Genomic Imprinting and the Regulation of Postnatal Neurogenesis
Abstract
Most genes required for mammalian development are expressed from both maternally and paternally inherited chromosomal homologues. However, there are a small number of genes known as “imprinted genes” that only express a single allele from one parent, which is repressed on the gene from the other parent. Imprinted genes are dependent on epigenetic mechanisms such as DNA methylation and post-translational modifications of the DNA-associated histone proteins to establish and maintain their parental identity. In the brain, multiple transcripts have been identified which show parental origin-specific expression biases. However, the mechanistic relationship with canonical imprinting is unknown. Recent studies on the postnatal neurogenic niches raise many intriguing questions concerning the role of genomic imprinting and gene dosage during postnatal neurogenesis, including how imprinted genes operate in concert with signalling cues to contribute to newborn neurons’ formation during adulthood. Here we have gathered the current knowledge on the imprinting process in the neurogenic niches. We also review the phenotypes associated with genetic mutations at particular imprinted loci in order to consider the impact of imprinted genes in the maintenance and/or differentiation of the neural stem cell pool in vivo and during brain tumour formation.
NEURAL STEM CELLS AND THE NEUROGENESIS PROCESS
Neurogenesis is the process of generating new neurons from neural stem cells (NSCs) and their progenitors. NSCs are characterized by the ability to self-renew and by multipotency, enabling the formation of astrocytes, oligodendrocytes and neurons upon differentiation [1]. In the embryonic mammalian brain, radial glia are the primary precursors of new neurons and astrocytes [2, 3]. These cells reside in the ventricular zone (VZ) but possess long processes that penetrate the brain parenchyma and contact the pial surface of the brain (Fig. 1a). The early postnatal VZ is largely composed of radial glial cell bodies that remain proliferative, display interkinetic nuclear migration and serve as progenitors of new neurons. In the adult lateral ventricles, a subpopulation of astrocytes derived from radial glia remain in contact with the ventricular lumen, into which they extend a single cilium similar to that found in neuroepithelial cells and radial cells (Fig. 1a). Taken together, current data suggest a continuum between embryonic and adult germinal brain centres[4, 5].
Fig.1
The radial glia nature of embryonic and adult neural stem cells: the role of imprinted genes. (a) There is a continuum of germinal activity that links neuroepithelial stem cells to radial glia and ultimately to the astrocytes that are stem cells in the adult brain. Radial glia in the neocortex produce several major brain cell classes, including neurons and astrocytes, via several rounds of proliferation and differentiation. The roles of the imprinted genes are indicated. Maternally and paternally expressed genes appear in pink and blue respectively. Imprinted genes that show biallelic expression are in black and bold. (b) Sagittal view showing the SVZ and the SGZ neurogenic niches in the adult mouse brain. In the SVZ, neuroblasts reach the olfactory bulb (OB) through the rostral migratory stream (rms). (c) Enlarged view of the SGZ: Type I stem cells (blue) show a radial single prolongation through the granular layer; type II precursors (purple) give rise to neuronal lineage-restricted progenitor type III cells (red) that differentiate into neurons, which in turn integrate into the granular layer (gr). (d) Enlarged view of the SVZ: type B1 stem cells (blue) contact the ventricle with a thin process extending between the ependymal cells (e; grey); transit-amplifying progenitors (TAP) or type C cells (purple) give rise to type A cells (red) and oligodendrocytes progenitors (yellow). Dividing stem cells and their TAP progeny are tightly apposed to blood vessels (bv); the choroid plexus-cerebrospinal fluid system (cp-CSF) is shown.
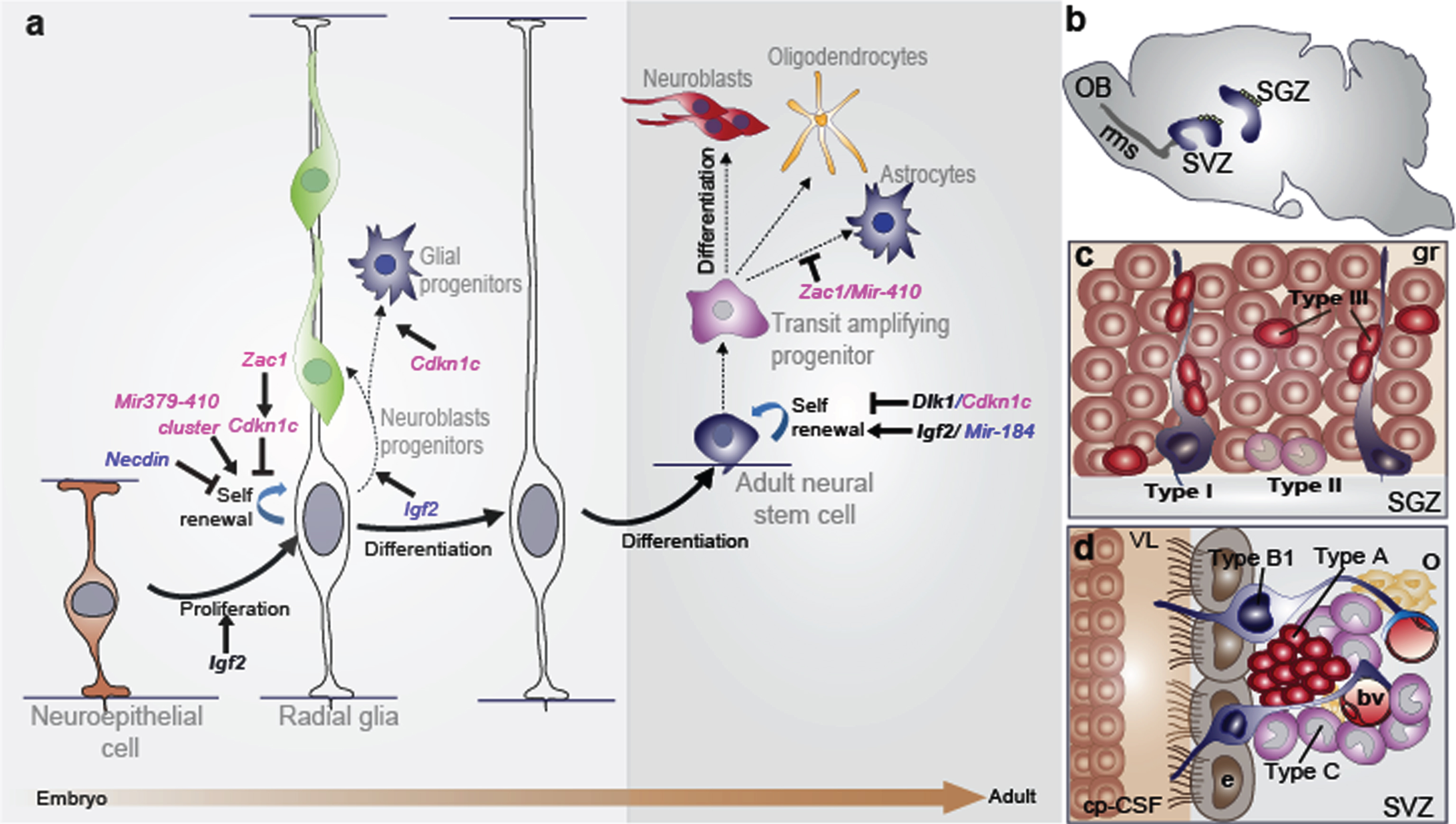
In the mammalian brain, two regions continue to generate new neurons throughout adulthood: the subventricular zone (SVZ) of the lateral ventricles’ wall and the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampus (Fig. 1b) [6, 7]. Adult NSCs originate from embryonic radial glia and thus express astrocytic markers such as the glial fibrillary acidic protein (GFAP) or the astrocyte-specific glutamate transporter known as excitatory amino acid transporters (GLAST) [8, 9]. Moreover, NSCs are relatively quiescent [10] and express the transcription factor Sox2 (SRY-related HMG box 2) and the neural progenitor markerNestin [11].
NSCs in the SVZ are called type B1 cells and give rise to rapid transit amplifying intermediate progenitors or type C cells, which in turn give rise to immature neurons called type A cells or neuroblasts (Fig. 1d). These latter cells migrate obliquely to the olfactory bulb (OB) via the rostral migratory stream (rms), where they integrate and mature (Fig. 1b) to generate different kinds of inhibitory interneurons that play an important role in rodent olfaction [12, 13]. Less frequently, NSCs also support oligodendrogenesis in this region. They are capable of producing oligodendroblasts that migrate to the corpus callosum and striatum, where they differentiate into myelinating and nonmyelinating oligodendrocytes [14]. However, in humans the functional role of the SVZ neurogenesis is not fully understood. In the SGZ, stem cells are located between the granular layer and the hilus of the DG, next to astrocytes that regulate their proliferation and differentiation. SGZ NSCs (type I cells) exhibit a radial process spanning the granule cell layer and give rise to type II cells, which are positive for the progenitor marker T-box gene-2 (Tbr2). Type II cells generate type III cells, which are neuronal precursors (Fig. 1c) and express markers of immature migrating neurons such as doublecortin (DCX) and the polysialylated-neural cell adhesion molecule (PSA-NCAM), which differentiate and integrate in the neuronal circuit in the CA3 region of the hippocampus [15]. A low percentage of SGZ stem cells also produce oligodendrocytes and astrocytes that migrate into the hilus and the molecular layer [16]. Integration of newborn neurons into the existing hippocampal neural circuitry plays a crucial role in learning and memory, as well as contributing significantly to hippocampal plasticity throughout the life span [6].
Given the absence of unique, decisive markers to identify the neural stem cell population in vivo, the analysis of NSCs has been widely based on the ex vivo behaviour of cells isolated from neurogenic niches. Individual cells dissected from either the postnatal SVZ or SGZ can proliferate in a medium containing basic fibroblast growth factor (FGF2) and/or epidermal growth factor (EGF) to produce multipotent clonal aggregates called “neurospheres” [17, 18]. In proliferating cultures, a NSC must produce a neurosphere that includes cells able to generate new aggregates upon clonal passage (self-renewal). The stem cell forming the initial clone undergoes several symmetrical self-renewing divisions before changing to an asymmetric division. The frequency of long-lived NSCs dissociated from the SVZ/SGZ is often estimated as the number of cells capable of generating such primary spheres. This property allows for the culture’s net expansion. Neurospheres can also be induced to differentiate in a culture so as to generate the three major cell types of the central nervous system (CNS): βIII-tubulin+ neurons, GFAP+ astrocytes and O4+ oligodendrocytes [17, 18]. There is still some controversy about the real functional relationship between neurosphere-forming cells and the stem cell population in vivo [19]. Nevertheless, this system represents a tractable model to investigate the contribution to cell fate specification made by signalling pathways, expression or epigenetic mechanisms, potentially applicable to neural stem cells for restorative neurogenesis in disease or trauma.
EPIGENETIC REGULATION AND MAMMALIAN GENOMIC IMPRINTING
Epigenetic mechanisms are heritable modifications to DNA and chromatin that do not involve changes in the DNA sequence itself but which can modulate gene expression and genome function [20, 21]. At the molecular level, epigenetic regulation involves chemical modifications to DNA such as methylation and hydroxymethylation, and to the histone proteins around which the DNA is wrapped, including acetylation, methylation, phosphorylation, and ubiquitylation and noncoding RNA (ncRNAs) action. It is becoming apparent that epigenetic modifications to developmental genes are very important cell-intrinsic programs that can interact with transcription factors and environmental cues to modulate cell maintenance and differentiation [22]. Indeed, epigenetic mechanisms seem to provide means for coordinately activating and repressing arrays of genes at specific steps in a differentiation pathway [23, 24].
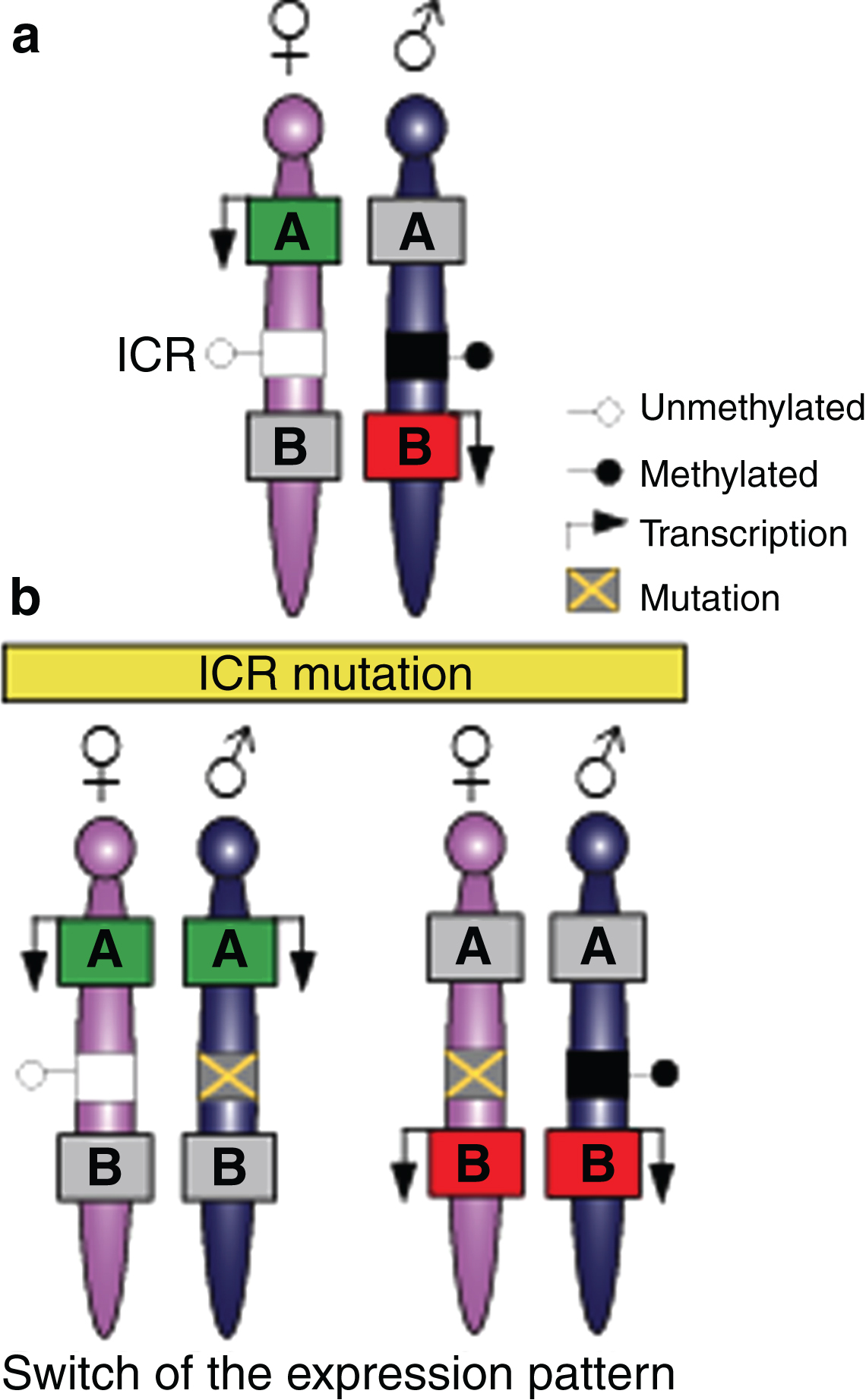
During mammalian development, the vast majority of genes are expressed or repressed from both alleles. However, there are a small number of genes known as imprinted genes that are expressed monoallelically from either the maternally or paternally inherited chromosome [25]. Approximately 150 imprinted genes have been described in mammals (for a complete list, see http://www.mousebook.org/mousebook-catalogs/imprinting-resource) and are generally organised in clusters, although examples of singleton imprinted genes do exist [26, 27]. These clusters typically contain at least one ncRNA and both maternally and paternally expressed imprinted genes. An imprinting cluster is usually under the control of a DNA element called the imprinting control region (ICR), which consists of a differentially DNA-methylated region (DMR) on the two parental chromosomes (Fig. 2a). Thus, ICRs can be divided into those which are methylated on the paternally-inherited copy and those with maternally-inherited methylation. Imprints are usually established in the germline [28] and the deletion of a germline-derived ICR results in a loss of imprinting of multiple genes in the cluster [25] (Fig. 2b).
Fig.2
Imprinted genes are located in clusters that are under the control of the imprinting control region (ICR). (a) Mammalian somatic cells contain a maternally-inherited chromosome (pink) and a paternally-inherited chromosome (blue). Imprinted genes are located in clusters and can be expressed in the maternally-inherited chromosome and repressed from the paternally-inherited chromosome (green box A), or expressed in the paternally-inherited chromosomes and repressed from the maternally inherited chromosome (red box B). Imprinting in the cluster is regulated by a DNA element called the imprinting control region (ICR) that has differentially-methylated regions (DMRs) on the two parental chromosomes. (b) Mutations in the ICR can modify imprinting, thus affecting several genes in the cluster and resulting in a switch in the expression patterns with the paternally-inherited chromosome, thereby acquiring an epigenotype typical in the maternally-inherited chromosome, or vice versa.
There are also somatic or secondary DMRs that acquire their DMR status after fertilization. Somatic DMRs directly depend on the presence of a germline DMR [28]. Mapping experiments and germline DMR deletion experiments in mice demonstrate that the primary ICRs are essential to establish monoallelic expression and the secondary somatic DMRs play important roles in maintaining imprints [29–31].
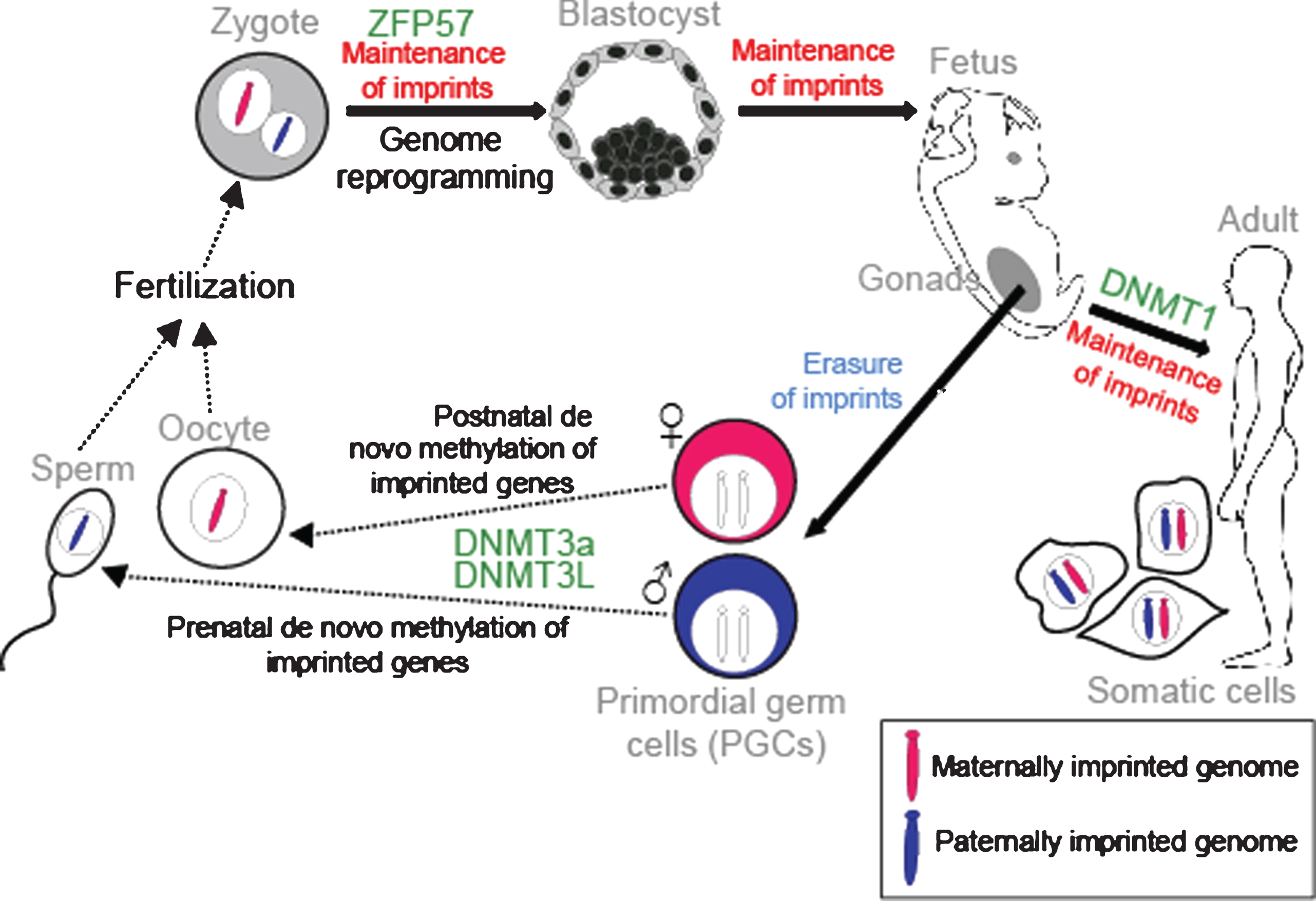
It is well established that these parental-specific marks are assigned in the germline. At this time, both genomes are in distinct compartments and the modifications can be performed according to the sex of the transmitting gametes. During embryogenesis, the primordial germ cells (PGCs), which will give rise to the gametes, have the methylation patterns characteristic of somatic cells. However, in the genital ridges the imprints are erased during gamete formation to allow for re-establishment of new parental-specific marks (Fig. 3). After demethylation and differentiation of the PGCs, methylation is established at the ICRs in an allele-specific manner depending on whether the developing gamete is in the male or female germline. This occurs by a de novo methylation process catalysed by the DNMT3a DNA methyltransferase. DNMT3L is a regulatory co-factor of DNMT3a and is also required [32]. Maternal germline DMRs are found in gene promoters, whereas paternal germline DMRs are found in intergenic regions [28]. In male and female germ cells, DNA re-methylation occurs at different times of development. Paternal-specific methylation occurs prenatally in prospermatogonia before meiosis in the germline [33]. In contrast, maternal-specific ICR methylation takes place postnatally in growing oocytes [34]. We now know that many differentially-methylated regions are established in the germline. However, unlike imprints, they are not sustained after fertilisation [35]. Following fertilisation, a rapid, extensive reprogramming of the parentally-inherited genomes occurs and most DNA methylation is lost. However, the parental-specific imprint must be maintained during this period of dynamic epigenetic change and a memory of parental origin is propagated into daughter cells during somatic cell division (Fig. 3). Critical factors such as the Kruppel-like zinc finger protein ZFP57 protect imprints during the post-fertilisation epigenetic reprogramming period [36]. Somatic heritability is conferred by the DNMT1 DNA methyltransferase. DNMT1 acts with a maintenance function that recognizes newly replicated hemi-methylated DNA and places methylation on the newly replicated strand [37]. Indeed, DNMT1 and ZFP57 are repressed in PGCs, allowing new imprints to be established during gamete formation [38].
Fig.3
Establishment and maintenance of imprints during development. DNA methylation is erased in PGCs in the genital ridge. However, imprints are maintained in somatic cells throughout the organism’s lifetime. Imprints are acquired in a sex-specific manner in the germline: maternally and paternally-methylated ICRs gain DNA methylation in oocytes and sperm respectively for transmission to the next generation. Following fertilisation, the parental-specific imprints are maintained in the developing organism despite genome-wide reprogramming elsewhere. ZFP57 protects imprints during the post-fertilization epigenetic reprogramming period. DNMT3a and DNMT3L catalyse the de novo methylation process and DNMT1 participates in maintaining imprints in somatic tissues.
GENOMIC IMPRINTING AND THE MAINTENANCE OF NEURAL STEM CELLS
Recent evidence suggests that the genomic imprinting process can be selectively lost or “switched off” in particular cell types or at specific developmental points in time to activate an allele that is usually repressed by imprinting [39, 40]. These heritable changes might be reversible and context-dependent, have an impact on stem cell plasticity and are likely to be essential for normal development and tissue regeneration [41]. Selective regulation of imprinting is probably a normal mechanism for modulating gene dosage to control stem cell potential in brain development and within the neurogenic niches throughout development and adult life [39, 42].
Genomic imprinting in early development
During cortical neurogenesis, radial glia highly express the paternally-expressed zinc finger protein Zac1 [43]. Elevated in vivo levels of ZAC1 promote cell cycle exit, disrupt neurogenesis and provoke a partial switch from neural to mesodermal/myogenic lineage with additional expression of pluripotency genes [43]. Moreover, overexpressing Zac1 induces the expression of other imprinted genes such as the maternally-expressed cyclin-dependent kinase inhibitor Cdkn1c, known to promote NSC cell cycle arrest and proglial differentiation [43–46]. Recent evidence suggests that some ncRNAs regulate imprinting by fine-tuning the gene expression of the protein-coding genes to maintain their dosage in cells [47]. For example, miR134, an imprinted miRNA belonging to the miR379–410 cluster, promotes cell proliferation and reduces cell migration in vivo [48]. Additionally, miR369-3p, miR496 and miR543, also belonging to the miR379–410 cluster, bind directly to the 3′UTR of the Ncad transcript and fine-tune the expression level of N-cadherin in VZ progenitors to control neuronal differentiation and migration [49]. Furthermore, miR369-3p has been shown to regulate the expression of Adam10 and TrappC8 during development of the neocortex [49]. Finally, the proliferative capacity of cortical progenitors in the embryonic mouse neocortex is repressed by paternal expression of Necdin, which is moderately expressed in the ventricular zone of the murine embryonic neocortex. Necdin suppresses neural progenitor proliferation by interacting with the polycomb protein Bmi1 [50]. These data show that imprinted genes regulate foetal neural stem cell activity in early neurodevelopment.
Imprinted genes and adult neurogenesis
In the adult brain, NSCs persistently generate new neurons in the SVZ and SGZ niches. In the SVZ, the paternally-expressed gene Delta-like homologue 1 (Dlk1), an atypical Notch ligand located on mouse chromosome 12 (human chromosome 14), plays an important dual function to regulate postnatal neurogenesis. Importantly, Dlk1 is a single gene that encodes for both a secreted factor (expressed by niche astrocytes) and a bound receptor (expressed by neural stem cells). Dlk1, which is a canonically imprinted gene elsewhere in the brain, shows a selective absence of imprinting in these cell types. Biallelic expression of Dlk1 is required for stem cell maintenance in the SVZ and final neurogenesis to the olfactory bulb [39]. This selective absence of Dlk1 imprinting in NSCs is associated with acquisition of DNA methylation in the germline-derived imprinting control region [39]. Notably, canonical imprinting of Dlk1 is maintained in foetal stem cells, indicating that imprinting relaxation occurs postnatally. The Dlk1 neighbouring gene, Gtl2, or the unlinked Snrpn located on chromosome 7C, keep their imprinting in the neurogenic zone, suggesting a selective gene-specific regulation of the absence of imprinting [39].
Insulin-like growth factor 2 (Igf2), which is canonically expressed from the paternally-inherited allele, is biallelically expressed in the choroid plexus and secreted to the cerebrospinal fluid to regulate NSC proliferation and to promote foetal cortical neurogenesis [51]. Igf2 is also biallelically expressed in the postnatal human and mouse choroid plexus epithelium and leptomeninges [52, 53]. As in the embryonic brain, biallelic expression in the adult choroid plexus and SVZ vasculature acts as a paracrine factor that regulates NSC homeostasis. In contrast, Igf2 is imprinted in the hippocampus and acts as an autocrine factor expressed in the SGZ neural stem cells solely from the paternal allele [40]. Hence, like Dlk1, selective absence of Igf2 imprinting is required for SVZ neurogenesis; however, in contrast to Dlk1, imprinted Igf2 is also required for SGZ neurogenesis. Furthermore, conditional mutagenesis of Igf2 in blood vessels has confirmed that endothelial-derived IGF2 contributes to NSC maintenance in the SVZ but not in the SGZ [40]. These data demonstrate that Igf2 is highly dosage-sensitive and that the distinct regulatory processes and mechanisms are unique to each neurogenic niche. In the SGZ, long-term deletion of the maternally expressed, cyclin-dependent kinase inhibitor (Cdkn1c) gene leads to NSC exhaustion and impaired neurogenesis in aged mice [54] showing that Cdkn1c also promotes NSCs quiescence.
Several micro RNAs have been described as having a role in adult NSCs. For example, the paternally-expressed miR-184, located at chromosome 9, promotes proliferation and inhibits differentiation of NSCs in the adult SGZ [55]. Mir-184 is directly repressed by the methyl-CpG binding protein 1 (MBD1) and it regulates the expression of Numblike by binding to the 3'-UTR of Numbl mRNA, affecting its translation [55]. The maternally expressed Mir-410, located at chromosome 12, is repressed by the neural inducer Noggin, causing the inhibition of neuronal and oligodendrocyte differentiation by promoting an astroglial fate [56].
Epigenetic regulation of imprinting in neurogenesis
The precise epigenetic modifications regulating the ICRs result in changes in the appearance and structure of DNA. DNA is methylated and then allele-specific histone modifications are recruited [57]. The enzymes that establish and maintain DNA methylation-specific patterns, including de novo DNA methyltransferases Dnmt3a and Dnmt3b and the maintenance Dnmt1, are highly expressed in postnatal NSCs and are required for neurogenesis and neuronal maturation [58, 59]. Furthermore, depletion of DNMT3b leads to deficient neuronal differentiation in vitro [60]. DNMT1 is highly expressed in the embryonic, perinatal and adult CNS in both dividing neural progenitors and mature neurons [58, 61, 62]. In support of this, mice lacking Dnmt1 specifically in neural precursor cells at E9–#x2013;#x2013;E10 exhibit deficits in neuronal function and die postnatally, suggesting a requirement for methylation in brain development [63]. A role for DNMT3a has also been demonstrated in postnatal neurogenesis [64], though the specific role of these molecules in the maintenance of imprinting marks in adult NSCs has not been well described. The N-terminal tails of histones are subject to post-translational modifications, including acetylation, methylation or phosphorylation; modifications that regulate the genomic accessibility and provide a scaffold to bind other factors to control the activation or repression of associated genes. Although several studies have implicated histone acetylation in regulating adult neurogenesis [65, 66], the role of histone modifications in imprinting control is less clear. Notably, DMRs are characteristic for the asymmetrical accumulation of different histone modifications in the two parental chromosomes and the identification of a ‘tri-mark’ comprising the trimethylation of H3K4, H3K9 and H4K20 at all known ICRs, which has reignited the debate about the role of histone modifications in imprintingcontrol [67].
ABERRATIONS OF GENOMIC IMPRINTING
Disruption of imprinting in neurodevelopment
The epigenetic imprints established in the germline are maintained throughout development and adult life. Abnormalities in imprints are associated with perturbations in growth, placental function, postnatal adaptations and neurobehavioural processes. Moreover, imprinted genes are expressed widely and highly during prenatal stages but predominantly down-regulate after birth. They are involved in multiple developmental processes capable of affecting organ development and growth. After birth, imprinted genes modulate postnatal neurological and metabolic functions. Their monoallelic expression makes these loci very vulnerable since mutation or loss of imprinting of the expressed allele can compromise expression and lead to severe developmental defects [68]. The majority of imprinted genes are expressed in the placenta and brain [69, 70]. Disruption of imprinting can result in a number of human congenital imprinting syndromes including Prader-Willi (PWS) and Angelman syndromes (AS), characterised by neurological and behavioural impairments and learning difficulties [71]. These distinct disorders map to the same imprinted domain on the human chromosome 15q [72]. Patients with PWS show mild mental retardation and behavioural problems, along with other symptoms. Paternally-expressed transcripts from the PWS/AS locus, including the protein-coding SNRPN gene, are not expressed in PWS and are implicated in the syndrome, although their precise contribution to the aetiology of the disorder is not known. Angelman Syndrome, in contrast, is associated with an absence of the UBE3A transcript, which is expressed from the maternally inherited chromosome. U be3a-maternal-deficient mice (AS model mice) show a significant impairment in adult hippocampal neurogenesis that could contribute to the cognitive disturbances observed in these mice [73]. However, little is known about how the absence or altered dosage of these genes in neural stem cell populations influences neuronal function. Other imprinted disorders such as Temple and Kagami-Ogawa Syndromes, associated with the altered dosage of imprinted genes on human chromosome 14, also exhibit neurological and behavioural impairments [74]. This further emphasizes the importance of correct regulation of imprinted genes during the neurogenesis process and in neural development.
Aberrations of genomic imprinting in brain cancer stem cells
The discovery of multipotent stem cells and restricted progenitors in the postnatal mammalian brain and the identification of stem-like cells in several brain tumours have led to the development of the “cancer stem cell” (CSC) theory of gliomas. This theory posits that gliomas are the result of the malignant transformation of cells deriving from the NSCs or lineage-restricted progenitor populations, rather than from the de-differentiation of mature cell populations [75]. The existence of cancer stem cells has been proven in brain tumours in which a small population of cells (less than 1%) shows the potential to give rise to the growing tumour. These cells have substantial stem cell characteristics such as self-renewal without loss of proliferation capacity or plasticity (capacity to change their fate and adopt a new identity). These behaviours are equally important in physiological regeneration processes and carcinogenesis. Gliomas have been shown to orchestrate a microenvironment by mimicking the normal neural stem cell niche, thereby helping to maintain the ‘glioma-initiating’ stem cell pool [76]. Furthermore, glioma-initiating cells are immortal, quite resistant to treatment and express markers typical of tumour stem cells.
Several epigenetic marks, especially DNA methylation and histone modification patterns, become altered in tumour progression and lead to changes in gene expression [77]. These changes affect the whole epigenome and thus cellular identity. However, whether the cancer stem cells originate from normal stem cells as a consequence of genetic and epigenetic changes and/or re-differentiation from somatic tumour cells to the stem-like cells remains to be investigated. Several imprinted genes have been shown to function as cellular mitogens or tumour suppressors [78, 79]. Indeed, disruption of imprinting can cause a predisposition to tumour formation and loss of imprinting in several genes, which has been observed in human cancers [80, 81], making aberrations in imprinting a potentially valuable tool for both diagnosis and treatment. Imprinted genes of maternal and paternal origin favour different degrees of neural stem cell proliferation. DLK1, essential for maintaining the stem cell pool in the SVZ, is more highly expressed in human glioma [82]. Loss of imprinting of other genes such as IGF2 has also been observed in human cancers [81] and abnormal regulation of the PEG3 gene is associated with several glioma subtypes [83]. Thus, an understanding of the mechanistic similarities between normal tissue stem cells and cancer stem cells in the CNS and their epigenetic signatures (including aberrations of imprinting) will be helpful in determining changes that might be involved in the development of human glioblastoma. Such analyses may prove fundamental in understanding stem cell regulation, the nature of cancer/tumour formation, and the future therapeutic use of stem cells in human disease.
CONCLUDING REMARKS
Loss or gain of genomic imprinting is used as a dynamic developmental mechanism to control gene dosage in NSCs. This implies that the epigenetic mechanisms controlling genomic imprinting may be adaptable to the environmental niche in which they are acting, contributing to the regulation of non-imprinted loci in the brain, too. It now becomes important to understand the epigenetic profile and some of the molecular players that participate in the regulation of the imprinting status in NSCs, as well as the role of these imprinting domains in the maintenance of the stem cell pool in the neurogenic niches in vivo. Dissecting the molecular players that participate in the preservation of the imprints at ICRs during postnatal neurogenesis under normal physiological conditions is also relevant and has the potential to provide insights into the wider epigenetic control of the neurogenic process. Moreover, the molecular mechanisms underlying normal NSC function may be the non-pathological counterparts to the tumoural processes in the adult brain. Thus, whether cancer stem cells originate from normal neural stem cells as a consequence of genetic and epigenetic changes in the brain, including aberrations of imprinting, remains to be investigated further.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
ACKNOWLEDGMENTS
We apologize to all those whose work could not be cited due to space limitations. We acknowledge Alexandra Ashcroft and Carol Edwards for their comments and suggestions for the manuscript. The work in the Ferguson-Smith lab is supported by grants from the MRC and Wellcome Trust and in the Ferrón lab by grants from the Ministerio de Economíca y Competitividad (SAF2016-78845), Generalitat Valenciana (ACOMP2014-258) and Fundación BBVA. S.R.F is a Ramón y Cajal researcher. A.L.U is funded by the Generalitat Valenciana fellowship programme and R.M.L is funded by the FPI fellowship programme from the Ministerio de Economía.
REFERENCES
[1] | Lim DA , Alvarez-Buylla A . The adult ventricular-subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harbor Perspectives in Biology. (2016) ;8: (5). |
[2] | Malatesta P , Hartfuss E , Gotz M . Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. (2000) ;127: (24):5253–63. |
[3] | Noctor SC , Flint AC , Weissman TA , Dammerman RS , Kriegstein AR . Neurons derived from radial glial cells establish radial units in neocortex. Nature. (2001) ;409: (6821):714–20. |
[4] | Tramontin AD , Garcia-Verdugo JM , Lim DA , Alvarez-Buylla A . Postnatal development of radial glia and the ventricular zone (VZ): A continuum of the neural stem cell compartment. Cereb Cortex. (2003) ;13: (6):580–7. |
[5] | Xu L , Tang X , Wang Y , Xu H , Fan X . Radial glia, the keystone of the development of the hippocampal dentate gyrus. Molecular Neurobiology. (2015) ;51: (1):131–41. |
[6] | Kempermann G , Song H , Gage FH . Neurogenesis in the Adult Hippocampus. Cold Spring Harbor perspectives in Biology. (2015) ;7: (9):a018812. |
[7] | Lim DA , Alvarez-Buylla A . Adult neural stem cells stake their ground. Trends in Neurosciences. (2014) ;37: (10):563–71. |
[8] | Doetsch F . The glial identity of neural stem cells. Nature Neuroscience. (2003) ;6: (11):1127–34. |
[9] | Merkle FT , Tramontin AD , Garcia-Verdugo JM , Alvarez-Buylla A . Radial glia give rise to adult neural stem cells in the subventricular zone. Proceedings of the National Academy of Sciences of the United States of America. (2004) ;101: (50):17528–32. |
[10] | Morshead CM , Reynolds BA , Craig CG , McBurney MW , Staines WA , Morassutti D , et al. Neural stem cells in the adult mammalian forebrain: A relatively quiescent subpopulation of subependymal cells. Neuron. (1994) ;13: (5):1071–82. |
[11] | Suh H , Consiglio A , Ray J , Sawai T , D’Amour KA , Gage FH . In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. (2007) ;1: (5):515–28. |
[12] | Bjornsson CS , Apostolopoulou M , Tian Y , Temple S . It takes a village: Constructing the neurogenic niche. Developmental Cell. (2015) ;32: (4):435–46. |
[13] | Bond AM , Ming GL , Song H . Adult mammalian neural stem cells and neurogenesis: Five decades later. Cell Stem Cell. (2015) ;17: (4):385–95. |
[14] | Menn B , Garcia-Verdugo JM , Yaschine C , Gonzalez-Perez O , Rowitch D , Alvarez-Buylla A . Origin of oligodendrocytes in the subventricular zone of the adult brain. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. (2006) ;26: (30):7907–18. |
[15] | Yu DX , Marchetto MC , Gage FH . How to make a hippocampal dentate gyrus granule neuron. Development. (2014) ;141: (12):2366–75. |
[16] | Encinas JM , Michurina TV , Peunova N , Park JH , Tordo J , Peterson DA , et al. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. (2011) ;8: (5):566–79. |
[17] | Ferron SR , Andreu-Agullo C , Mira H , Sanchez P , Marques-Torrejon MA , Farinas I . A combined ex/in vivo assay to detect effects of exogenously added factors in neural stem cells. Nature Protocols. (2007) ;2: (4):849–59. |
[18] | Reynolds BA , Weiss S . Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. (1992) ;255: (5052):1707–10. |
[19] | Singec I , Knoth R , Meyer RP , Maciaczyk J , Volk B , Nikkhah G , et al. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nature Methods. (2006) ;3: (10):801–6. |
[20] | Bird A . Perceptions of epigenetics. Nature. (2007) ;447: (7143):396–8. |
[21] | Jaenisch R , Bird A . Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nature Genetics. (2003) ;33: (Suppl):245–54. |
[22] | Yao B , Christian KM , He C , Jin P , Ming GL , Song H . Epigenetic mechanisms in neurogenesis. Nature Reviews Neuroscience. (2016) . |
[23] | Hsieh J , Gage FH . Epigenetic control of neural stem cell fate. Current Opinion in Genetics & Development. (2004) ;14: (5):461–9. |
[24] | Surani MA , Hayashi K , Hajkova P . Genetic and epigenetic regulators of pluripotency. Cell. (2007) ;128: (4):747–62. |
[25] | Ferguson-Smith AC . Genomic imprinting: The emergence of an epigenetic paradigm. Nature reviews Genetics. (2011) ;12: (8):565–75. |
[26] | Choi JD , Underkoffler LA , Wood AJ , Collins JN , Williams PT , Golden JA , et al. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Molecular and Cellular Biology. (2005) ;25: (13):5514–22. |
[27] | Kagitani F , Kuroiwa Y , Wakana S , Shiroishi T , Miyoshi N , Kobayashi S , et al. Peg5/Neuronatin is an imprinted gene located on sub-distal chromosome 2 in the mouse. Nucleic Acids Research. (1997) ;25: (17):3428–32. |
[28] | Edwards CA , Ferguson-Smith AC . Mechanisms regulating imprinted genes in clusters. Current Opinion in Cell Biology. (2007) ;19: (3):281–9. |
[29] | Sutcliffe JS , Nakao M , Christian S , Orstavik KH , Tommerup N , Ledbetter DH , et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nature Genetics. (1994) ;8: (1):52–8. |
[30] | Thorvaldsen JL , Duran KL , Bartolomei MS . Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes & Development. (1998) ;12: (23):3693–702. |
[31] | Williamson CM , Turner MD , Ball ST , Nottingham WT , Glenister P , Fray M , et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nature Genetics. (2006) ;38: (3):350–5. |
[32] | Bourc’his D , Xu GL , Lin CS , Bollman B , Bestor TH . Dnmt3L and the establishment of maternal genomic imprints. Science. (2001) ;294: (5551):2536–9. |
[33] | Davis TL , Yang GJ , McCarrey JR , Bartolomei MS . The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Human Molecular Genetics. (2000) ;9: (19):2885–94. |
[34] | Lucifero D , Mann MR , Bartolomei MS , Trasler JM . Gene-specific timing and epigenetic memory in oocyte imprinting. Human Molecular Genetics. (2004) ;13: (8):839–49. |
[35] | Smallwood SA , Kelsey G . De novo DNA methylation: A germ cell perspective. Trends in Genetics: TIG. (2012) ;28: (1):33–42. |
[36] | Takahashi N , Gray D , Strogantsev R , Noon A , Delahaye C , Skarnes WC , et al. ZFP57 and the targeted maintenance of postfertilization genomic imprints. Cold Spring Harbor Symposia on Quantitative Biology. (2015) ;80: :177–87. |
[37] | Goll MG , Bestor TH . Eukaryotic cytosine methyltransferases. Annual Review of Biochemistry. (2005) ;74: :481–514. |
[38] | Kagiwada S , Kurimoto K , Hirota T , Yamaji M , Saitou M . Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. The EMBO Journal. (2013) ;32: (3):340–53. |
[39] | Ferron SR , Charalambous M , Radford E , McEwen K , Wildner H , Hind E , et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. (2011) ;475: (7356):381–5. |
[40] | Ferron SR , Radford EJ , Domingo-Muelas A , Kleine I , Ramme A , Gray D , et al. Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nature Communications. (2015) ;6: :8265. |
[41] | Kar S , Parbin S , Deb M , Shilpi A , Sengupta D , Rath SK , et al. Epigenetic choreography of stem cells: The DNA demethylation episode of development. Cellular and Molecular Life Sciences: CMLS. (2014) ;71: (6):1017–32. |
[42] | Perez JD , Rubinstein ND , Dulac C . New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain. Annual Review of Neuroscience. (2016) ;39: :347–84. |
[43] | Rraklli V , Sodersten E , Nyman U , Hagey DW , Holmberg J . Elevated levels of ZAC1 disrupt neurogenesis and promote rapid in vivo reprogramming. Stem cell Research. (2016) ;16: (1):1–9. |
[44] | Adnani L , Langevin LM , Gautier E , Dixit R , Parsons K , Li S , et al. Zac1 Regulates the Differentiation and Migration of Neocortical Neurons via Pac1. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. (2015) ;35: (39):13430–47. |
[45] | Joseph B , Andersson ER , Vlachos P , Sodersten E , Liu L , Teixeira AI , et al. p57Kip2 is a repressor of Mash1 activity and neuronal differentiation in neural stem cells. Cell Death and Differentiation. (2009) ;16: (9):1256–65. |
[46] | Schmidt-Edelkraut U , Daniel G , Hoffmann A , Spengler D . Zac1 regulates cell cycle arrest in neuronal progenitors via Tcf4. Molecular and Cellular Biology. (2014) ;34: (6):1020–30. |
[47] | Kanduri C . Long noncoding RNAs: Lessons from genomic imprinting. Biochimica et Biophysica Acta. (2016) ;1859: (1):102–11. |
[48] | Gaughwin P , Ciesla M , Yang H , Lim B , Brundin P . Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. (2011) ;21: (8):1857–69. |
[49] | Rago L , Beattie R , Taylor V , Winter J . miR379-410 cluster miRNAs regulate neurogenesis and neuronal migration by fine-tuning N-cadherin. The EMBO Journal. (2014) ;33: (8):906–20. |
[50] | Minamide R , Fujiwara K , Hasegawa K , Yoshikawa K . Antagonistic interplay between necdin and Bmi1 controls proliferation of neural precursor cells in the embryonic mouse neocortex. PloS One. (2014) ;9: (1):e84460. |
[51] | Lehtinen MK , Zappaterra MW , Chen X , Yang YJ , Hill AD , Lun M , et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. (2011) ;69: (5):893–905. |
[52] | DeChiara TM , Robertson EJ , Efstratiadis A . Parental imprinting of the mouse insulin-like growth factor II gene. Cell. (1991) ;64: (4):849–59. |
[53] | Giannoukakis N , Deal C , Paquette J , Goodyer CG , Polychronakos C . Parental genomic imprinting of the human IGF2 gene. Nature Genetics. (1993) ;4: (1):98–101. |
[54] | Furutachi S , Matsumoto A , Nakayama KI , Gotoh Y . p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. The EMBO Journal. (2013) ;32: (7):970–81. |
[55] | Liu C , Teng ZQ , Santistevan NJ , Szulwach KE , Guo W , Jin P , et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. (2010) ;6: (5):433–44. |
[56] | Tsan YC , Morell MH , O’Shea KS . miR-410 controls adult SVZ neurogenesis by targeting neurogenic genes. Stem Cell Research. (2016) ;17: (2):238–47. |
[57] | Sun J , Ming GL , Song H . Epigenetic regulation of neurogenesis in the adult mammalian brain. The European Journal of Neuroscience. (2011) ;33: (6):1087–93. |
[58] | Feng J , Zhou Y , Campbell SL , Le T , Li E , Sweatt JD , et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature Neuroscience. (2010) ;13: (4):423–30. |
[59] | Okano M , Bell DW , Haber DA , Li E . DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. (1999) ;99: (3):247–57. |
[60] | Bai S , Ghoshal K , Datta J , Majumder S , Yoon SO , Jacob ST . DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Molecular and Cellular Biology. (2005) ;25: (2):751–66. |
[61] | Brooks PJ , Marietta C , Goldman D . DNA mismatch repair and DNA methylation in adult brain neurons. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. (1996) ;16: (3):939–45. |
[62] | Goto K , Numata M , Komura JI , Ono T , Bestor TH , Kondo H . Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation; Research in Biological Diversity. (1994) ;56: (1-2):39–44. |
[63] | Fan G , Beard C , Chen RZ , Csankovszki G , Sun Y , Siniaia M , et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. (2001) ;21: (3):788–97. |
[64] | Wu H , Coskun V , Tao J , Xie W , Ge W , Yoshikawa K , et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. (2010) ;329: (5990):444–8. |
[65] | Kim HJ , Leeds P , Chuang DM . The HDAC inhibitor, sodium butyrate, stimulates neurogenesis in the ischemic brain. Journal of Neurochemistry. (2009) ;110: (4):1226–40. |
[66] | Merson TD , Dixon MP , Collin C , Rietze RL , Bartlett PF , Thomas T , et al. The transcriptional coactivator Querkopf controls adult neurogenesis. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. (2006) ;26: (44):11359–70. |
[67] | McEwen KR , Ferguson-Smith AC . Distinguishing epigenetic marks of developmental and imprinting regulation. Epigenetics & Chromatin. (2010) ;3: (1):2. |
[68] | Cleaton MA , Edwards CA , Ferguson-Smith AC . Phenotypic outcomes of imprinted gene models in mice: Elucidation of pre- and postnatal functions of imprinted genes. Annual Review of Genomics and Human Genetics. (2014) ;15: :93–126. |
[69] | Coan PM , Burton GJ , Ferguson-Smith AC . Imprinted genes in the placenta–a review. Placenta. (2005) ;26: (Suppl A):S10–20. |
[70] | Wilkinson LS , Davies W , Isles AR . Genomic imprinting effects on brain development and function. Nature Reviews Neuroscience. (2007) ;8: (11):832–43. |
[71] | Hirasawa R , Feil R . Genomic imprinting and human disease. Essays in Biochemistry. (2010) ;48: (1):187–200. |
[72] | Horsthemke B , Wagstaff J . Mechanisms of imprinting of the Prader-Willi/Angelman region. American Journal of Medical Genetics Part A. (2008) ;146A: (16):2041–52. |
[73] | Godavarthi SK , Dey P , Sharma A , Jana NR . Impaired adult hippocampal neurogenesis and its partial reversal by chronic treatment of fluoxetine in a mouse model of Angelman syndrome. Biochemical and Biophysical Research Communications. (2015) ;464: (4):1196–201. |
[74] | Eggermann T , Perez de Nanclares G , Maher ER , Temple IK , Tumer Z , Monk D , et al. Imprinting disorders: A group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clinical Epigenetics. (2015) ;7: :123. |
[75] | Galli R , Binda E , Orfanelli U , Cipelletti B , Gritti A , De Vitis S , et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Research. (2004) ;64: (19):7011–21. |
[76] | Calabrese C , Poppleton H , Kocak M , Hogg TL , Fuller C , Hamner B , et al. A perivascular niche for brain tumor stem cells. Cancer Cell. (2007) ;11: (1):69–82. |
[77] | Esteller M . Epigenetics in cancer. The New England Journal of Medicine. (2008) ;358: (11):1148–59. |
[78] | Hao Y , Crenshaw T , Moulton T , Newcomb E , Tycko B . Tumour-suppressor activity of H19 RNA. Nature. (1993) ;365: (6448):764–7. |
[79] | Lynch CA , Tycko B , Bestor TH , Walsh CP . Reactivation of a silenced H19 gene in human rhabdomyosarcoma by demethylation of DNA but not by histone hyperacetylation. Molecular Cancer. (2002) ;1: :2. |
[80] | D’Angelo RC , Wicha MS . Stem cells in normal development and cancer. Progress in Molecular Biology and Translational Science. (2010) ;95: :113–58. |
[81] | Jelinic P , Shaw P . Loss of imprinting and cancer. The Journal of Pathology. (2007) ;211: (3):261–8. |
[82] | Yin D , Xie D , Sakajiri S , Miller CW , Zhu H , Popoviciu ML , et al. DLK Increased expression in gliomas and associated with oncogenic activities. Oncogene. (2006) ;25: (13):1852–61. |
[83] | Otsuka S , Maegawa S , Takamura A , Kamitani H , Watanabe T , Oshimura M , et al. Aberrant promoter methylation and expression of the imprinted PEG3 gene in glioma. Proceedings of the Japan Academy Series B, Physical and Biological Sciences. (2009) ;85: (4):157–65. |



