Research data management at Elsevier: Supporting networks of data and workflows
Abstract
Sharing research data has the potential to make research more reproducible and efficient. Scientific research is a complex process and it is crucial that at the different stages of this process, researchers handle data in a way that will allow sharing and reuse. In this paper, we present a framework for the different steps involved in managing research data: a hierarchy of research data needs, and describe some of our own ongoing efforts to support these needs.
Creating a good data ecosystem that supports each of these data needs requires collaboration between all parties that are involved in the generation, storage, retrieval and use of data: researchers, librarians, institutions, government offices, funders, and also publishers. We are actively collaborating with many other participants in the research data field, to develop a data ecosystem that enables data to be more useful, and reusable, throughout science and the humanities.
1.Introduction
With the increased digitization of research, as well as the increased possibilities to store and preserve research data, awareness of the importance of research data preservation, storage and sharing has grown: a key goal has been to preserve data to enable reuse [17]. The concept of Research Object has been coined to describe complex objects that combine data, software, and workflow components [4], and discussions on the best way to coordinate efforts for data and storage are carried out in domains as varied as psychiatry [1], geography [26], hydrology [25], asthma studies [7], and condensed matter physics [28].
Funding bodies are also actively taking steps to encourage data sharing. Under Horizon 2020, the European Commission has launched an ‘Open Access to Data Pilot’ [18] where in several core areas researchers are asked to share data, unless they have a reason to opt out. Similarly, the National Institutes of Health has announced that ‘NIH intends to make public access to digital scientific data the standard for all NIH-funded research’ [19]. At a national level, there are increasingly requirements that researchers submit data management plans when applying for grants to enable data sharing and where possible, reuse [10,21,22].
A survey carried out by the Publishing Research Consortium in 2010 [23] showed that even though researchers are aware that it is important to have access to research data, they do not find it easy to access data. Some reasons for this are that data storage opportunities are very fragmented, and that there are not always clear data management practices in place. Given how much time researchers spent designing, modifying, and recording results, it is crucial that they do this in a way that will allow reuse and sharing of data to ensure maximum yields. To make this investment it will be necessary to put reward systems in place for researchers who take the time and effort to make their data sharable and reusable [29].
2.A hierarchy of research data needs
The main goal of data sharing is that other researchers will be able to reuse the shared data. It is therefore important that reusability is constantly taken into account when designing systems that store and create data. All parties interested and involved in handling research data should care about how research data gets stored in a way that makes it optimally usable downstream. Following up on earlier work, e.g. [8,12,24]. We propose that a better alignment of the nine aspects listed in Fig. 1 supports optimal data reuse pyramid can function as a roadmap for the development of data management better processes and systems throughout the data lifecycle.
Fig. 1.
A “pyramid” of requirements for reusable data, indicating that in order to be reused, it needs to be saved, shared, and trusted. For details on each of the ‘layers’ of the Pyramid, see the corresponding section of the paper.
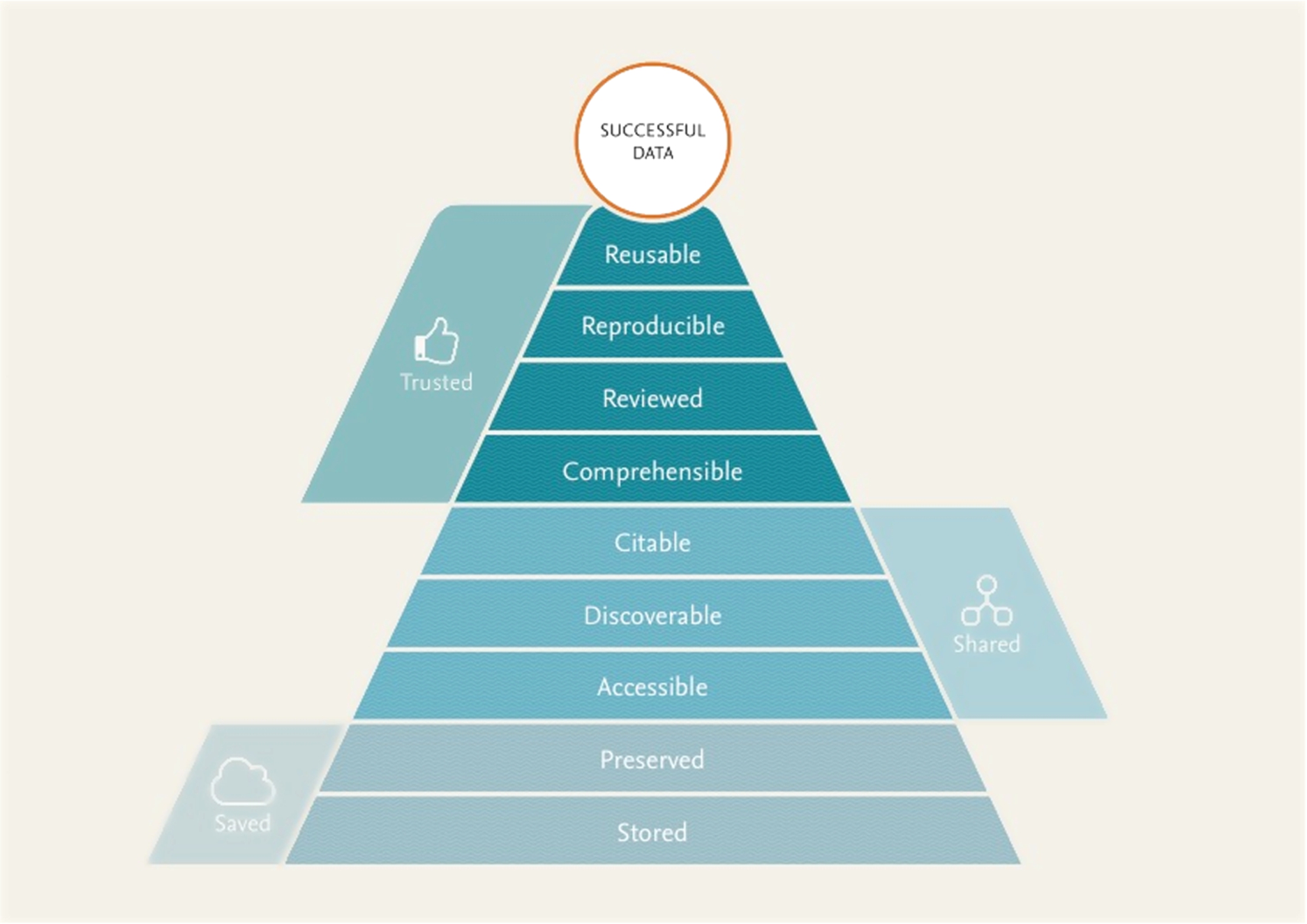
There is an intended hierarchy to these aspects, akin to the Maslow hierarchy of human needs [16]: each builds on, adds value to, and in many cases requires the aspects preceding them. We strongly support the vision set forth in earlier work, such as the ODE report the integration of data and publications [9]: in particular, in Table 2 of that paper, a very similar list of issues and opportunities for publishers is mentioned. Similarly, the FAIR Principles (which Elsevier contributed to) are to make data Findable, Accessible, Interoperable and Reproducible, and describe technical and practical steps to make this happen [33].
2.1.Data should be stored
First of all it is imperative that data not be thrown away. Data management plans aim to ensure that research groups define the ways to store their datasets in advance of their experiments. Use or simple programs like Word or Excel will allow researchers to store information about their experiment. Additionally, new technologies such as electronic lab notebooks present a viable option for storing the observations and results of experiments.
As an example of efforts to encourage electronic data storage, we worked with a neuroscience lab at Carnegie Mellon University to develop a tailored Electronic Lab Notebook to allow metadata to be stored during data creation [30]. Similarly, the Hivebench Electronic Lab notebook (http://hivebench.com) was developed with the goal of sharing protocols in life science settings (note: Hivebench was acquired by Elsevier on June 1, 2016).
2.2.Data should be preserved
Information can only be valuable when it is in a format that we can use in the long term. Even if there are no measures in place to ensure that data were properly preserved, data can in some cases still be rescued.
Elsevier collaborated on a number of different projects to ‘rescue’ data sets and save them from oblivion. In a collaboration with Carnegie Mellon University, we helped pilot the Olive Executable Archive project [27] to bring a number of arcane operating systems back to life as Virtual Machines. The user can load these locally and view the software and data created within the old systems, ensuring that these historical collections which were created in an obsolete operating system can still be accessed. As a further example of an effort in data preservation, we worked together with the Lamont–Doherty Research Center at Columbia University in New York to run the International Data Rescue Award in the Geosciences, an initiative that encourages and rewards efforts to preserve data that were not initially saved for the long [14]. Organizations such as the Data Archiving and Networked Services, DANS (http://www.dans.knaw.nl/en) in the Netherlands, provide an infrastructure for data preservation. Mendeley Data (http://data.mendeley.com) has teamed up with DANS to ensure long-term preservation of their Datasets (https://blog.mendeley.com/2015/11/09/put-your-research-data-online-with-mendeley-data/).
2.3.Data should be accessible
If data is stored and preserved, this does not necessarily mean that it is automatically accessible. Both researchers and machines may want to access the data, for example for meta-analyses or other kinds of re-use. Researchers are increasingly being required by their institution or funder to make their data accessible, which has caused researchers to start thinking about solutions.
In Mendeley Data (http://data.mendeley.com) researchers create private data sharing spaces which can be opened to larger communities, or opened up to the wider public. Also, we recently launched an open data pilot (http://www.elsevier.com/about/open-science/research-data/open-data) to make raw research data (as submitted with an article) openly-accessible alongside the article for any web user.
2.4.Data should be discoverable
Elsevier and other publishers support various mechanisms to support the discoverability of datasets, for instance through inclusion of data DOI’s that link to associated data in public databases (an overview is given in http://www.elsevier.com/databaselinking). Recent funding proposals encourage the development of data discoverability. The first of the data Principles drawn up by the FAIR Data group are to make data Findable (as well as, Accessible, Interoperable and Reproducible – see Ref. [33]). Initiatives such as the National Data Service and the Data Discovery Index (http://grants.nih.gov/grants/guide/rfa-files/RFA-HL-14-031.html) aim to provide a data discovery layer over disparate data collections.
In a project co-funded by a National Science Foundation EAGER Grant, Elsevier collaborated with the Carnegie Mellon School of Computer Science to develop superior ways by which to access and query tabular content extracted from articles and imported from research databases (see http://boston.lti.cs.cmu.edu/eager/deusre/ for an overview).
Working within the Research Data Alliance (RDA) Publishing Data Services Working Group (https://rd-alliance.org/groups/rdawds-publishing-data-services-wg.html) we worked to bring together different stakeholders to agree on common standards, combine links from disparate sources, and create the Data-Literature Interlinking Service, a universal, open service for collecting and sharing such links. A prototype of this service is now available (http://dliservice.research-infrastructures.eu/) that demonstrates the population of and access to a graph of dataset–literature links collected from a variety of major data centers, publishers, and research organizations. Next to this prototype, the Working Group has developed ‘Scholix’ (http://www.scholix.org/) see also [5,6], – a high-level interoperability framework for exchanging information about the links between scholarly literature and data. It aims to build an open information ecosystem to understand systematically what data underpins literature and what literature references data, and developed a set of guidelines for these practices, which are aimed to be developed and implemented in a subsequent RDA working group.
2.5.Data should be citable
If datasets are to be promoted to be first-class knowledge assets (and, for example, be counted for researcher’s assessments), better methods of citation of datasets are needed.
FORCE11 has developed a set of principles [15] to describe how data should be cited. Elsevier has been one of the first publishers to implement these FORCE11 principles. In several data repositories datasets now get their own DOIs which can be used to cite the dataset.
2.6.Data should be comprehensible
To enable data reuse it is essential that user understands what units of measurements were used, how the data was collected, and what abbreviations and parameters were used. One way to make sure that the future user understands how a dataset was created is by publishing an extensive description of how the data was collected, analyzed, and otherwise manipulated One way of delivering this description is by writing a so-called ‘data article,’ describing exactly that.
Several publishers now publish specific data journals. Elsevier has developed a new article type, called ‘Research Elements,’ which contain data and software articles. Research Elements can be a part of an existing journal, but we have also started several Open Access journals specifically devoted to data papers. One of these is ‘Data in Brief’ (http://www.journals.elsevier.com/data-in-brief) which allows the author(s) to provide a thorough description of their datasets. For data already published within the article, we have developed a suite of tools to improve data comprehension, such as in-article data visualizations, like interactive plots (http://www.elsevier.com/books-and-journals/content-innovation/iplots). These interactive content elements allow readers to manipulate datasets by for example, hovering over a plot to see the value of a data point or by switching from a graphical to a tabular view to inspect the data in greater detail (for other examples see https://www.elsevier.com/books-and-journals/content-innovation).
2.7.Data should be reviewed
To be trusted (a necessary step towards being reusable) it is important that research data is viewed, at least, and ideally, reviewed. This review can take different forms: a data set might be manually checked for complying with domain standards or having the proper metadata appended to it; and it might be checked automatically to make sure it renders to a standard format or methodology. In other cases, the data might be validated for having a proper description attached as metadata – with which the data can be fully understood and re-used.
In the Open Data Pilot (http://www.elsevier.com/about/open-science/research-data/open-data), reviewers are asked to check that the submitted files are raw data that can be parsed and are commonly used within the relevant domain; for the data journals described in the previous section, data are more thoroughly checked by reviewers of the paper.
2.8.Data should be reproducible
It has been argued that there is a ‘reproducibility crisis’ in research, as many domains find that experimental results are difficult or impossible to reproduce [11,20]. Irreproducibility often originates from missing elements to research data, which are needed in order to achieve the same research results. As an example of this, there is often inadequate reporting of key elements of a dataset or research method. For instance, Vasilevsky et al. found that for example antibodies, model organisms, and other data resources reported in the biomedical literature often lack sufficient detail to enable reproducibility or reuse [32].
We have contributed to, and are actively implementing the guidelines established by the Force11 Resource Identification Initiative, which aims to enable resource identification within the biomedical literature through a pilot study promoting the use of unique Research Resource Identifiers (RRIDs) [3]. By active participation in the Reproducibility Initiative (http://validation.scienceexchange.com/#/), we support efforts to validate key experimental results via independent replication. The group has obtained funding to actually reproduce a series of key experiments and report on the statistical significance of a result when it combines its data with that of the original experiments [31]. Another way in which we explored reproducibility is through the Executable Paper Challenge: an open challenge for projects that showed the execution of a piece of software inside a paper [13].
2.9.Data should be reusable
Only if research data is comprehensible, trusted, and reproducible will other researchers consider reusing it. But one further factor that inhibits reuse is uncertainty of the legal or copyright status of the dataset. Ideally, the legal status of a dataset should be identified at the moment of storage or sharing.
In line with recommendations by, for example, the UK’s Digital Curation Centre [2], we therefore ask all researchers using Mendeley Data (https://data.mendeley.com/) to select a Creative Commons license (https://creativecommons.org/licenses/by/4.0/) under which their data is made available. Similarly, our data and software journals allow for selection of a data license during submission (see for example, https://www.elsevier.com/journals/data-in-brief/2352-3409/open-access-journal).
3.Discussion
Creating an efficient and effective ecosystem for data requires collaboration between all parties that are involved in the creation, storage, retrieval, and use of this data: researchers, institutions, government offices and funders, as well as publishers and software developers. Cross-stakeholder groups bringing all of these parties together are essential to setting the pace of change towards better sharing of data and methods, more transparency, and a more effective way of scholarly communication. Through our active participation in panels, at conferences, and through working groups for, among others, the Research Data Alliance, ICSU World Data System, CASRAI, JISC and Force11, we aim to make significant contributions to increasing the transparency and accountability of data to accelerate scientific discovery and scholarly conduct. We hope that the hierarchy presented in this paper may help provide a framework to support these conversations, moving forward.
About the author
Anita de Waard has a background in physics and joined Elsevier in 1988. Her goal is to improve science by improving scholarly communication. She performs research, collaborates with academic groups, and cofounded Force11.org. In her current remit, she is developing cross-disciplinary frameworks to store, share, and search experimental research outputs.
References
[1] | Psychiatric Genomics Consortium, A framework for interpreting genome-wide association studies of psychiatric disorders, Molecular Psychiatry 14: (1) ((2009) ), 10–17. doi:10.1038/mp.2008.126. |
[2] | A. Ball, How to license research data, http://www.dcc.ac.uk/resources/how-guides/license-research-data#fn3x0. |
[3] | A. Bandrowski, M. Brush, J.S. Grethe, M.A. Haendel, D.N. Kennedy, S. Hill, P.R. Hof, M.E. Martone, M. Pols, S.C. Tan, N. Washington, E. Zudilova-Seinstra, N. Vasilevsky and RINL Resource Identification Initiative, The Resource Identification Initiative: A cultural shift in publishing, Brain and Behavior 6: (1) ((2016) ), e00417. doi:10.1002/brb3.417. |
[4] | S. Bechhofer, I. Buchan, D. De Roure, P. Missier, J. Ainsworth, J. Bhagat, P. Couch, D. Cruickshank, M. Delderfield, I. Dunlop, M. Gamble, D. Michaelides, S. Owen, D. Newman, S. Sufi and C. Goble, Why linked data is not enough for scientists, Future Generation Computer Systems 29: (2) ((2013) ), 599–611, ISSN 0167-739X. doi:10.1016/j.future.2011.08.004. |
[5] | A. Burton, H. Koers, P. Manghi, S. La Bruzzo, A. Aryani, M. Diepenbroek and U. Schindler, Metadata and Semantics Research, Springer, Dordrecht, (2015) , pp. 324–335. |
[6] | H. Cousijn, W. Haak and H. Koers, Finding better ways to connect research data with scientific literature: The Scholix initiative is building an interoperability framework that will make it easier to share, exchange and aggregate data, https://www.elsevier.com/connect/finding-better-ways-to-connect-research-data-with-scientific-literature. |
[7] | A. Custovic, J. Ainsworth, H. Arshad, C. Bishop, I. Buchan, P. Cullinan, G. Devereux, J. Henderson, J. Holloway, G. Roberts, S. Turner, A. Woodcock and A. Simpson, The study team for early life asthma research (STELAR) consortium ‘Asthma e-lab’: Team science bringing data, methods and investigators together, Thorax 70: (8) ((2015) ), 799–801. doi:10.1136/thoraxjnl-2015-206781. |
[8] | A. De Waard, Ten habits of highly effective data, discovery informatics, in: Papers from the AAAI-14 Workshop, online at https://www.aaai.org/ocs/index.php/WS/AAAIW14/paper/viewFile/8846/8346. |
[9] | A. De Waard, H. Cousijn and I.J. Aalbersberg, 10 aspects of highly effective research data: Good research data management makes data reusable, Elsevier Connect, posted on 11 December 2015. |
[10] | European Commission, Guidelines on Open Access to scientific publications and research data in horizon 2020, http://ec.europa.eu/research/participants/data/ref/h2020/grants_manual/hi/oa_pilot/h2020-hi-oa-pilot-guide_en.pdf. |
[11] | J. Hall, 1,500 scientists speak out on science’s reproducibility crisis, ExtremeTech Blog, June 3, 2016, http://www.extremetech.com/extreme/229519-scientists-speak-out-on-reproducibility-in-science. |
[12] | H. Koers, How do we make it easy and rewarding for researchers to share their data? A publisher’s perspective, Journal of Clinical Epidemiology 70: ((2016) ), 261–263. ISSN 0895-4356. doi:10.1016/j.jclinepi.2015.06.016. |
[13] | H. Koers, A. Gabriel and R. Capone, Executable papers in computer science go live on ScienceDirect, Elsevier Connect, 2013, available at: http://www.elsevier.com/connect/executable-papers-in-computer-science-go-live-on-sciencedirect. |
[14] | D. Lovegrove and K. Lehnert, The 2015 International Data Rescue Award in the Geosciences, Elsevier, http://www.journals.elsevier.com/marine-geology/news/british-macrofossils-online-wins-data-rescue-award/. |
[15] | M. Martone, Data Citation Synthesis Group: Joint Declaration of Data Citation Principles, Force11, 2014, https://www.force11.org/datacitation. |
[16] | A.H. Maslow, A theory of human motivation, Psychological Review 50: (4) ((1943) ), 370–396. doi:10.1037/h0054346. |
[17] | J.P. Mesirov, Accessible reproducible research, Science 327: (5964) ((2010) ), 415–416. doi:10.1126/science.1179653. |
[18] | T. Narock, R. Arko, S. Carbotte, A. Krisnadhi, P. Hitzler, M. Cheatham, A. Shepherd, C. Chandler, L. Raymond, P. Wiebe and T. Finin, The OceanLink project, in: Proceedings – 2014 IEEE International Conference on Big Data, IEEE Big Data 2014, (2015) , article number 7004347, pp. 14–21. |
[19] | National Institutes of Health Plan for Increasing Access to Scientific Publications and Digital Scientific Data from NIH Funded Scientific Research, February 2015, http://grants.nih.gov/grants/NIH-Public-Access-Plan.pdf. |
[20] | Nature Special Issue, Challenges in Irreproducible Research, Nature Special Issue, October 7, 2014, http://www.nature.com/news/reproducibility-1.17552. |
[21] | NWO, Start pilot data management, Netherlands Organisation for Scientific Research, January 2015, http://www.nwo.nl/en/policies/open+science/data+management. |
[22] | Policy on data management and sharing, Wellcome Trust, 2010, http://www.wellcome.ac.uk/about-us/policy/policy-and-position-statements/wtx035043.htm. |
[23] | Publishing Research Consortium (PRC), Access vs. Importance: A global study assessing the importance of and ease of access to professional and academic information. Phase I Results, PRC, 2010, http://publishingresearchconsortium.com/index.php/prc-projects/research-reports. |
[24] | S. Reilly, W. Schallier, S. Schrimpf, E. Smit and M. Wilkinson, Report on Integration of Data and Publications. Opportunities for Data Exchange, ODE, 2011, http://www.stm-assoc.org/2011_12_5_ODE_Report_On_Integration_of_Data_and_Publications.pdf. |
[25] | J.M. Sadler, D.P. Ames and S.J. Livingston, Extending HydroShare to enable hydrologic time series data as social media, Journal of Hydroinformatics 18: (2) ((2016) ), 198–209. |
[26] | A.D. Singleton, S. Spielman and C. Brunsdon, Establishing a framework for Open Geographic Information Science, International Journal of Geographical Information Science 30: (8) ((2016) ), 1507–1521. doi:10.1080/13658816.2015.1137579. |
[27] | G. St. Clair and D. Ryan, Olive: A digital archive for executable content, Coalition for Networked Information, Washington, D.C., December 2011, http://works.bepress.com/gloriana_stclair/20. |
[28] | B. Stvilia, C.C. Hinnant, S. Wu, A. Worrall, D.J. Lee, K. Burnett, G. Burnett, M.M. Kazmer and P.F. Marty, Research project tasks, data, and perceptions of data quality in a condensed matter physics community, Journal of the Association for Information Science and Technology 66: (2) ((2015) ), 246–263. doi:10.1002/asi.23177. |
[29] | C. Tenopir, S. Allard, K. Douglass, A.U. Aydinoglu, L. Wu, E. Read, M. Manoff and M. Frame, Data sharing by scientists: Practices and perceptions, PLoS ONE 6: ((2011) ), e21101. doi:10.1371/journal.pone.0021101. |
[30] | S.J. Tripathy, J. Alder, S.D. Burton, M. Harviston, D. Marques, N.N. Urban and A. De Waard, The UrbanLegend Project: a system for cellular neurophysiology data management and exploration, Frontiers in Neuroinformatics, Conference Abstract: Neuroinformatics 2014. doi:10.3389/conf.fninf.2014.18.00077. |
[31] | R. Van Noorden, Sluggish data sharing hampers reproducibility effort, Nature News, 03 June 2015, http://www.nature.com/news/sluggish-data-sharing-hampers-reproducibility-effort-1.17694. |
[32] | N.A. Vasilevsky, M.H. Brush, H. Paddock, L. Ponting, S.J. Tripathy, G.M. Larocca et al., On the reproducibility of science: Unique identification of research resources in the biomedical literature, PeerJ 1: ((2013) ), e148. doi:10.7717/peerj.148. |
[33] | M.D. Wilkinson et al., The FAIR guiding principles for scientific data management and stewardship, Sci. Data 3: ((2016) ), 160018. doi:10.1038/sdata.2016.18. |



