to the findings in the PD CSF samples, the levels of LAMP-1 and LAMP-2 were unchanged, while LC3 levels were significantly increased in 4-repeat taupathy patients, corresponding to a value 92% higher, compared with control CSF samples (Fig. 2A and B). EEA1 and lysozyme were present at detectable levels in the CSF, but no significant difference was observed between CSF samples from control and 4-repeat tauopathy patients. The complete western blots containing all CSF samples are demonstrated in Supplementary 1B.
Different CSF level profiles of lysosomal network proteins EEA-1, LAMP-1, LAMP-2, LC3 and lysozyme in pathologically confirmed CBD and PSP CSF
To confirm our findings, and to further explore the differences in lysosomal network protein expression profiles between PD, CBS and PSP, we investigated the levels of the lysosomal network proteins in CSF from clinically diagnosed CBD and PSP patients where the diagnosis had been pathologically confirmed post-mortem. LAMP-1, LAMP-2, LC3 and lysozyme levels were significantly increased in pathologically confirmed CBD compared with control samples (Fig. 3A and B); LAMP-1 by 66%, LAMP-2 and LC3 by 280% and lysozyme by 456%. A different pattern emerged in the pathologically confirmed PSP patients, where the EEA1 level was significantly decreased by 29% and the lysozyme level was significantly increased by 211% in PSP patient CSF compared with control samples (Fig. 3C and D). The complete Western blots containing all CSF samples are demonstrated in Supplementary 1C.
Patterns of lysosomal network proteins showed high diagnostic accuracy
Analyses combining data from the three sets was used to test the diagnostic accuracy of the lysosomal network protein expression levels. The patterns of LAMP-1 and LAMP-2 expression were useful for separating controls from patients with PD (X2 = 10.2, p = 0.006) with sensitivity of 94.9%, specificity of 44.4% and accuracy of 78.9%. The effect size was 0.17. A profile including LAMP-2, lysozyme and LC-3 was useful at discriminating controls from CBD with a sensitivity of 100%, specificity of 60% and accuracy of 91.8% (X2 = 32.1, p < 0.001). The effect size was 0.50. The expression of lysozyme was different in controls and PSP patients. Lysozyme expression identified PSP patients with a sensitivity of 97.4%, specificity of 31.3% and accuracy of 78.2% (X2 = 12.1, p < 0.001). The effect size was 0.20. The expression patterns of LAMP-1, LAMP-2, lysozyme and LC-3 were useful at discriminating patients with PD from patients with 4-repeat tauopathies (CBD and PSP combined) with a sensitivity of 77.8%, specificity of 73.1% and accuracy of 75% (X2 = 10.4, p = 0.03). The effect size was 0.23. A model testing simultaneous discrimination between controls, PD and 4-repeat tauopathies showed that a composite of LAMP-2, lysozyme and LC-3 had a sensitivity of 92.3% but low specificity for both PD (5.6%) and 4-repeat tauopathies (38.5%) (X2 = 32.3 p < .001). The accuracy of this composite model was 56.6%, with an effect size of 0.28(Fig. 4).
DISCUSSION
We have discovered that parkinsonian syndromes display different patterns of CSF select lysosomal network proteins, compared to their appropriate controls. The lysosomal proteins LAMP-1 and LAMP-2 were significantly decreased in PD compared with control; LC3 was increased in clinically diagnosed 4-repeat tauopathy patients (CBS and PSP) compared with controls; pathologically diagnosed CBD patients had significantly increased LAMP-1, LAMP-2, LC3 and lysozyme levels compared with controls; whilst pathologically diagnosed PSP patients had significantly decreased EEA1 level and increased lysozyme levels compared with controls. A pool factorial analysis confirmed that combined patterns of lysosomal patterns are sensitive at discriminating controls from the different parkinsonian diseases. These results support that the lysosomal protein network is activated in parkinsonian syndromes. Furthermore, a combination of LAMP-1, LAMP-2, LC3 and lysozyme levels was able to distinguish patients with PD, from patients with 4-repeat tauopathies. It can thus be hypothesized that profiles of lysosomal network proteins could be different among parkinsonian diseases and could have value as biomarkers useful in the differential diagnosis of PD, CBD and PSP. These data are consistent with previous results from AD CSF samples, where no general increase of lysosomal network proteins was observed, but a select subset of lysosomal proteins was changed [12]. Based on these data showing differential expression profiles of select CSF lysosomal network proteins, further validating studies that also address their specificity in parkinsonian syndromes should be pursued. Whether other parkinsonian syndromes with different associated pathology, such as multiple system atrophy, or other non-related parkinsonian conditions with predominant tau pathology, such as behavioral variant frontotemporal dementia, share some of the observed CSF lysosomal protein changes should be also furtherexplored.
Some of the earliest abnormalities observed in AD and PD are connected to malfunctioning protein clearance in the lysosomal network [11, 18, 19]. Our data indicate that it is highly likely that changes to lysosomal network proteins are involved in the pathogenesis of CBD and PSP, in one or more of the lysosomal network compartments; lysosomes, autophagosomes or endosomes. The pathologies of AD, PD, PSP and CBD have in common the accumulation of misfolded and aggregation-prone proteins; Aβ, α-synuclein, and tau, which are not degraded efficiently but instead may negatively affect degradation pathways, such as the lysosomal network [20]. It has been previously described that levels of LAMP-2 are decreased early in the disease progression in the PD brain [10, 21]. in particular, the lysosomal membrane protein isoform LAMP-2A is believed to be primarily involved in PD-related neurodegeneration, since the levels of the isoforms LAMP-2B and LAMP-2C are unchanged [22] and LAMP-2A is known to be involved in chaperone mediated autophagy. The decreased level of LAMP-2 correlates with a decreased level and activity of glucocerebrosidase and high levels of α-synuclein in PD [21]. Our study demonstrates decreased levels of LAMP-2 in CSF from PD patients, which may reflect the levels of glucocerebrosidase and α-synuclein in the PD brain. The levels of the lysosomal membrane protein LAMP-1 are not altered in substantia nigra pars compacta or amygdala in the PD brain [10, 23], however, we detected a decreased level of LAMP-1 in PD CSF which might indicate that other brain regions and different cellular subtypes could be implicated in PD-related neurodegeneration. Overexpressing α-synuclein in a microglia cell line shows, among other things, decreased LAMP-1 levels [24]. In the present study, CBD cases showed increased CSF LAMP-1 and 2 levels, mirroring what is seen in AD [12]. We found that the level of the lysosomal glycoside hydrolase and innate immunity protein lysozyme was increased in CBD but unchanged in PD and PSP. The increase of lysozyme in CBD could reflect an astrocytic response towards the tau pathology seen in astrocytes in this disorder [4]. LC3 is involved in formation of the autophagosome and earlier reports show increased levels of autophagic vesicles containing LC3 in PD brains as well as mice and cell models [9]. LC3 and impaired autophagy have previously been reported in a cellular model of CBD and PSP [8], as well as in a mouse model for tauopathies [25]. We could not detect any differences in LC3 levels in CSF from PD or pathologically diagnosed PSP patients, but the protein was found at higher levels in CSF in pathologically confirmed CBD patients. Why the previously reported increased LC3 levels in the PD brain is not mirrored in the CSF could be explained by that LC3 was reported to be sequestered in Lewy bodies in PD [10, 23], and that Lewy bodies are also detected in some PSP patients [26]. The endosomal protein EEA1, which is involved in fusion and delivery of endosomal cargo, was significantly decreased in CSF from PSP cases, but to our knowledge there are no prior studies on EEA1 in PD,CBD or PSP.
The difference in results between the clinically diagnosed CBS and PSP samples versus the pathologically diagnosed CBD and PSP samples could be caused by the propensity of clinically diagnosed patients to be misdiagnosed [27], highlighting the need for reliable biomarkers, but could also indicate that the levels of certain proteins changes during the disease progression, like prior reports that sAPPα levels decreases longitudinally in APS patients [7]. It is noteworthy that the lysosomal marker profile of pathologically-confirmed CBD patients resembles that of the one previously reported for AD patients [12, 13], more than PD patients, even though not identical, where CBD and AD patients have increased levels of LAMP-1, LAMP-2, lysozyme and LC3. This finding indicates that there may be overlapping pathophysiological changes in lysosomal function between these disorders and that therapies directed towards optimizing lysosomal functions in AD may turn out to be beneficial also in CBD. A recent study investigating the diagnostic power of combining the nine CSF proteins T-Tau, NFL, monocyte chemoattractant protein-1, YKL-40, sAPPα, sAPPβ, Aβ1-42, P-Tau and α-synuclein to distinguish between e.g. PD, APS, AD and DLB gave promising results [7]. To combine the levels and patterns of these proteins with the lysosomal network proteins, LAMP-1, LAMP-2, lysozyme, LC3 and EEA1 might boost the diagnostic accuracy of this diagnostic panel.
There are several limitations to this study. The number of analyzed cases is small, which likely resulted in underestimation of specificity and low accuracy. This in turn compromises the external validity of the findings. The predictive value of the current findings is contingent upon their replication in larger patient sets. Samples were obtained from different centers, which possibly contributed to case heterogeneity, thus affecting sensitivity estimates. Also, disease duration at the time of CSF sampling significantly varied between patient diagnoses and analyte levels could be significantly affected by the disease stage. The cross-sectional nature of this analysis presents a reduced dimension of analyte expression and further longitudinal studies will be needed to examine the consistency of the observed changes and the prognostic utility of the profiles. A strength of this study is the use of age-matched controls in each recruitment center. This may overcome potential bias related to sample processing. In addition, no differences were observed in control values across patient sets. This study also included more than one diagnostic group, which explores the specificity of the diagnostic tests. Results observed in clinically diagnosed APS cases had a replication in pathologically-diagnosed counterparts. The use of pathologically confirmed cases aids in the specificity of findings and strengthens the findings in light of our previous report on AD patients.
In summary; this study provides proof of principle that the levels and patterns of the select lysosomal network proteins LAMP-1, LAMP-2, lysozyme, LC3 and EEA1 differ between PD, CBD and PSP CSF as compared to their appropriate controls. These proteins have the potential as tools in investigating the disease mechanisms for AD, parkinsonian disorders and other neurodegenerative conditions featuring abnormal protein degradation and aggregation, as potential biomarkers to distinguish between the diseases; and perhaps even as future targets for novel treatments. Further validation studies on the role of lysosomal network protein expression profiles are indicated.
CONFLICT OF INTEREST
The authors have no conflict of interest.
ACKNOWLEDGMENTS
The authors thank Karin & Sten Mortstedt CBD Solutions for support, and the patients whose CSF was used in this study.
This work was supported by the Swedish Alzheimer foundation, the Swedish Dementia foundation, Linköping University Neurobiology Center, Karin & Sten CBD Solutions AB, AZ-KI TSC, ALF, US National Institutes of Health R01AG038791 and U54NS092089, the Tau Consortium, the Hellman Family Foundation.
Appendices
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/JPD-150759.
REFERENCES
[1] | McCann H , Stevens CH , Cartwright H , & Halliday GM ((2014) ) alpha-Synucleinopathy phenotypes, Parkinsonism Relat Disord 20: (Suppl 1), S62–S67. |
[2] | Kouri N , Whitwell JL , Josephs KA , Rademakers R , & Dickson DW ((2011) ) Corticobasal degeneration: A pathologically distinct 4R tauopathy, Nat Rev Neurol 7: , 263–272. |
[3] | Dickson DW , Ahmed Z , Algom AA , Tsuboi Y , & Josephs KA ((2010) ) Neuropathology of variants of progressive supranuclear palsy, Curr Opin Neurol 23: , 394–400. |
[4] | Hattori M , Hashizume Y , Yoshida M , Iwasaki Y , Hishikawa N , Ueda R , & Ojika K ((2003) ) Distribution of astrocytic plaques in the corticobasal degeneration brain and comparison with tuft-shaped astrocytes in the progressive supranuclear palsy brain, Acta Neuropathol 106: , 143–149. |
[5] | Backstrom DC , Eriksson Domellof M , Linder J , Olsson B , Ohrfelt A , Trupp M , Zetterberg H , Blennow K , & Forsgren L ((2015) ) Cerebrospinal fluid patterns and the risk of future dementia in early, incident parkinson disease, JAMA Neurol 72: , 1175–1182. |
[6] | Magdalinou N , Lees AJ , & Zetterberg H ((2014) ) Cerebrospinal fluid biomarkers in parkinsonian conditions: An update and future directions, J Neurol Neurosurg Psychiatry 85: , 1065–1075. |
[7] | Magdalinou NK , Paterson RW , Schott JM , Fox NC , Mummery C , Blennow K , Bhatia K , Morris HR , Giunti P , Warner TT , de Silva R , Lees AJ , & Zetterberg H ((2015) ) A panel of nine cerebrospinal fluid biomarkers may identify patients with atypical parkinsonian syndromes, J Neurol Neurosurg Psychiatry 86: , 1240–1247. |
[8] | Leyk J , Goldbaum O , Noack M , & Richter-Landsberg C ((2015) ) Inhibition of HDAC6 modifies tau inclusion body formation and impairs autophagic clearance, J Mol Neurosci 55: , 1031–1046. |
[9] | Lynch-Day MA , Mao K , Wang K , Zhao M , & Klionsky DJ ((2012) ) The role of autophagy in Parkinson’s disease, Cold Spring Harb Perspect Med 2: , e009357. |
[10] | Alvarez-Erviti L , Rodriguez-Oroz MC , Cooper JM , Caballero C , Ferrer I , Obeso JA , & Schapira AH ((2010) ) Chaperone-mediated autophagy markers in Parkinson disease brains, Arch Neurol 67: , 1464–1472. |
[11] | Ihara Y , Morishima-Kawashima M , & Nixon R ((2012) ) The ubiquitin-proteasome system and the autophagic-lysosomal system in Alzheimer disease, Cold Spring Harb Perspect Med 2: ,pii. a006361 |
[12] | Armstrong A , Mattsson N , Appelqvist H , Janefjord C , Sandin L , Agholme L , Olsson B , Svensson S , Blennow K , Zetterberg H , & Kagedal K ((2014) ) Lysosomal network proteins as potential novel CSF biomarkers for Alzheimer’s disease, Neuromolecular Med 16: , 150–160. |
[13] | Helmfors L , Boman A , Civitelli L , Nath S , Sandin L , Janefjord C , McCann H , Zetterberg H , Blennow K , Halliday G , Brorsson AC , & Kagedal K ((2015) ) Protective properties of lysozyme on beta-amyloid pathology: Implications for Alzheimer disease, Neurobiol Dis 83: , 122–133. |
[14] | Hughes AJ , Daniel SE , Kilford L , & Lees AJ ((1992) ) Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases, J Neurol Neurosurg Psychiatry 55: , 181–184. |
[15] | Litvan I , Agid Y , Calne D , Campbell G , Dubois B , Duvoisin RC , Goetz CG , Golbe LI , Grafman J , Growdon JH , Hallett M , Jankovic J , Quinn NP , Tolosa E , & Zee DS ((1996) ) Clinical research criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome): Report of the NINDS-SPSP international workshop, Neurology 47: , 1–9. |
[16] | Armstrong MJ , Litvan I , Lang AE , Bak TH , Bhatia KP , Borroni B , Boxer AL , Dickson DW , Grossman M , Hallett M , Josephs KA , Kertesz A , Lee SE , Miller BL , Reich SG , Riley DE , Tolosa E , Troster AI , Vidailhet M , & Weiner WJ ((2013) ) Criteria for the diagnosis of corticobasal degeneration, Neurology 80: , 496–503. |
[17] | Gallyas F ((1971) ) Silver staining of Alzheimer’s neurofibrillary changes by means of physical development, Acta Morphol Acad Sci Hung 19: , 1–8. |
[18] | Son JH , Shim JH , Kim KH , Ha JY , & Han JY ((2012) ) Neuronal autophagy and neurodegenerative diseases, Exp Mol Med 44: , 89–98. |
[19] | Brockmann K , & Berg D ((2014) ) The significance of GBA for Parkinson’s disease, J Inherit Metab Dis 37: , 643–648. |
[20] | Xilouri M , & Stefanis L ((2015) ) Chaperone mediated autophagy to the rescue: A new-fangled target for the treatment of neurodegenerative diseases, Mol Cell Neurosci 66: , 29–36. |
[21] | Murphy KE , Gysbers AM , Abbott SK , Tayebi N , Kim WS , Sidransky E , Cooper A , Garner B , & Halliday GM ((2014) ) Reduced glucocerebrosidase is associated with increased alpha-synuclein in sporadic Parkinson’s disease, Brain 137: , 834–848. |
[22] | Murphy KE , Gysbers AM , Abbott SK , Spiro AS , Furuta A , Cooper A , Garner B , Kabuta T , & Halliday GM ((2015) ) Lysosomal-associated membrane protein 2 isoforms are differentially affected in early Parkinson’s disease, Mov Disord 30: , 1639–1647. |
[23] | Dehay B , Bove J , Rodriguez-Muela N , Perier C , Recasens A , Boya P , & Vila M ((2010) ) Pathogenic lysosomal depletion in Parkinson’s disease, J Neurosci 30: , 12535–12544. |
[24] | Rojanathammanee L , Murphy EJ , & Combs CK ((2011) ) Expression of mutant alpha-synuclein modulates microglial phenotype, J Neuroinflammation 8: , 44. |
[25] | Schaeffer V , & Goedert M ((2012) ) Stimulation of autophagy is neuroprotective in a mouse model of human tauopathy, Autophagy 8: , 1686–1687. |
[26] | Uchikado H , DelleDonne A , Ahmed Z , & Dickson DW ((2006) ) Lewy bodies in progressive supranuclear palsy represent an independent disease process, J Neuropathol Exp Neurol 65: , 387–395. |
[27] | Ling H , O’Sullivan SS , Holton JL , Revesz T , Massey LA , Williams DR , Paviour DC , & Lees AJ ((2010) ) Does corticobasal degeneration exist? A clinicopathological re-evaluation, Brain 133: , 2045–2057. |
Figures and Tables
Fig.1
The lysosomal network proteins LAMP-1 and LAMP-2 are downregulated in the CSF of PD patients. A) Representative samples from one Western blot of CSF from controls (C) and Parkinson’s disease (PD) patients analyzed for the lysosomal network proteins EEA1, LAMP-1, LAMP-2, LC3 and lysozyme. B) Mean densitometric quantification of the scanned Western blots from C (n = 18) and PD (n = 18) patients. The protein levels are normalized to a standard CSF sample loaded on each gel. C) Equal sample loading was verified by Ponceau S staining of total protein in each lane on the membranes. The bars represent the mean±SD, *p < 0.05.
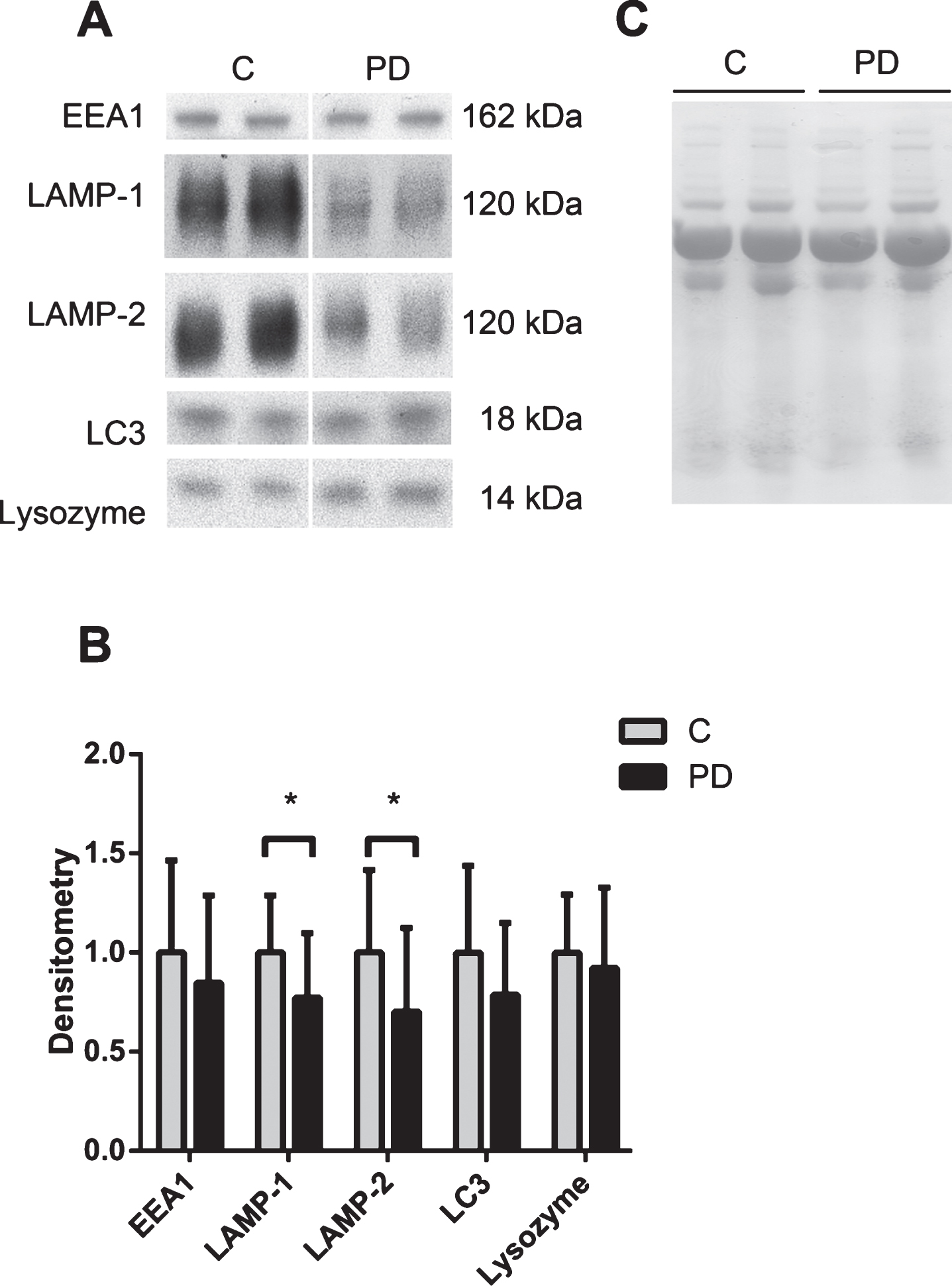
Fig.2
The lysosomal network protein LC3 is upregulated in the CSF of clinically diagnosed 4-repeat tauopathy patients. A) Representative samples from one Western blot of CSF from controls (C) and clinically diagnosed 4-repeat tauopathy patients (CBS/PSP), analyzed for the lysosomal network proteins EEA1, LAMP-1, LAMP-2, LC3 and lysozyme B) Mean densitometric quantification of the scanned Western blots from C (n = 11), clinically diagnosed 4-repeat tauopathy patients (CBS, n = 6 and PSP, n = 5). The protein levels are normalized to a standard CSF sample loaded on each gel. C) Equal sample loading was verified by Ponceau S staining of total protein in each lane on the membranes. The bars represent the mean±SD, ***p < 0.001.
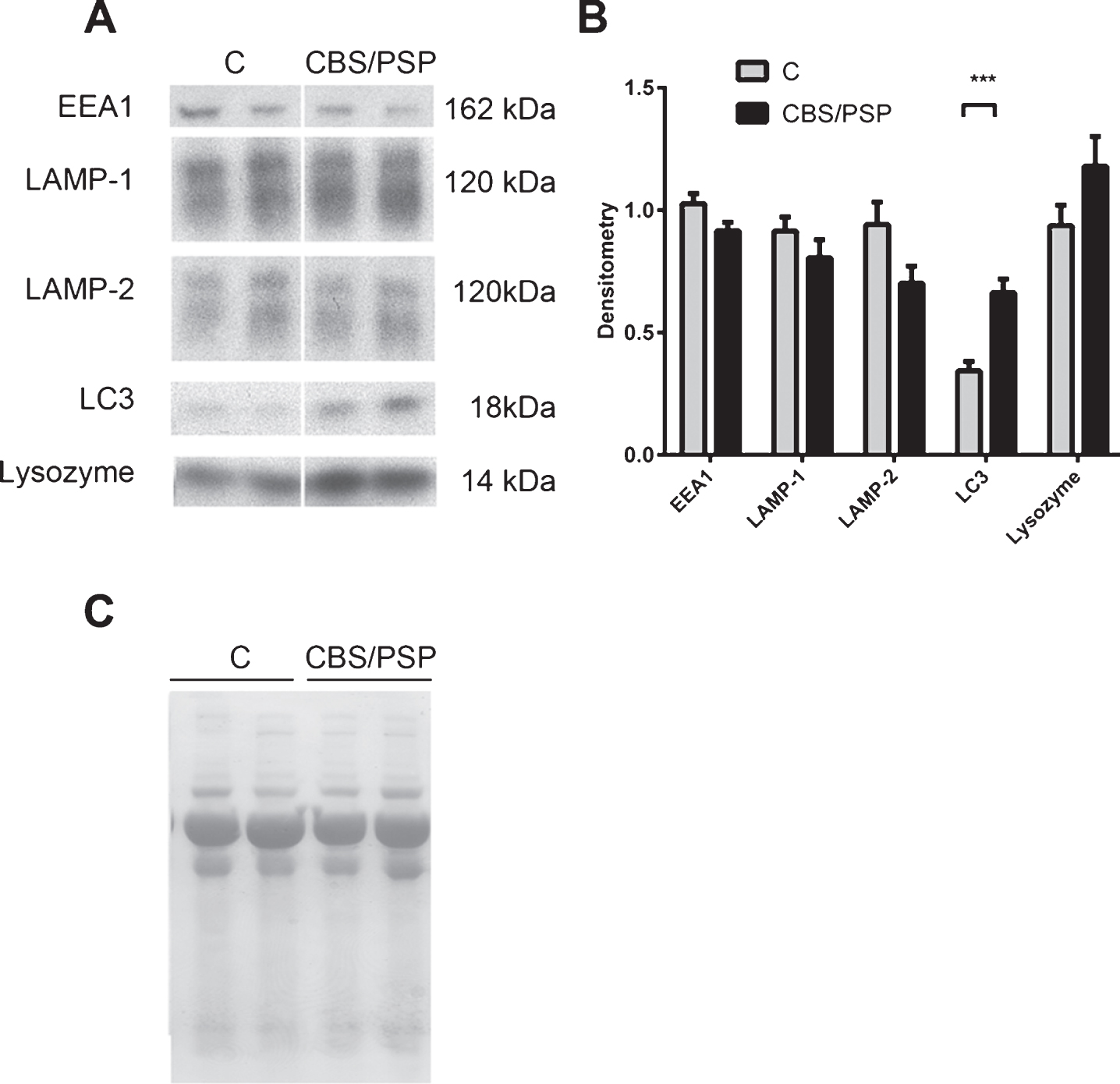
Fig.3
The lysosomal network proteins LAMP-1, LAMP-2, LC3 and lysozyme are upregulated in the CSF of pathologically confirmed CBD patients, whilst EEA1 is decreased and lysozyme is upregulated in the CSF of pathologically confirmed PSP patients. A) Representative samples from one Western blot of CSF from controls (C) and pathologically confirmed corticobasal degeneration (CBD) patients analyzed for the lysosomal network proteins EEA1, LAMP-1, LAMP-2, LC3 and lysozyme. B) Mean densitometric quantification of the scanned Western blots from C (n = 10) and pathologically confirmed CBD (n = 7) patients. C) Representative Western blots of CSF from controls (C) and pathologically confirmed progressive supranuclear palsy patients (PSP) analyzed for the lysosomal network proteins EEA1, LAMP-1, LAMP-2, LC3 and lysozyme. D) Densitometric quantification of the scanned Western blots from C (n = 10) and pathologically confirmed PSP patients (n = 8). The protein levels are normalized to a standard CSF sample loaded on each gel. E and F) Equal sample loading was verified by Ponceau S staining of total protein in each lane on the membranes in A and C respectively. The bars represent the mean±SD, *p < 0.05, **p < 0.01, ***p < 0.001.
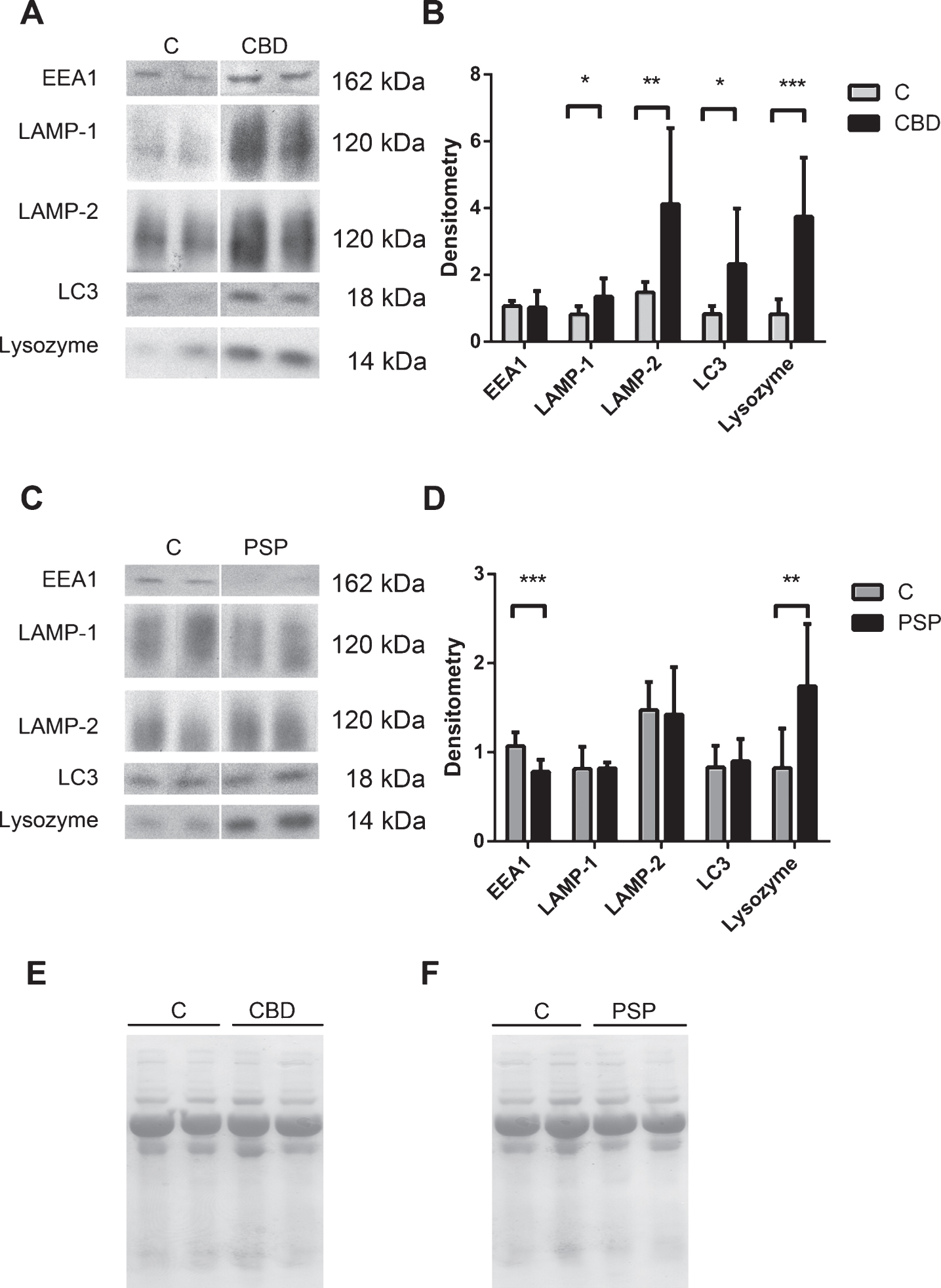
Fig.4
Patterns of lysosomal network protein expression showed differences between control subjects, patients with Parkinson’s disease and 4-repeat tauopathies (CBD and PSP combined). Function 1 (canonical coefficients: lysozyme 1.4, LAMP-2 -0.15 and LC3 -0.39) showed a higher segregation of 4-repeat tauopathies (4RTau) patients. In turn, function 2 (canonical coefficients: LAMP-2 1.7, lysozyme –0.8 and LC3 –1.4, respectively) showed discrimination of PD patients. Control subjects featured comparatively lower LC3 with variable lysozyme and LC3. Conversely, PD featured low LC3 and lysozyme, whereas 4-repeat tauopathies showed high lysozyme score with variable LAMP-2 and LC3.
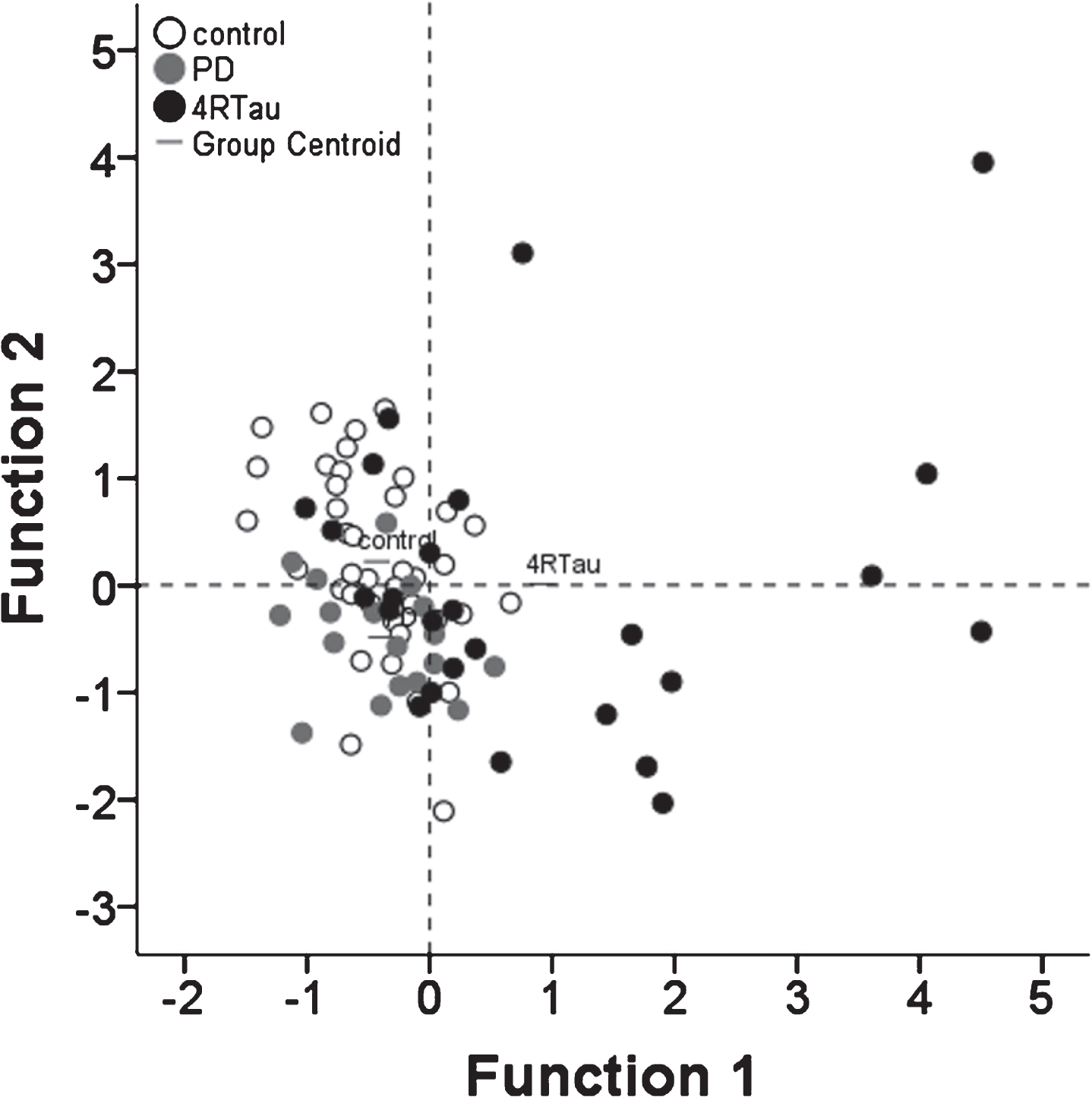
Table 1
Cerebrospinal fluid (CSF) study demographics of Parkinson’s Disease (PD) and controls (C) (CSF set 1), clinically confirmed 4-repeat tauopathy (CBS and PSP) patients and C (CSF set 2), clinically and pathologically confirmed CBD and PSP patients and C (CSF set 3). Sample collection and CSF measurements for sets 1 and 2 were performed at Karolinska University Hospital and at UCSF for set 3. Data are presented as the mean and (range). There was no overlap between control samples used in the different sets.
| Age (years) | Gender | Disease duration | Levodopa equivalent | |
| (M:F) | (years) | dose (LED) | ||
| CSF set 1 | ||||
| PD (n = 18) | 61 (39–80) | 15 : 3 | 04 (1–14) | 114 (0–2012) |
| C (n = 18) | 62 (46–83) | 14 : 4 | n/a | n/a |
| CSF set 2 | ||||
| Clinically confirmed 4-repeat tauopathy (n = 11) | 71 (63–81) | 07 : 4 | 3 (1–6) | 143 (0–764) |
| C (n = 11) | 64 (54–73) | 06 : 5 | n/a | n/a |
| CSF set 3 | ||||
| Pathologically confirmed CBD (n = 7) | 64 (61–70) | 04 : 3 | 6 (5–8) | 0 |
| Pathologically confirmed PSP (n = 8) | 69 (55–88) | 02 : 6 | 10 (4–17) | 150 (0–600) |
| C (n = 10) | 59 (52–68) | 04 : 6 | n/a | n/a |




