BcCluster: A Bladder Cancer Database at the Molecular Level
Abstract
Background:
Bladder Cancer (BC) has two clearly distinct phenotypes. Non-muscle invasive BC has good prognosis and is treated with tumor resection and intravesical therapy whereas muscle invasive BC has poor prognosis and requires usually systemic cisplatin based chemotherapy either prior to or after radical cystectomy. Neoadjuvant chemotherapy is not often used for patients undergoing cystectomy. High-throughput analytical omics techniques are now available that allow the identification of individual molecular signatures to characterize the invasive phenotype. However, a large amount of data produced by omics experiments is not easily accessible since it is often scattered over many publications or stored in supplementary files.
Objective:
To develop a novel open-source database, BcCluster (http://www.bccluster.org/), dedicated to the comprehensive molecular characterization of muscle invasive bladder carcinoma.
Materials:
A database was created containing all reported molecular features significant in invasive BC. The query interface was developed in Ruby programming language (version 1.9.3) using the web-framework Rails (version 4.1.5) (http://rubyonrails.org/).
Results:
BcCluster contains the data from 112 published references, providing 1,559 statistically significant features relative to BC invasion. The database also holds 435 protein-protein interaction data and 92 molecular pathways significant in BC invasion. The database can be used to retrieve binding partners and pathways for any protein of interest. We illustrate this possibility using survivin, a known BC biomarker.
Conclusions:
BcCluster is an online database for retrieving molecular signatures relative to BC invasion. This application offers a comprehensive view of BC invasiveness at the molecular level and allows formulation of research hypotheses relevant to this phenotype.
INTRODUCTION
Urothelial bladder carcinoma (BC) is a common malignancy of the urinary tract system presenting an estimate of 468,351 new bladder cancer casesand 179,753 deaths predicted for the year 2015 by the WHO GLOBOCAN (http://globocan.iarc.fr/Default.aspx) [1, 2]. Albeit variable for individual bladder cancer patients, initial symptoms include hematuria and flank pain [3, 4]. Cystoscopy is the gold standard diagnostic procedure with a reported sensitivity of 62–84% and specificity of 43–98% . This wide variability in sensitivity and specificity indicates a significant inter-operator variability [5]. Non muscle-invasive BC (CIS, Ta, T1) is treated withresection and intravesical therapy whereas muscle-invasive tumors (T2, T3, T4) require radical cystectomy usually combined with systemic cisplatin-based chemotherapy [6–8]. Non-invasive BC comprises two forms: papillary urothelial carcinoma (Ta) and carcinoma in-situ (CIS), both of which can progress to invasive disease of the lamina propria (T1) or the detrusor muscle (T2) [6]. Muscle invasive tumors have been distinguished into three distinct molecular subtypes, basal, luminal, and “p53-like” [9, 10]. Another classification system for the molecular subtypes of muscle invasive BC was carried out using data stored in The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) and resulted in the report of four clusters termed TCGA I, II, III, and IV as described by Weinstein JN et al. [11]. In addition, the Lund classification reports six different BC molecular subtypes [12].A recent review compares the different classifications of muscle invasive BC subtypes and concludes muscle invasive BC exhibits significant heterogeneity compared to other tumor types [13]. Moreover, a recent report highlights significant errors in clinical staging of patients with bladder cancer that underwent cystectomy [14]. Obtaining an accurate staging diagnosis is particularly crucial in patient selection for surgical treatments (i.e. cystectomy) and the choice of chemotherapy. Due to the invasive procedure of cystoscopy and in order to improve accuracy in the phenotype detection, blood or urine biomarkers could support clinical assessment [15]. Biomarkers measured at the DNA, RNA and/or protein levels provide the potential to choose optimum surveillance measures and treatment regimens for specific patient populations [16]. A better understanding of muscle invasive BC could be achieved by combining information obtained from individual biomarkers. Such a comprehensive view could help in predicting novel molecular targets and improve disease management.
High-throughput experimental platform technologies range from genomic sequencing to epigenomic, transcriptomic, proteomic and metabolomic profiling to characterize the molecular aspects of clinical phenotypes [17–24]. The advent of these approaches that generate a comprehensive view of the molecular landscape for a biological sample has introduced a paradigm shift in the way diseases are perceived [17, 18, 25]. A variety of datasets for such characterizations have become available for e.g. in Array Express/Gene Expression Omnibus (GEO) for transcriptomics, Human Proteinpedia for proteomics, Human Protein Atlas (http://www.proteinatlas.org/) for immunohistochemically validated proteins, or in large data consolidation platforms such as GeneCards [26]. In regard to disease specific omics data, valuable general sources in oncology include TCGA, Oncomine [27], and Online Mendelian Inheritance in Man (OMIM) [28]. The TCGA oncology portal currently lists SNPs, methylation data, mutations, mRNAs, miRNAs and proteins relevant to BC. A recent report presents a systems biology approach for the analysis of the MIBC dataset contained in TCGA [11]. Another database for BC that provides molecular features in regard to miRNAs identified in literature (http://bladder.pparser.net/MIRMarkers.php) is also available [29, 30]. In addition, a user-friendly tool called BC-BET is also available and allows the evaluation of gene expression profiles determined by microarray studies across bladder cancer patients[31].
Though omics profiling has provided an abundance of data, technical boundaries involving incompleteness of the individual molecular datasets together with the static representation of cellular activity limit the insights on molecular processes and their interaction dynamics [32–34]. A large number of biological pathway analysis tools are available, including KEGG [35], PANTHER [36], REACTOME [37] and AmiGO [38] described in PathGuide (http://www.pathguide.org/), and allow detection of significant metabolic and signaling pathways. Previous omics studies report biomarkers associated with bladder cancer, and therapeutic targets that could allow development of personalized therapies [39–43].
Currently abundant datasets are readily available, but the information gathered from these large number of omics experiments is not fully exploited, as they are either scattered in many publications and databases or held in supplementary data files. In order to allow efficient retrieval of information and offer a global view of BC invasion at the molecular level, we developed BcCluster an open-source Bladder Cancer database (http://www.bccluster.org/).
The purpose of the publication is to describe the database and offer an example on how it can be used in the context of MIBC research with a specific example of its application (survivin).
MATERIALS AND METHODS
Data sources for characterizing bladder cancer pathophysiology
In order to retrieve molecular features associated with muscle invasive bladder cancer, NCBI PubMed, Web of Science, Google Scholar and the omics repositories Gene Expression Omnibus (GEO) [44] and ArrayExpress [45] were queried. The keywords for the literature search included “bladder OR urothelial OR transitional cell” AND “neoplasm OR tumor OR carcinoma” AND “muscle” AND “invas* OR aggress* OR progress* OR inflammation” (Database version of June, 2015). The list of publications relevant to muscle invasion in BC was isolated from the complete list of retrieved papers. Publications were further screened for adequacy in sample size (at least 50 samples included in study design), magnitude of differential abundance (>2-fold change for proteomics, transcriptomics, metabolomics and miRNAs), FDR <0.1 for mutations, p values <0.05 for methylation and –omics studies, in addition to the specific phenotypic conditions; T2a/b, T3a/b, T4a/b. The MIBC-specific molecular features retrieved from the publications comprised of various sources such as DNA-mutations, DNA-methylation, mRNAs, miRNAs, proteins (IHC validated and proteomics) and metabolites. The features were then combined for further systems biology analysis.
Systems biology analysis –protein-protein interactions (PPI) and pathway enrichment
For performing systems biology analysis that includes integration of molecular features from various resources, certain filtering steps for the total number of features were taken into account. For the protein-protein interaction and pathway enrichment analysis, microRNA and metabolomic data, although they are an integral part of the database, were not taken into consideration (i.e. official gene symbols mapped from metabolomics and miRNA data, by querying HMDB and miRBase). This choice was imposed by the inability to correlate metabolite and microRNA data with specific proteins. Metabolites are part of biochemicalpathways composed of several reactions that are catal-yzed by many different enzymes, hence it is not possible to match one metabolite with a specific enzyme. Moreover, each microRNA has many target genes and these genes are regulated by different microRNAs. Thus, it is not possible to map a single microRNA to a specific gene. In regard to genomics and epigenetics, we only incorporated those genes that contained information on the protein/mRNA abundance levels. In addition, the total set of features was screened to remove duplicated proteins caused by combining molecular features from various–omics sources. This new filtered set reduced the total feature set and was then subjected to PPI analysis in the context of muscle invasive bladder cancer.
In order to retrieve protein-protein interaction information for the muscle invasive BC associated proteins, IntAct [46], BioGRID [47], STRING [48] and Reactome [37] were queried. All available human proteins along with the PPI information were downloaded into Cytoscape [49] to yield the human interactome based on experimental evidence. Then, the proteins relative to MIBC were put on a separate list. MIBC proteins that had at least one binding partner in the list of MIBC specific proteins were retained to generate the muscle invasive BC specific interactome.
To retrieve molecular pathway information for MIBC, proteins from the muscle invasive BC interactome were subjected to pathway enrichment analysis. This analysis used two additional sub-applications from Cytoscape; ClueGO and CluePedia [50–52]. The statistical criterion used in generating molecular pathways included a two-sided hypergeometry test [51]. Information from KEGG [35] and Reactome [37] databases was used in retrieving significant pathways associated to MIBC with a Bonferroni corrected p-value <0.05. In addition, the list of pathways was inspected manually and redundant pathway-terms (names) were combined. The filtered list of pathway-terms was then divided into previously known pathways and novel findings in the context of muscle invasive bladder carcinoma [53].
Development of BcCluster database and query interface
BcCluster database has been built by selecting the MySQL database system (http://www.mysql.com/), to store all comprehensive information regarding published papers, proteins, protein-interactions and molecular pathways associated specifically to the muscle invasive phenotype. The query interface for BcCluster is a web application developed in the Ruby programming language and using the framework Rails (http://rubyonrails.org/). This framework was opted as it provides an advantage to update the BcCluster database on regular intervals and allow data incorporation for newly published articles relevant in BC research. Supplementary Fig. 1, generated by the Dia software, illustrates the architecture of the BcCluster database. The query interface using the Ruby on Rails framework is detailed in supplementary Fig. 2.
RESULTS
Data mining
Database sources (Pubmed, Google scholar and Web of Science) and omics profiles (GEO and ArrayExpress) provided a total of 112 unique references for manual screening. These references yielded a total of 1,559 molecular features being significantly associated to muscle invasive bladder cancer (Fig. 1). The features from individual studies included 113 proteins from IHC studies, 424 mRNAs and 280 miRNAs (targeted to official gene symbols, by querying miRBase [54]). 49 genes were derived from DNA mutations (distinguished into 15 oncogenes and 34 tumor suppressor genes), metabolomics experiments provided 464 molecular features (associated to an enzyme in gene symbols, by querying HMDB [55]). In addition 79 genes were obtained from DNA methylation studies and 150 proteins were derived from proteomics studies. The list of 112 articles collected in this analysis is provided in Supplementary Table 1.
Systems biology analysis –PPI and pathway enrichment
The dataset associated with miRNAs and metabolites was not used for pathway enrichment analysis. This choice was opted due to the inability to correlate metabolite and microRNA data with specific proteins. In addition, the available bioinformatics tools (Cytoscape) do not allow efficient integration of these datasets with other omics data. Subsequently, the obtained dataset of significant MIBC features was processed in order to eliminate duplicate entries. The new filtered set resulted in a total of 737 proteins that were then subjected to PPI analysis in the context of muscle invasive bladder cancer. To obtain all possible protein interaction information for the 737 muscle invasive bladder cancer specific proteins, PPI databases including IntAct, BioGRID, String and Reactome were queried. Figure 2a, illustrates the steps taken in producing the muscle invasive bladder cancer interactome. In the first step, all available PPIs for the human proteome were downloaded into Cytoscape to form the human interactome. The PPIs relevant to the 737 MIBCproteins were retrieved from the human interactome and only proteins with at least one binding partner were retained (Fig. 2a). This step yielded 435 proteins along with 4,768 PPIs that were used in the pathway enrichment analysis. 302 proteins that did not have a binding partner among the full list of 737 relevant to MIBC were not retained for further analysis.
The 435 proteins that were obtained from the muscle invasive BC interactome (Fig. 2a) were incorporated into ClueGO and CluePedia to identify significant pathways in the context of MIBC. A total of 292 molecular pathways were retrieved in this analysis. This pathway list was manually screened in order to combine redundant pathway-names. This screening step reduced the list of pathways to a total of 92 pathways that were then distinguished into 75 previously known in literature and 17 novel pathway findings in the context to muscle invasive BC (Fig. 2b).
BcCluster database and web interface
In order to efficiently store and retrieve information in regard to the 112 published articles, 1,559 molecular features, 435 proteins, 4,768 PPIs and 92 molecular pathways associated to muscle invasive bladder carcinoma, a database supported by a web user-interface was developed. The BcCluster web portal (http://bccluster.org/biomolecules/study_type) lists all the bladder cancer associated molecular features based on individual study type and is presented in Fig. 3. The application allows the user to browse specific omics traits to gain information regarding features associated to bladder cancer at different stages and grades of the disease. In addition the PubMed reference ids for these retrieved data are also provided and the application can be queried based on the keywords as described in Table 1.
For selecting proteins based on literature mined IHC validated experiments, each muscle invasive BC-associated protein was evaluated for being a member of the 92 muscle invasive BC enriched pathways. The evidence of identified pathways and extracted proteins involved in muscle invasive BC was assessed based on the number of individual bladder cancer studies that described such proteins. The quality of publications obtained for each protein was assessed by manually reviewing these articles. Only papers where a direct relevance for the protein to bladder cancer muscle invasion was proven were retained. For the entire 92 muscle invasive BC enriched pathways, each relevant pathway was ranked based on the statistical significance (p value). Pathways that appeared at least once in a bladder cancer study were grouped as literature known pathways (containing 75 enriched pathways), while those pathways that were not previously reported in muscle invasive bladder cancer were categorized as novel findings (containing 17 enriched pathways).
In-silico hypothesis generation: Survivin as an example
Previous studies by Shariat SF et al. [56] report survivin to be a promising biomarker for cancer diagnosis, prognosis and prediction of response to intravesical or systemic therapies. Urinary survivin tests in BC have been shown to perform better than urinary cytology with a sensitivity of 0.77 (95% confidence interval [CI] 0.75–0.80) and specificity of 0.92 (95% CI 0.90–0.93) [57–60]. Survivin is a protein involved in the inhibition of apoptosis and has been reported to control mitotic progression, in addition to inducing changes in the gene expression of caspases 3, 7 and 9 that are associated with cell invasion in BC [61–63]. Hence, we selected survivin as an example to query BcCluster. The workflow of a simple query to the BcCluster database via the web application is presented in Fig. 4. The search term used was BIRC5. The results to the search provide a list of the data available for the molecular feature in different published studies. The search also displays other important information such as the type of study (e.g. IHC, transcriptomics, etc.), the clinical samples used for the comparison and the relative amount of the gene/mRNA/protein present in these samples. Additionally, the application also displays, if available, a list of the protein-binding partners based on experimental evidence and muscle invasive BC pathways significantly enriched for the specific query term.
Survivin was found to be significantly associated to 3 molecular pathways namely cell cycle, hippo signaling and signal transduction pathways. The 3 molecular pathways presented in Table 2 are sorted according to the corrected p-values. Signal transduction (p-value 2.46 10 - 14) pathway was found to be the most significant followed by Hippo signaling (p-value 4.93 10 - 09), and cell cycle (p-value 2.55 10 - 04). From our analysis, we predict the involvement of survivin to be significantly associated to Hippo signaling pathway in the context of muscle invasive bladder cancer.
DISCUSSION
The main aim of this study was to develop a database specific for invasive bladder cancer. This was achieved by integrating all possible information publically available from literature and sequencing platforms, PPIs and pathway enrichment resources in the context of MIBC. An example of the database utility is given by extracting information relevant to survivin in BC.
Aggressive MIBC progresses rapidly to metastatic diseases and generates high patient mortality [66]. Radical cystectomy combined with systemic cisplatin-based chemotherapy (either before or after cystectomy) is the current standard of care for high-risk MIBC [6]. Treatment selection depends particularly on clinico-pathologic features. Unfortunately current staging systems are not optimum and yield high rates of clinical under-staging leading to inadequate treatment [9, 14]. Moreover, distinct molecular subtypes in MIBC have also been reported including basal, luminal and interestingly “p53-like” that are resistant to chemotherapy [9]. Understanding the molecular pathophysiology of muscle-invasive bladder carcinoma and elucidating the network of pathways involved in invasion could lead to targeted therapies. In addition, discovery of specific de-regulated pathways linked to progressive disease holds the promise of supporting an improved, biomarker-based risk assessment followed by appropriate clinical intervention. High throughput omics platforms have provided a wealth of information in describing the molecular status of bladder carcinoma [11, 24, 67–69]. In order to contribute to the molecular characterization of invasive BC, we integrated omics data into a database called BcCluster. A database system at the level of networks and pathways was created in order to provide efficient retrieval of information for a single molecular feature (such as mutations, DNA methylation, miRNAs, mRNAs, metabolomics and IHC). Thus, a comprehensive view on the involvement of a feature in BC invasiveness can be obtained. Moreover, researchers can access a list of available high quality data from individual omics platforms (for e.g. transcriptomics data) and perform systems biology analysis (protein-protein interactions and pathway enrichment) in order to characterize bladder cancer at the molecular level. In addition, users can download the data from each study type that is useful to their research. We are willing to offer all the BcCluster data in a compressed format to interested users on request.
An application of the database is formulation of scientific hypotheses that can be validated experimentally, illustrated by the example of survivin. There is ample experimental evidence supporting the role of survivin in the context of BC pathology [70–72]. Our approach in integrating PPI and pathway databases yielded three significant pathways for survivin (cell cycle, signal transduction, and hippo signaling). The expression of survivin positively correlates with the levels of signal transduction proteins EGFR (Epidermal growth factor receptor) and VEGFA (Vascular endothelial growth factor A) in muscle invasive BC [64]. In Reactome, the term “signal transduction pathway” is described as a hierarchy of signaling events that elicit changes in cell state and activity due to the result of extracellular signals. Some of the types of signal transduction pathways include, signaling by RTKs, PDGF, NGF, TGF-beta, JNK and NF-kB, RAF/MAPK and Rho GTPases. In order to determine which of these signal transduction pathways were specifically associated to survivin, we manually queried survivin in the Reactome database (http://www.reactome.org/). The most prominent pathway involving survivin was signaling by Rho GTPases. A transcriptomics study determined that the majority of the Rho family small GTPases (RhoA, RhoB, RhoC, Rac1 and Cdc42) are significantly over-expressed in BC, hence providing experimental validation of our in silico prediction [73]. The role of survivin in cell cycle has previously been reported for MIBC, whereas hippo signaling is a novel finding. Down-regulation of survivin is associated with cell cycle arrest and apoptosis in BC cell lines [65]. The experimental evidence confirms our in silico analysis for the involvement of survivin in BC pathways reported in the literature. Hippo signaling is a conserved pathway that regulates organ size and is implicated in cancer development [74]. Survivin expression was proven to be inversely correlated with hippo signaling in vascular tumors of the lungs and liver [75]. The available information indicates that survivin is likely involved in hippo signaling pathway in the context of BC and experimental validation for this in silico generated hypothesis could be attempted.
In summary, data retrieval from the literature and omics studies resulted in the collection of molecular features associated with muscle invasive BC. Using these features allowed the generation of the muscle invasive bladder cancer interactome and pathways highly associated to the invasive phenotype. The pathway enrichment analysis yielded significant muscle invasive BC pathways that have been previously reported in literature including PIK3 signaling, MAPK, focal adhesion, cell cycle, EGFR and ErbB signaling pathways [11, 14, 76–78]. These studies confirm the validity of our systems biology analysis. With the BcCluster application, we aim towards supporting a better understanding of complex molecular mechanisms in BC invasion. Similar web-based databases on individual molecular disorders such as for lung cancer [79], breast cancer [80], liver diseases [81], kidney disorders [82] or specifically for personalized medicine [83] are currently available. These databases provide information on features extracted from various omics studies (ranging from Single Nucleotide Polymorphisms, to transcriptomics, miRNAs, metabolomics, transcriptional binding-factor motifs, PPI networks and pathways). In addition, the features from the individual studies are integrated in a manner to allow researchers to efficiently retrieve molecular information for a specific gene or protein search. In regard to BC, there are limited database resources that allow the molecular characterization of the invasive disease. Publically available BC sources exist, either within the TCGA portal or in databases focused on miRNAs [29–31]. All the molecular features in regard to mutations, mRNAs, miRNAs and proteins from the TCGA-MIBC specific database were incorporated in the BcCluster database. In addition, when comparing the features stored in HLungDb [79] and BcCluster, the lung cancer database contains 2,797 molecular features by integrating genes and miRNAs, whereas the BcCluster contains 1,559 molecular features comprising of molecular information ranging from DNA-methylation, DNA-mutations, mRNAs, proteins (retrieved from proteomics and IHC experiments) and metabolites. In addition, our database also holds 4,768 PPIs and 92 BC significant pathways enriched from our systems biology analysis.
In the future, we plan to further expand the BcCluster database by updating our dataset with published studies and large-scale omics profiles in regular time intervals. This updated version will allow to refine the generated muscle invasive protein interactome leading to a comprehensive molecular characterization and identification of novel modules in the context of muscle invasive BC.
FUNDING SOURCES
The research leading to these results has received funding from the Marie Curie Actions –BCMolMed under grant agreement no. FP7-PEOPLE-2012-ITN-EID and the European Community’s Seventh Framework Programme under grant agreement no. 306157.
DISCLOSURE
Harald Mischak is the founder and co-owner of Mosaiques Diagnostics, Germany, and Akshay Bhat is employed by Mosaiques Diagnostics. All authors declare that they have no competing interests.
ACKNOWLEDGMENTS
HM, VJ and AV designed and coordinated the study, JZ and MM provided support in data retrieval, and AB performed the analysis and drafted the manuscript. All authors contributed to the interpretation of the results and drafted the publication along with reading and approving the final manuscript.
Appendices
The supplementary table and figure are available in the electronic version of this article: http://dx.doi.org/10.3233/BLC-150024.
REFERENCES
[1] | Vishnu P , Mathew J , Tan WW . Current therapeutic strategies for invasive and metastatic bladder cancer. Onco Targets Ther (2011) ;4: :97–113. |
[2] | Ferlay J , Soerjomataram II , Dikshit R , Eser S , Mathers C , Rebelo M , et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer (2014) ;136: :E359–E386. |
[3] | Henning A , Wehrberger M , Madersbacher S , Pycha A , Martini T , Comploj E , et al. Do differences in clinical symptoms and referral patterns contribute to the gender gap in bladder cancer? BJU Int (2013) ;112: :68–73. |
[4] | Raman JD , Shariat SF , Karakiewicz PI , Lotan Y , Sagalowsky AI , Roscigno M , et al. Does preoperative symptom classification impact prognosis in patients with clinically localized upper-tract urothelial carcinoma managed by radical nephroureterectomy? Urol Oncol (2011) ;29: :716–723. |
[5] | Cauberg Evelyne CC , de la Rosette JJMCH , de Reijke TM . Emerging optical techniques in advanced cystoscopy for bladder cancer diagnosis: A review of the current literature. Indian J Urol (2011) ;27: :245–251. |
[6] | Knowles MA , Hurst CD . Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat Rev Cancer (2014) ;15: :25–41. |
[7] | Niegisch G , Lorch A , Droller MJ , Lavery HJ , Stensland KD , Albers P . Neoadjuvant chemotherapy in patients with muscle-invasive bladder cancer: Which patients benefit? Eur Urol (2013) ;64: :355–357. |
[8] | Sternberg CN , Apolo AB . Everything old is new again! Neoadjuvant chemotherapy in the treatment of muscle-invasive bladder cancer. J Clin Oncol (2014) ;32: :1868–18670. |
[9] | Choi W , Porten S , Kim S , Willis D , Plimack ER , Hoffman-Censits J , et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell (2014) ;25: :152–165. |
[10] | Damrauer JS , Hoadley KA , Chism DD , Fan C , Tiganelli CJ , Wobker SE , et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A (2014) ;111: :3110–3115. |
[11] | Weinstein JN , Akbani R , Broom BM , Wang W , Verhaak RGW , McConkey D , et al. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature (2014) ;507: :315–322. |
[12] | Sjödahl G , Lauss M , Lövgren K , Chebil G , Gudjonsson S , Veerla S , et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res (2012) ;18: :3377–3386. |
[13] | Aine M , Eriksson P , Liedberg F , Höglund M , Sjödahl G . On Molecular Classification of Bladder Cancer: Out of One, any Eur Urol (2015) ;68: :921–923. |
[14] | Kim PH , Cha EK , Sfakianos JP , Iyer G , Zabor EC , Scott SN , et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol (2015) ;67: :198–201. |
[15] | Emmert-Streib F , Abogunrin F , de Matos Simoes R , Duggan B , Ruddock MW , Reid CN , et al. Collectives of diagnostic biomarkers identify high-risk subpopulations of hematuria patients: Exploiting heterogeneity in large-scale biomarker data. BMC Med (2013) ;11: :12. |
[16] | Ru Y , Dancik GM , Theodorescu D . Biomarkers for prognosis and treatment selection in advanced bladder cancer patients. Curr Opin Urol (2011) ;21: :420–7. |
[17] | Mayer P , Mayer B , Mayer G . Systems biology: Building a useful model from multiple markers and profiles. Nephrol Dial Transplant (2012) ;27: :3995–4002. |
[18] | Kreeger PK , Lauffenburger DA . Cancer systems biology: A network modeling perspective. Carcinogenesis (2010) ;31: :2–8. |
[19] | Chihara Y , Kanai Y , Fujimoto H , Sugano K , Kawashima K , Liang G , et al. Diagnostic markers of urothelial cancer based on DNA methylation analysis. BMC Cancer (2013) ;13: :275. |
[20] | Weiss GJ , Liang WS , Demeure MJ , Kiefer JA , Hostetter G , Izatt T , et al. A pilot study using next-generation sequencing in advanced cancers: Feasibility and challenges. PLoS Onee (2013) ;8: :76438. |
[21] | Mischak H , Schanstra JP . CE-MS in biomarker discovery, validation, and clinical application. Proteomics Clin Appl (2011) ;5: :9–23. |
[22] | Alberice JV , Amaral AFS , Armitage EG , Lorente JA , Algaba F , Carrilho E , et al. Searching for urine biomarkers of bladder cancer recurrence using a liquid chromatography-mass spectrometry and capillary electrophoresis-mass spectrometry metabolomics approach. J Chromatogr A (2013) ;1318: :163–170. |
[23] | Pignot G , Cizeron-Clairac G , Vacher S , Susini A , Tozlu S , Vieillefond A , et al. microRNA expression profile in a large series of bladder tumors: Identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer (1318) ;132: :2479–2491. |
[24] | Liu Y , Noon AP , Aguiar Cabeza E , Shen J , Kuk C , Ilczynski C , et al. Next-generation RNA sequencing of archival formalin-fixed paraffin-embedded urothelial bladder cancer. Eur Urol (2014) ;66: :982–986. |
[25] | Hanash S . Disease proteomics. Nature (2003) ;422: :226–232. |
[26] | Stelzer G , Dalah I , Stein TI , Satanower Y , Rosen N , Nativ N , et al. In-silico human genomics with GeneCards. Hum Genomics (2011) ;5: :709–17. |
[27] | Rhodes DR , Yu J , Shanker K , Deshpande N , Varambally R , Ghosh D , et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia (2004) ;6: :1–6. |
[28] | Hamosh A , Scott AF , Amberger JS , Bocchini CA , McKusick VA . Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids ResD (2005) ;33: :514–517. |
[29] | Zabolotneva AA , Zhavoronkov AA , Shegay PV , Gaifullin NM , Alekseev BY , Roumiantsev SA , et al. A systematic experimental evaluation of microRNA markers of human bladder cancer. Front Genet (2013) ;4: :247. |
[30] | Zabolotneva AA , Zhavoronkov A , Garazha AV , Roumiantsev SA , Buzdin AA . Characteristic patterns of microRNA expression in human bladder cancer. Front Genet (2012) ;3: :310. |
[31] | Dancik GM . An online tool for evaluating diagnostic and prognostic gene expression biomarkers in bladder cancer. BMC Urol (2015) ;15: :59. |
[32] | Mayer G , Heinze G , Mischak H , Hellemons ME , Heerspink HJL , Bakker SJL , et al. Omics-bioinformatics in the context of clinical data. Methods Mol Biol (2011) ;719: :479–497. |
[33] | Abu-Asab MS , Chaouchi M , Alesci S , Galli S , Laassri M , Cheema AK , et al. Biomarkers in the age of omics: Time for a systems biology approach. OMICS (2011) ;15: :105–112. |
[34] | De Las Rivas J , Fontanillo C . Protein-protein interactions essentials: Key concepts to building and analyzing interactome networks. PLoS Comput Biol (2010) ;6: :e1000807. |
[35] | Kanehisa M , Goto S , Kawashima S , Okuno Y , Hattori M . The KEGG resource for deciphering the genome. Nucleic Acids Res (2004) ;32: :D277–D280. |
[36] | Mi H , Muruganujan A , Thomas PD . PANTHER in Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res (2013) ;41: :D377–D386. |
[37] | Croft D , O’Kelly G , Wu G , Haw R , Gillespie M , Matthews L , et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res (2011) ;39: :D691–D697. |
[38] | Carbon S , Ireland A , Mungall CJ , Shu S , Marshall B , Lewis S . AmiGO: Online access to ontology and annotation data. Bioinformatics (2009) ;25: :288–289. |
[39] | Forbes SA , Bindal N , Bamford S , Cole C , Kok CY , Beare D , et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res (2011) ;39: :D945–D950. |
[40] | Gui Y , Guo G , Huang Y , Hu X , Tang A , Gao S , et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet (2011) ;43: :875–878. |
[41] | Hurst CD , Platt FM , Taylor CF , Knowles MA . Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis. Clin Cancer Res (2012) ;18: :5865–5877. |
[42] | Lindgren D , Sjödahl G , Lauss M , Staaf J , Chebil G , Lövgren K , et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One (2012) ;7: :e38863. |
[43] | Williams SV , Hurst CD , Knowles MA . Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet (2013) ;22: :795–803. |
[44] | Barrett T , Wilhite SE , Ledoux P , Evangelista C , Kim IF , Tomashevsky M , et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res (2013) ;41: :D991–D995. |
[45] | Rustici G , Kolesnikov N , Brandizi M , Burdett T , Dylag M , Emam I , et al. ArrayExpress update–trends in database growth and links to data analysis tools. Nucleic Acids Res (2013) ;41: :D987–D990. |
[46] | Kerrien S , Aranda B , Breuza L , Bridge A , Broackes-Carter F , Chen C , et al. The IntAct molecular interaction database in Nucleic Acids Res (2012) ;40: :D841–D846. |
[47] | Stark C , Breitkreutz B-J , Chatr-Aryamontri A , Boucher L , Oughtred R , Livstone MS , et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res (2011) ;39: :D698–D704. |
[48] | Franceschini A , Szklarczyk D , Frankild S , Kuhn M , Simonovic M , Roth A , et al. STRING v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res (2013) ;41: :D808–D815. |
[49] | Shannon P , Markiel A , Ozier O , Baliga NS , Wang JT , Ramage D , et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res (2003) ;13: :2498–2504. |
[50] | Saito R , Smoot ME , Ono K , Ruscheinski J , Wang P-L , Lotia S , et al. A travel guide to Cytoscape plugins. Nat Methods (2012) ;9: :1069–1076. |
[51] | Bindea G , Mlecnik B , Hackl H , Charoentong P , Tosolini M , Kirilovsky A , et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics (2009) ;25: :1091–1093. |
[52] | Bindea G , Galon J , Mlecnik B . CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics (2013) ;29: :661–663. |
[53] | Bhat A , Heinzel A , Mayer B , Perco P , Mühlberger I , Husi H , et al. Protein interactome of muscle invasive bladder cancer. PLoS One (2015) ;10: :e0116404. |
[54] | Kozomara A , Griffiths-Jones S . miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res (2011) ;39: :D152–D157. |
[55] | Wishart DS , Tzur D , Knox C , Eisner R , Guo AC , Young N , et al. HMDB: The Human Metabolome Database. Nucleic Acids Res (2007) ;35: :D521–D526. |
[56] | Margulis V , Lotan Y , Shariat SF . Survivin: A promising biomarker for detection and prognosis of bladder cancer. World J Urol (2008) ;26: :59–65. |
[57] | Ku JH , Godoy G , Amiel GE , Lerner SP . Urine survivin as a diagnostic biomarker for bladder cancer: A systematic review. BJU Int (2012) ;110: :630–636. |
[58] | Srivastava AK , Singh PK , Srivastava K , Singh D , Dalela D , Rath SK , et al. Diagnostic role of survivin in urinary bladder cancer. Asian Pac J Cancer Prev (2013) ;14: :81–85. |
[59] | Fristrup N , Ulhøi BP , Birkenkamp-Demtröder K , Mansilla F , Sanchez-Carbayo M , Segersten U , et al. Cathepsin E, maspin, Plk1, and survivin are promising prognostic protein markers for progression in non-muscle invasive bladder cancer. Am J Pathol (2012) ;180: :1824–1834. |
[60] | Johnen G , Gawrych K , Bontrup H , Pesch B , Taeger D , Banek S , et al. Performance of survivin mRNA as a biomarker for bladder cancer in the prospective study UroScreen. PLoS One (2012) ;7: :e35363. |
[61] | Li F , Ling X . Survivin study: An update of “what is the next wave”? J Cell Physiol (2006) ;208: :476–486. |
[62] | Altieri DC . Molecular circuits of apoptosis regulation and cell division control: The survivin paradigm. J Cell Biochem (2004) ;92: :656–663. |
[63] | Deveraux QL , Reed JC . IAP family proteins–suppressors of apoptosis. Genes Dev (1999) ;13: :239–252. |
[64] | Płuciennik E , Nowakowska M , Stępien A , Wołkowicz M , Stawiński A , Różański W , et al. Alternating expression levels of WWOX tumor suppressor and cancer-related genes in patients with bladder cancer. Oncol Lett (2014) ;8: :2291–2297. |
[65] | Yuan X , Li T , Xiao E , Zhao H , Li Y , Fu S , et al. Licochalcone B inhibits growth of bladder cancer cells by arresting cell cycle progression and inducing apoptosis. Food Chem Toxicol (2014) ;65: :242–251. |
[66] | Shah JB , McConkey DJ , Dinney CPN . New strategies in muscle-invasive bladder cancer: On the road to personalized medicine. Clin Cancer Res (2011) ;17: :2608–2612. |
[67] | Mischak H , Allmaier G , Apweiler R , Attwood T , Baumann M , Benigni A , et al. Recommendations for biomarker identification and qualification in clinical proteomics. Sci Transl Med (2010) ;2: :46ps42. |
[68] | Vlahou A . Network views for personalized medicine. Proteomics Clin Appl (2013) ;7: :384–387. |
[69] | Frantzi M , Latosinska A , Flühe L , Hupe MC , Critselis E , Kramer MW , et al. Developing proteomic biomarkers for bladder cancer: Towards clinical application. Nat Rev Urol (2015) ;12: :317–330. |
[70] | Gou X , Yang H-A , He W-Y , Xioa M-C , Wang M . Gene silence-induced downregulation of survivin inhibits bladder cancer cells. Oncol Res (2011) ;19: :535–541. |
[71] | Ku JH , Seo SY , Kwak C , Kim HH . Cytotoxicity and apoptosis by survivin small interfering RNA in bladder cancer cells. BJU Int (2010) ;106: :1812–1816. |
[72] | Wuttig D , Kunze D , Fuessel S , Toma M , Stade J , Kotzsch M , et al. Are overexpressed alternative survivin transcripts in human bladder cancer suitable targets for siRNA-mediated in vitro inhibition? Int J Oncol (2007) ;30: :1317–1324. |
[73] | Volanis D , Zaravinos A , Kadiyska T , Delakas D , Zoumpourlis V , Spandidos DA . Expression profile of Rho kinases in urinary bladder cancer. J BUON (2011) ;16: :511–521. |
[74] | Zeng Q , Hong W . The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell (2008) ;13: :188–92. |
[75] | Tsuneki M , Madri JA . Adhesion molecule-mediated hippo pathway modulates hemangioendothelioma cell behavior. Mol Cell Biol (2014) ;34: :4485–4499. |
[76] | Korkolopoulou P , Levidou G , Trigka E-A , Prekete N , Karlou M , Thymara I , et al. A comprehensive immunohistochemical and molecular approach to the PI3K/AKT/mTOR (phosphoinositide 3-kinase/v-akt murine thymoma viral oncogene/mammalian target of rapamycin) pathway in bladder urothelial carcinoma. BJU Int (2012) ;110: :E1237–E1248. |
[77] | Naik DSL , Sharma S , Ray A , Hedau S . Epidermal growth factor receptor expression in urinary bladder cancer. Indian J Urol (2011) ;27: :208–214. |
[78] | Colquhoun AJ , Mellon JK . Epidermal growth factor receptor and bladder cancer. Postgrad Med J (2002) ;78: :584–589. |
[79] | Wang L , Xiong Y , Sun Y , Fang Z , Li L , Ji H , et al. HLungDB: An integrated database of human lung cancer research. Nucleic Acids Res (2010) ;38: :D665–D669. |
[80] | Raju R , Paul AM , Asokachandran V , George B , Radhamony L , Vinaykumar M , et al. The Triple-Negative Breast Cancer Database: An omics platform for reference, integration and analysis of triple-negative breast cancer data. Breast Cancer Res (2014) ;16: :490. |
[81] | Zhang Y , Yang C , Wang S , Chen T , Li M , Wang X , et al. LiverAtlas: A unique integrated knowledge database for systems-level research of liver and hepatic disease. Liver Int (2013) ;33: :1239–1248. |
[82] | Klein J , Jupp S , Moulos P , Fernandez M , Buffin-Meyer B , Casemayou A , et al. The KUPKB: A novel Web application to access multiomics data on kidney disease. FASEB J (2012) ;26: :2145–2153. |
[83] | Bauer C , Stec K , Glintschert A , Gruden K , Schichor C , Or-Guil M , et al. BioMiner: Paving the Way for Personalized Medicine. Cancer Inform (2015) ;14: :55–63. |
Figures and Tables
Fig.1
Data retrieval flow for the muscle invasive bladder cancer articles and significant molecular features.
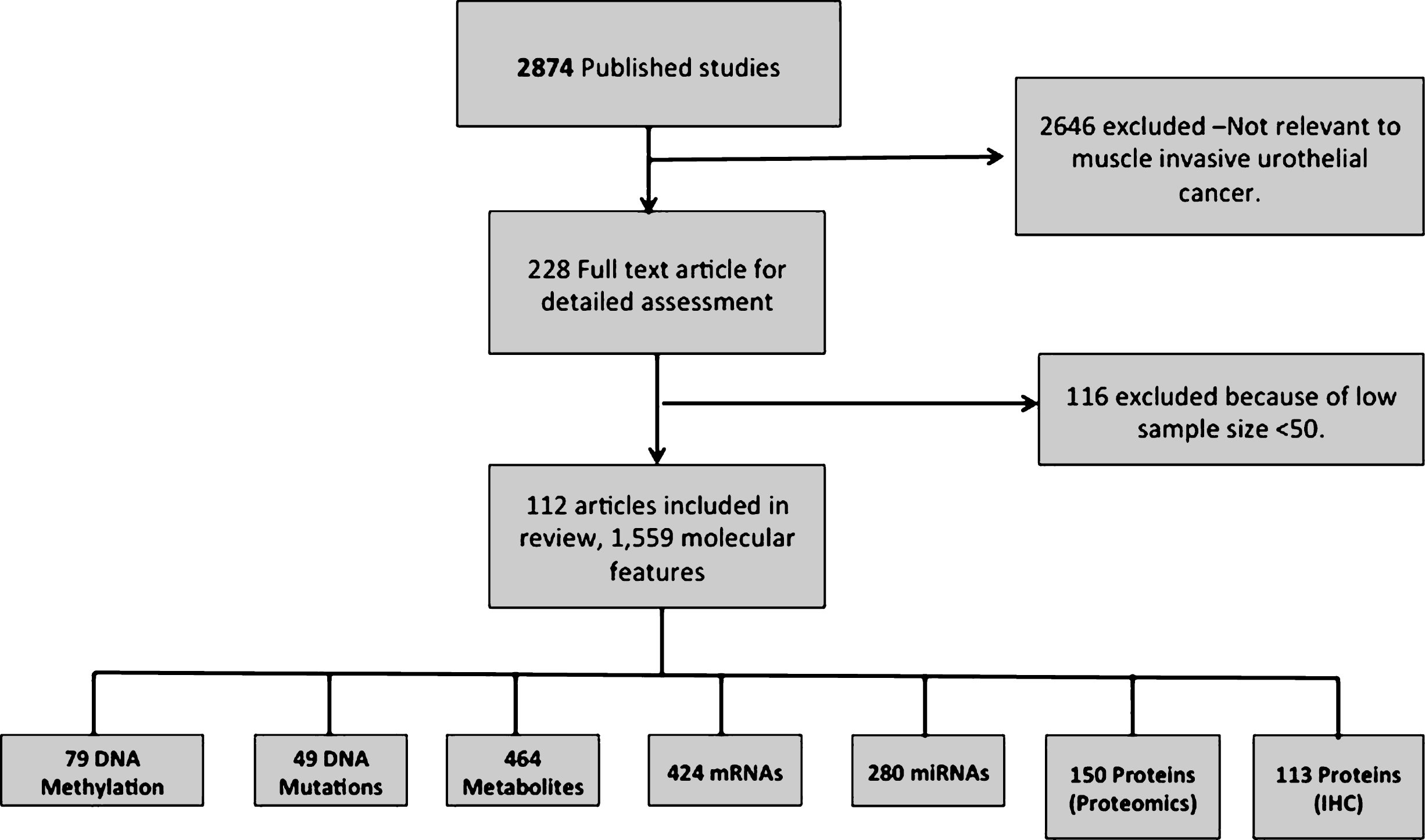
Fig.2
a. Generating the muscle invasive bladder cancer interactome. b. Pathway enrichment using the 435 MIBC specific proteins. Each circle represents a pathway and dots denote proteins. Filled circles represents down-regulation of the pathway in the context of MIBC, while open circles denotes up-regulation. Pathways are connected through common proteins.
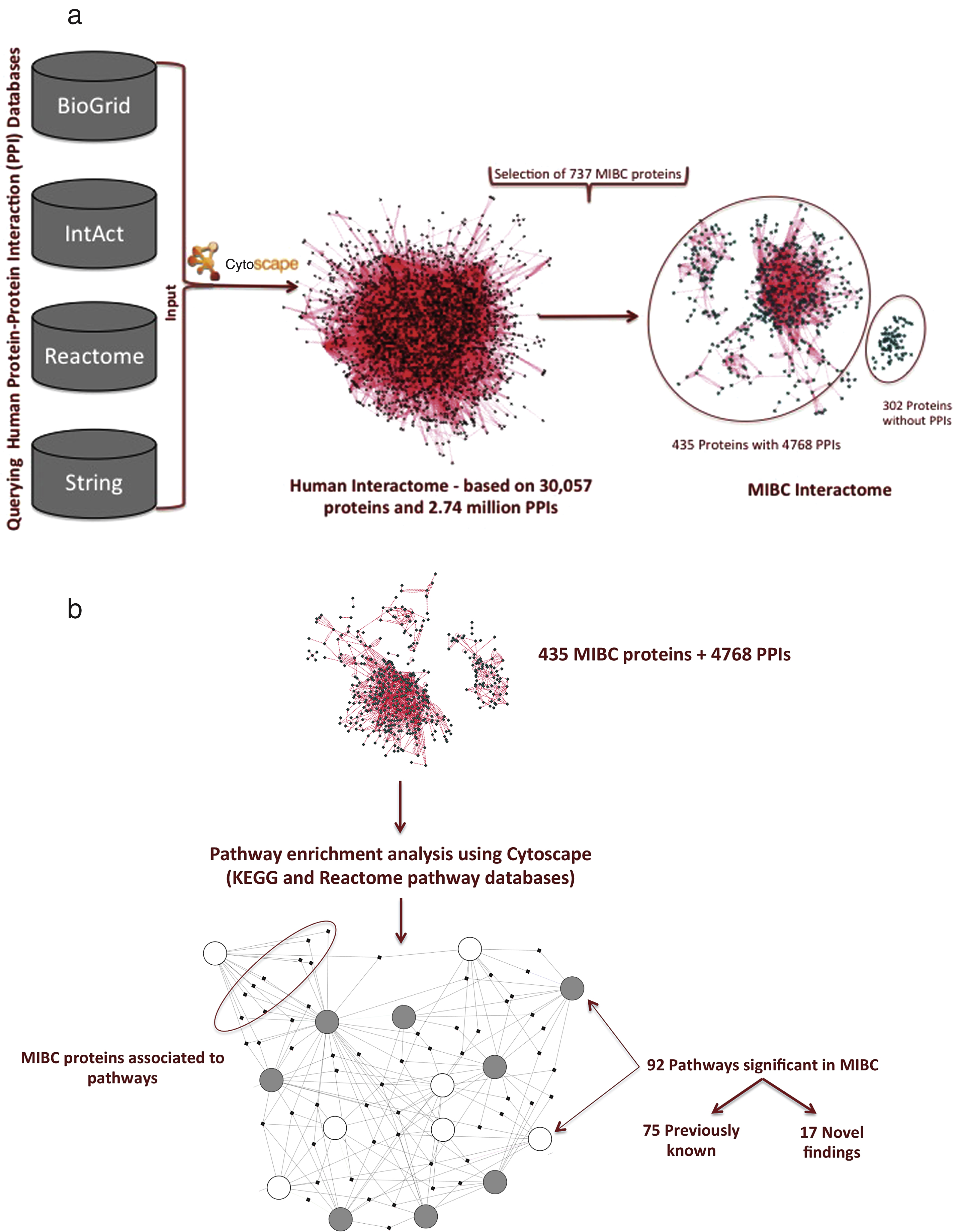
Fig.3
Display of molecular features based on study type from the BcCluster database. Molecular features significant in muscle invasive BC are organized and displayed based on the study type. Users can access these molecular features and retrieve biological information by clicking any of the listed study types. Additionally, all the results displayed in the application can be downloaded in various file formats (.csv,.txt and.xls).
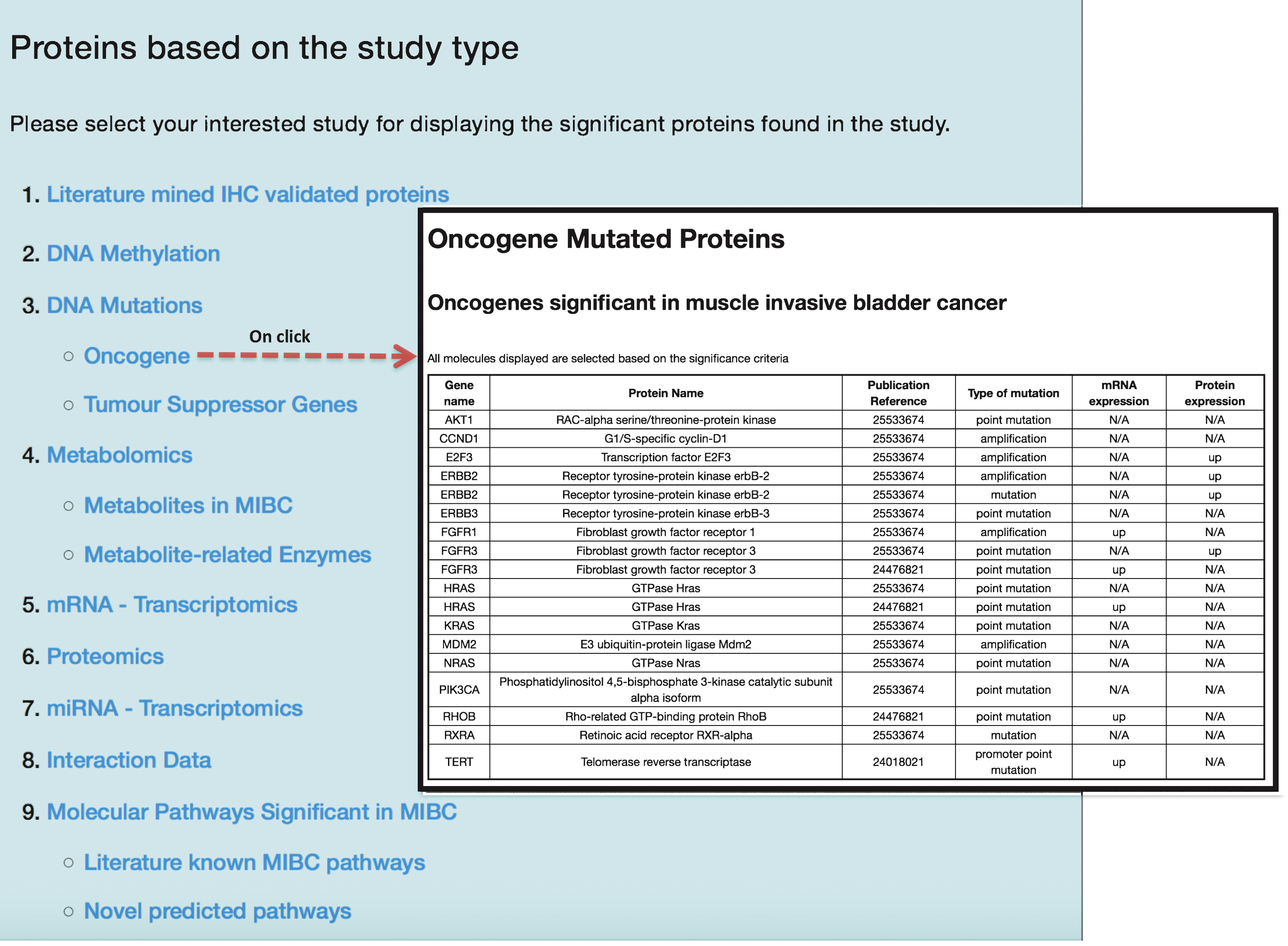
Fig.4
Example workflow for Survivin. The query for the protein-coding gene BIRC5 provides a list of publications, the type of studies, the clinical samples used for the comparison and the relative amount of the gene/mRNA/protein present in these samples. The query term also provides information on physical protein interactions and molecular pathways since the query term (i.e. protein-coding gene) is part of protein complexes relevant to muscle invasive BC.
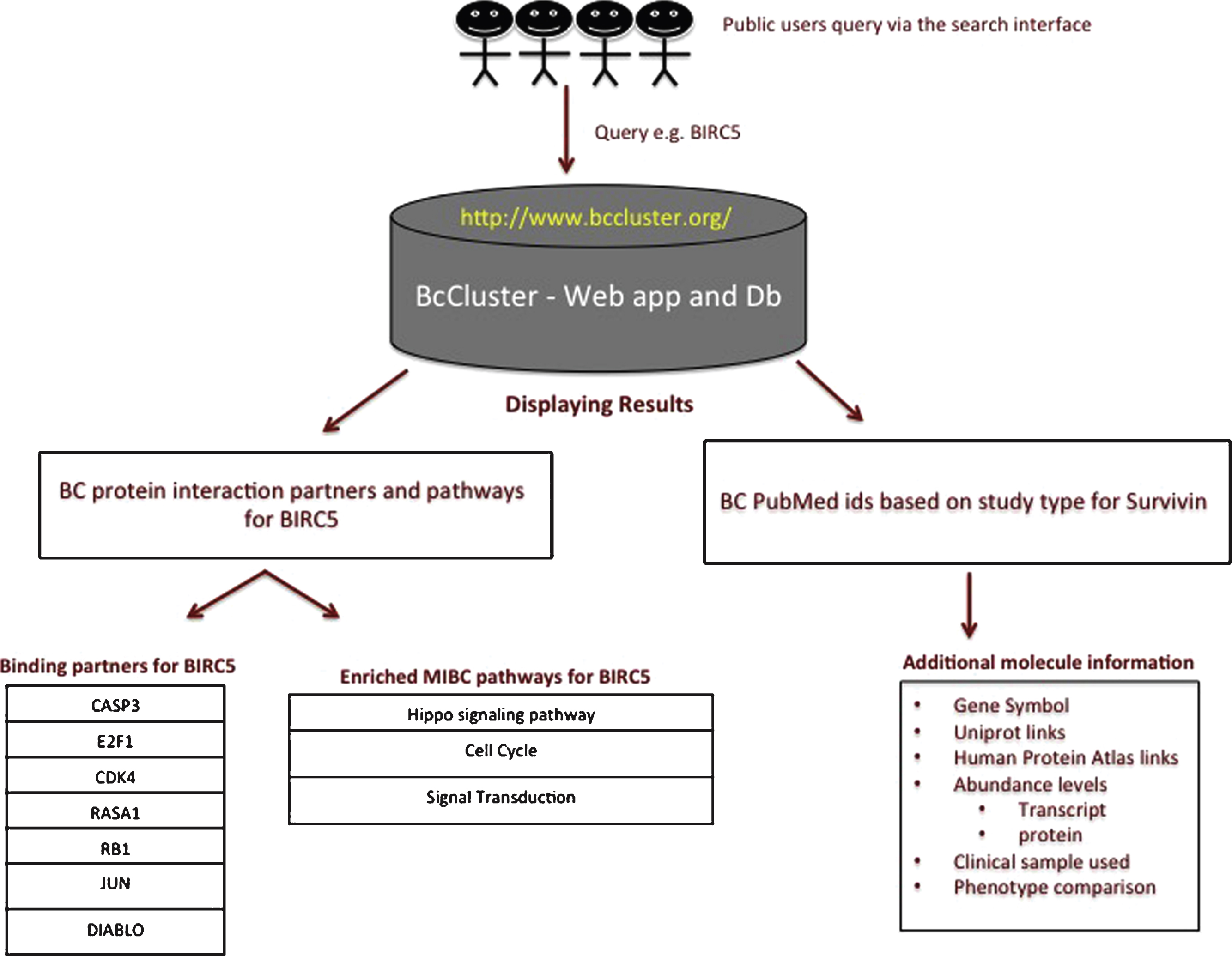
Table 1
Query terms that can be used in BcCluster. Listing the types of entities that can be used for querying the BcCluster database with a few examples
| Search entity | Example of search |
| Protein coding gene | TP53, RB1, ERBB2, etc. |
| UniProt accession | P04637, P06400, P04626, etc. |
| miRNA id | hsa-miR-21, hsa-miR-106a, |
| hsa-miR-200b, etc. | |
| Metabolite name | palmitic acid, Lysine, |
| S-Adenosylmethionine, etc. | |
| Chromosome/ | chr5q32-q34, chr12q13, chr22q11.23 |
| Methylation site | |
| MedLine id (PMID) | 24476821, 18840072, etc. |
N.B. A combination of query terms can also be used. For e.g. “TP53 and 24476821”, would provide results for the protein-coding gene TP53 that was reported in the specific article 24476821.
Table 2
List of molecular pathways displayed for survivin. Muscle invasive bladder cancer pathways that were enriched for survivin have been listed and sorted based on the p-value
| Enriched pathway | p-value | MIBC proteins* | Pathway source | Previous reports in MIBC |
| Signal transduction | 2.46 10 - 14 | 153 | Reactome:111102 | Available [64] |
| Hippo signaling pathway | 4.93 10 - 09 | 27 | Kegg:04390 | Novel |
| Cell cycle | 2.55 10 - 04 | 45 | Reactome:115566 | Available [65] |
*Number of proteins from the 435 entries forming the MIBC interactome enriched in the pathway.



