Analysis of the expression of β-actin, RuANS and RuMYB10 genes involved in the biosynthesis of anthocyanin using RT-qPCR in Rubus niveus in the Rumiñahui Canton
Abstract
BACKGROUND:
Berries such as Rubus has caught the attention of scientists and consumers because of their nutritional quality. Furthermore, the presence of phenolic compounds specifically anthocyanins have added potential of antiaging, anti-carcinogenic and antioxidant properties to the fruit. Gene expression studies have demonstrated the importance of flavonoids and their biosynthetic pathways, further increasing the research interests alternative and natural sources for these compounds.
OBJECTIVE:
The aim of this work was to analyze the level of expression of β-Actin as a control gene, RuANS gene encoding for anthocyanin synthase and RuMYB10 gene encoding for a transcription factor involved in the biosynthesis of anthocyanins in Rubus niveus.
METHODS:
To investigate the expression of the genes, the polymerase chain reaction with reverse transcription (RT-qPCR) was performed and the Qubit® fluorometry equipment for standard curve data.
RESULTS:
The ANOVA at 95% classified the genes into three homogeneous groups whose means are not statistically different in the gene expression, with an F = 0.46, a p = 0.6348. The results were generated in average: gene β-Actin: 174,65 ng/mL, which has greater concentration, followed by the RuMYB10 gene: 167.43 ng/mL and finally the gene RuANS with 163,55.
CONCLUSIONS:
The Rubus niveus specie presents a level of similar expression among the three analyzed genes.
1Introduction
The last decades have seen an increase interest in the properties of anthocyanins, according to Miyazawa and co-workers [1], (1999), these compounds have healthy effects such as the reduction of heart diseases and improvement of eyesight, as well as antitumoral, anticancer and antiaging effects [2] as a result of its properties [3].
Zhang and co-workers [4] report that anthocyanins are generated from the phenylalanine and acetate precursors, the anthocyanins are synthesised in the cytoplasm and transported in the vacuole where they join with a matrix protein and form anthocyanic vacuolar inclusions (AVI’s), which are considerated the storage sites of anthocyanins.
Quinton [5] notes that Rubus niveus is native to Asia, however it is now present all around the world. The diversity of this genus forms a wide variety of fruits, that contain chemopreventive compounds such as anthocyanins [6]. It is as a prennial plant, but inzones with severe winters it adopts an annual behaviour, the fruit’s composition is aproximately about 7,8% sugar, 0,13% protein and 0,77% ashes [7].
Deighton [8] reports that the juice of Rubus niveus has a high content of anthocyanins (1.186 g/mL) which increase with maturity [9].
Tamara [10] reports that the blackberry is extremely sought after in the national and international market and its consumption has increased in recent years.
PCR is a qualitative test that provides affirmative and negative answers to a specific question. The RT-qPCR allows the measurement of specific genes in a given sample [11]. For the purpose of this analysis, β-actin has been evaluated as a reference gene, as it is one of the most widely used due to its high concentrations. This allows for the normalisation of a standard curve and generating reliable data in the analysis.
In this work, we compare the genes most directly associated with anthocyanin biosynthesis. The level of transcription of the gene that specifically controls this compound is anthocyanin synthase (RuANS); RuMYB10 encodes a transcription factor and is used for role-specific regulation in the anthocyanin pathway [9].
The knowledge and analysis of genes, their expression and functions, mark a new era in biology and biotechnology in Ecuador. The current aim is to research for a more complete and detailed knowledge of molecular bases, allowing genomics to move into the more practical field of improving food and healthtechnologies [12].
2Materials and methods
2.1Sample location and size
The present research was conducted in the Life Sciences Laboratories (CIVABI) of the Salesian Polytechnic University in Quito.
The number of samples was decided with the formula proposed by Yánez [13]:
Where the value of the standard deviation is 0,05 that results in a simple size of 15 extractions per gene.
2.2Sample collection for analysis
The sample consisted of a selection of ripe Rubus niveus fruits from 3 climatically and geographically important areas within the Valle de Los Chillos (Rumiñahui Canton in the Province of Pichincha): San Rafael, Capelo and San Pedro de Taboada.
Two field trips took place during September and October 2014 and 1 kg of fruit per sector was aseptically collected, considering the organoleptic characteristics. The samples were taken to the Molecular Biology Laboratory of Salesian Polytechnic University for the relevant preservation and maintenance treatments to protect them from changes in temperature, lighting and handling.
2.3RNA extraction
Disinfection with UV rays was performed for 15 minutes in the laminar flow chamber to prevent cross-contamination.
The PureLink® RNA Mini Kit (Ambion, Life) was used to obtain samples of nucleic acids.
2.4Reverse transcription
To obtain the cDNA samples analyzed in the RT-qPCR the reverse transcription the Transcriptor First Strand cDNA synthesis Kit (Roche) was used.
2.5Standard curve plotting
Rubus glaucus was used to plot the standard curve with the β-Actin gene. β-Actin is selected as a reference gene as a result of its constant level of transcription in fruits and leaves [14].
To obtain the reference values serial solutions on a logarithmic scale of the previously obtained cDNA were prepared. The samples were quantified in the Qubit® 2.0 Fluorometer, and the Qubit® dsDNA HS Assay Kit (Life) was used to measure fluorescence.
2.6Quantification through RT-qPCR amplification
Light Cycler® Fast Start DNA Master PLUS SYBR Green I (Roche) was previously prepared with 14 μl of 1a (Polymerase Enzyme) in the 1b tube (Reaction Mix).
For initial denaturation consisting of one cycle, the target temperature was set to 95°C.
After denaturation, a process consisting of 60 cycles with three different target temperatures (95, 60 and 72°C) was carried out. The melting process was conducted which consists of a single cycle with target temperatures of 95, 65 and 95°C is conducted. Lastly, the cooling cycle consisting of one cycle set to a target temperature of 40°C was carried out, thereby yielding the graphical results.
3Results
For the quantification of the analyzed samples, the standard curve points were obtained from the concentration values obtained in the Qubit®.
In Fig. 1 the standard curve displays an efficiency of 1.922 and a very low error close to 0, in order to validate the test results.
Fig.1
β-Actin standard curve for RuANS and RuMYB10 gene quantification.
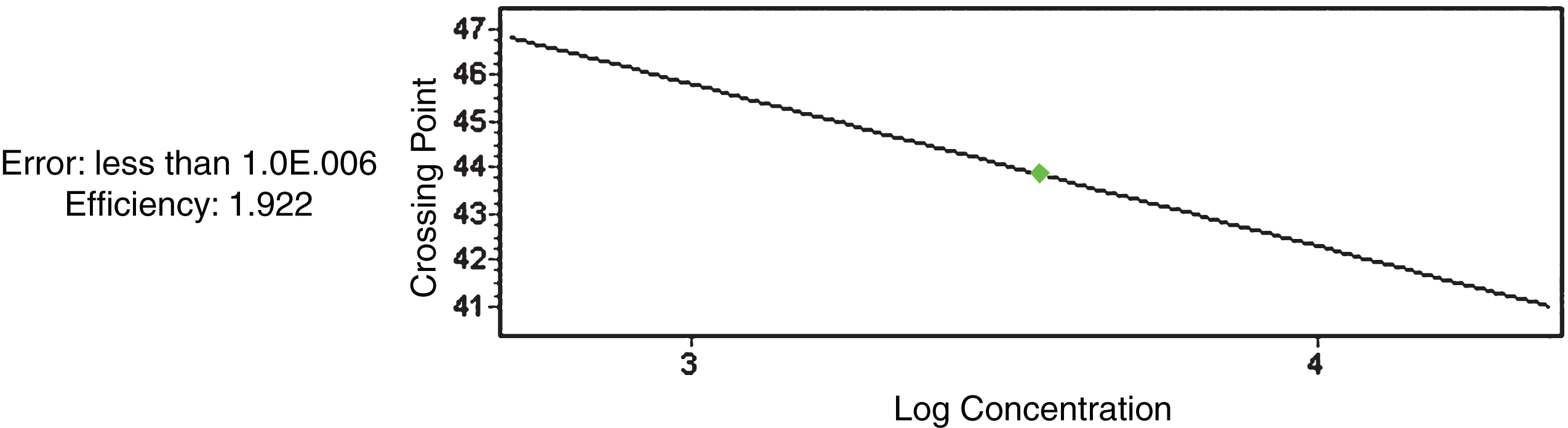
The Shapiro-Wilks test confirmed that the data obtained is normally distributed, thus allowing a second statistical test to analyze the expression level and determine the relation between the β-Actin, RuANS and RuMYB10 genes in Rubus niveus by means of RT- qPCR.
The variance analysis (ANOVA), classified the genes into three homogeneous groups whose means are not statistically different, with an F = 0.46 and a p = 0.6348. This analysis was conducted at a confidence level of 95%. To corroborate the results obtained from the variance analysis the Tukey test was used and the conclusion that all 3 genes are involved in the same manner in anthocyanin biosynthesis was reached. The group that differs from the three gene groups is the negative control, as shown in Fig. 2.
Fig.2
Tukey test data for β-Actin, RuANS and RuMYB10 genes present in Rubus niveus quantification.
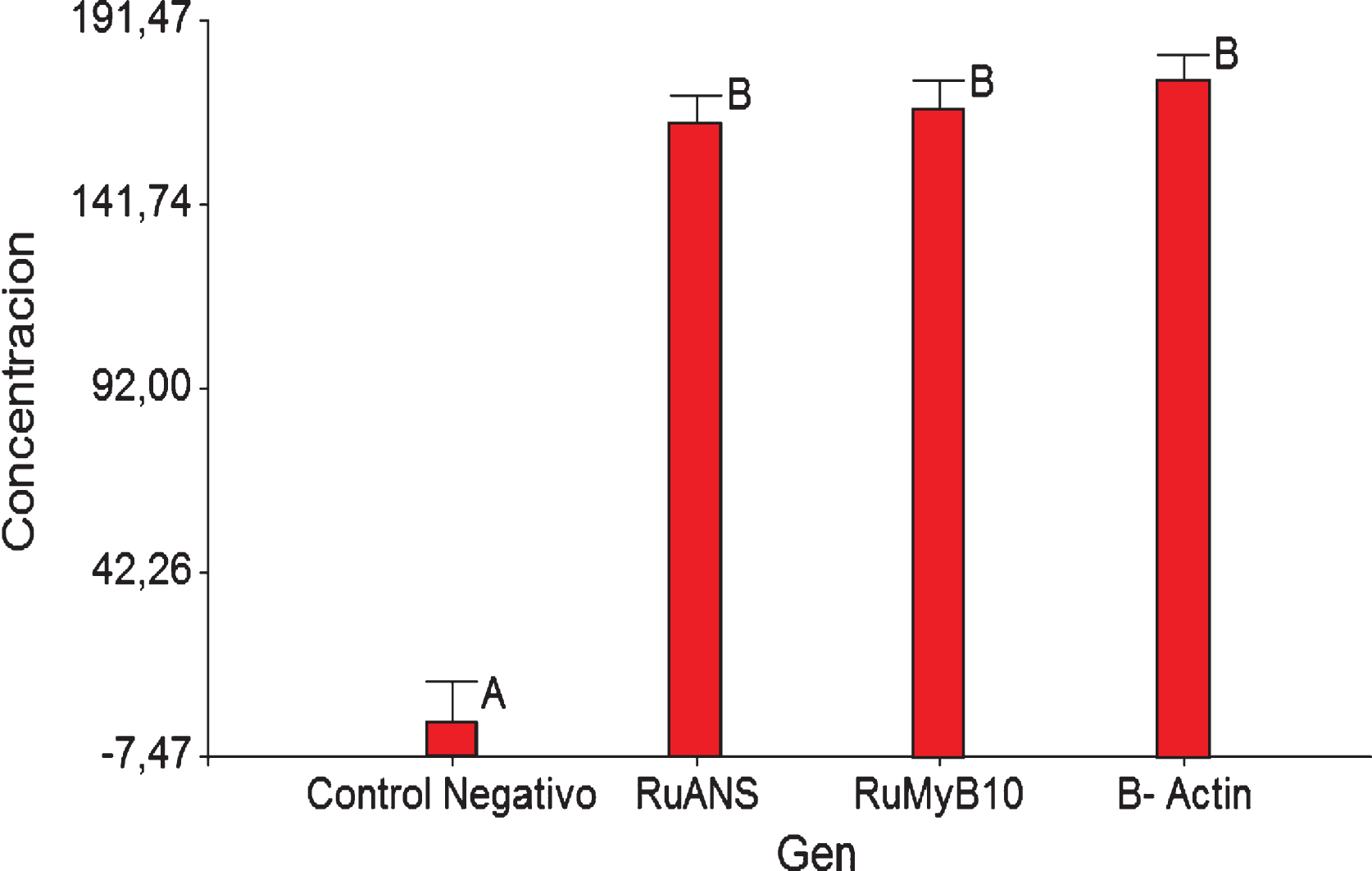
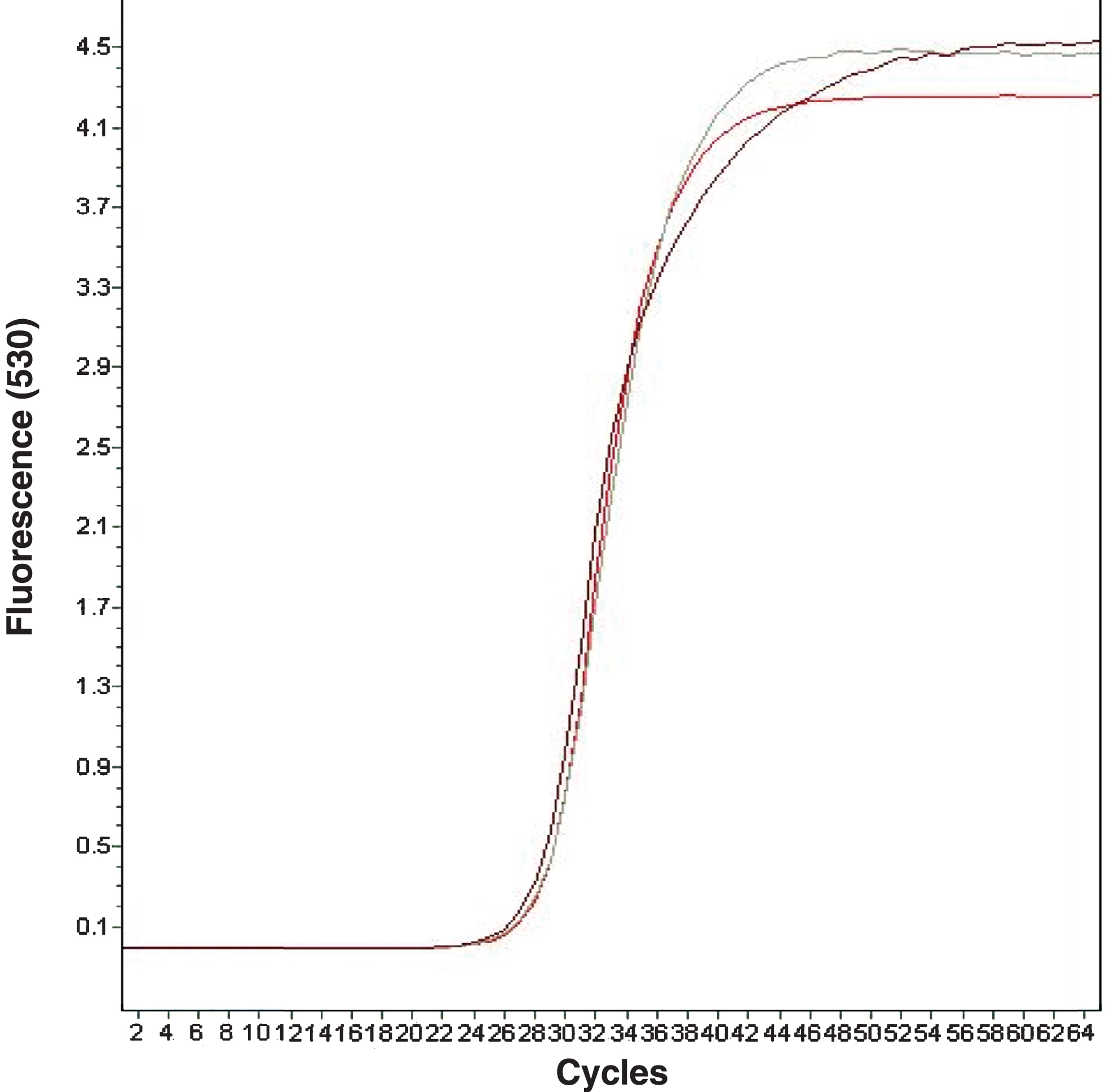
Figure 3 shows three of the curves obtained for the β-Actin, RuANS and RuMYB10 gene expression analysis, thus confirming that all three are similar, as they present a Cp close to around 28 points; concentration is always inversely proportional to Cp.
Fig.3
Amplification curves for β-Actin, RuANS and RuMYB10 genes: Fluorescence vs. cycles.
The average of the obtained concentrations is as follow: for the control gene 174,65 ng/mL, the gene expression presents with 167.43 ng/mL for RuMYB10 and 163,55 for the RuANS gene. The metabolic pathway leading to the formation of anthocyanins and various plant species such as Rubus sp, and others, is regulated at a level of expression of the gene encoding the enzyme anthocyanidin synthase (ANS).
RuMYB10 and RuANS were gradually up-regulated, reaching the maximum level when anthocyanin accumulated in large quantities, if it is at the highest level of maturity with its optimum harvest points. MYB transcription factors had been viewed as one of the most important regulators involving the flavonoid biosynthesis.
4Discussion and conclusions
The results obtained indicate that the expression of the β-Actin, RuANS and RuMYB10 genes, do not present statistically significant differences. Table 1 displays the distribution of the number of repetitions for each gene with the averages of their respective concentrations in ng/mL.
Table 1
β-Actin, RuANS and RuMYB10 gene expression concentration (ng/mL) in Rubus niveus species
| Concentration (ng/mL) | |||
| Gene | β- Actin | RuANS | RuMYB10 |
| Number | |||
| 1 | 180 | 93 | 189 |
| 2 | 211.6 | 193 | 165 |
| 3 | 141.8 | 143 | 142.6 |
| 4 | 179.98 | 211.8 | 219.2 |
| 5 | 173.9 | 167.8 | 176 |
| 6 | 148.3 | 151 | 149 |
| 7 | 166.2 | 172 | 222.4 |
| 8 | 137 | 174 | 162 |
| 9 | 162 | 156 | 180 |
| 10 | 168 | 138 | 177 |
| 11 | 194 | 162 | 160 |
| 12 | 147 | 184 | 197.6 |
| 13 | 136 | 208 | 129 |
| 14 | 217 | 167.2 | 132.4 |
| 15 | 257 | 132.4 | 110.2 |
| Average | 174.65 | 163.55 | 167.43 |
It can, therefore, be concluded that the species present a similar expression level between all three genes analyzed and that it is a good source of anthocyanins.
The high levels of anthocyanins are present as a result of internal and external factors such as the ripening degree, humidity, temperature and light, which may be promoters or inhibitors of the transcription factors involved in anthocyanin biosynthesis [9, 14].
Real-time PCR for detection and quantification displayed higher sensitivity and specificity in the analysis of gene expression. The methodology employed during research demonstrated extreme swiftness due to the short time necessary to analyze all three genes. It is worth mentioning that real-time PCR is much more efficient than the conventional method.
We are pioneers in Ecuador for this type of research and analyses. With the experience gained from working with Rubus we continue our efforts into the understanding and protecting our own endangered endemic species. Our further aim is to understand genetic enhancement in plant species and our discoveries of the gene expression of Rubus niveus further shed light on the physiological and metabolic pathways in the development of fruits in the Valley region of Ecuador and beyond. Thus, assisting in developing technologies that can assist in future attempts to understand other pathways of biosynthesis.
Conflict of interest
None to report.
Acknowledgments
To Daniel Acurio for his valuable contribution to the data analysis and team that conducted this research.
References
[1] | Miyazawa T , Nakagawa K , Kudo M , Muraishi K , Someya K . Directintestinal absorption of red fruit anthocyanins,cyanidin-3-glucoside and cyanidin-3,5-diglucoside, into rats andhumans. Journal Agricultural and Food Chemistry. (1999) ;1083–1091. DOI: 10.1021/jf9809582 |
[2] | Del Carpio C , Serrano C , Giusti M . Caracterización de lasantocianinas de los frutos de Berberis boliviana Lechler. Revista de la Sociedad Química del Perú. Rev SocQuím Perú. (2009) ;75: (1):76–86. Lima ene./mar. |
[3] | Aguilera M , Reza M , Chew R , Meza J . Propiedades funcionales de lasantocianinas. Revista de Ciencias Biológicas y de la Salud. (2011) ;16–22. |
[4] | Zhang W , Franco C , Curtin C , Conn S . To stretch the boundary ofsecondary metabolite production in plant cell-based bioprocessing:Anthocyanin as a case study. Journal of Biomedicine andBiotechnology. (2004) ;264–71. DOI: 10.1155/S1110724304404148 |
[5] | Quinton J , Fay MF , Ingrouille M , Faull J . Characterization ofRubus niveus: A prerequisite to its biological control inoceanic islands. Biocontrol Science and Technology. (2011) ;21: :733–52. DOI: 10.1080/09583157.2011.570429 |
[6] | Martínez N , Arévalo K , Verde M , Rivas C , Oranday A , Núñez M , Morales M . Antocianinas y actividad antiradicales libres de Rubus adenotrichus Schltdl (zarzamora). Revista Mexicana de Ciencias Farmacéuticas. (2011) 66–71. |
[7] | Morton J . Fruits of warm climates. Mysore Raspberry. (1987) 109–10. |
[8] | Deighton N , Brennan R , Finn C , Davies H . Antioxidant properties ofdomesticated and wild Rubus species. Journal of the Scienceof Food and Agriculture. (2000) ;1307–13. DOI: 10.1002/1097-0010(200007)80:9<1307::AID-JSFA638>3.0.CO;2-P |
[9] | Chen Q , Yu H , Tang H , Wang X . Identification and expressionanalysis of genes involved in anthocyanin and proanthocyanidinbiosynthesis in the fruit of blackberry. Scientia Horticulturae. (2012) ;61–8. DOI: 10.1016/j.scienta.2012.04.025 |
[10] | Tamara LM , Vallejo I . Perfil de Mora. Centro de Información e Inteligencia Comercial – CICO de CORPEI. (2009) ;1–23. |
[11] | Sethi M . qPCR y RT - qPCR con garantia de éxito. Farmespaña Industrial. (2011) ;63–5. |
[12] | Garzón R , Riesgo L , Orfa M . Bioquímica I. Lapregunta como base de la inquietud científica. Bogotá–Colombia: Centro Editorial Rosarista, (2006) . |
[13] | Yánez P . Biometría y Bioestadística: Analizandola Estructura Numérica de la Información en ProyectosBiológicos y Ecológicos. Quito - Ecuador: UniversidadPolitécnica Salesiana. (2007) . |
[14] | Lin-Wang K , Bolitho K , Grafton K , Kortstee A , Karunairetnam S , McGhie T , Espley R , Hellens R , Allan A . An R2R3 MYB transcriptionfactor associated with regulation of the anthocyanin biosyntheticpathway in Rosaceae. BMC Plant Biology. (2010) ;10–50. DOI: 10.1186/1471-2229-10-50 |




