A combined approach of surface passivation and specific immobilization to study biomolecules by ATR-FTIR spectroscopy1
Abstract
Attenuated total reflection Fourier-transform infrared (ATR-FTIR) spectroscopy is a surface-sensitive and label-free technique, which is applied to obtain dynamic structural information of biomolecules. The study of proteins by ATR-FTIR spectroscopy can be impeded by their tendency to adsorb to solid surfaces. Furthermore, the adsorption process of proteins is often accompanied with conformational changes, which can interfere with the intended structural analysis. We efficiently modified a silicon ATR crystal surface with polyethylene glycol and thereby create a protein-repellent surface. To achieve a high sensitivity, which enables the study of small conformational changes of biomolecules, we combine surface passivation with specific immobilization. This is accomplished via the biotin-streptavidin interaction, which is one of the strongest known non-covalent protein-ligand interactions. As a proof of concept we present the specific immobilization of DNA. The modified surface is stable against elevated temperatures and 8 M urea and can therefore be used to study a wide range of biochemical systems and reactions. The surface chemistry is simple and performed under mild conditions, which leads to a high applicability of the presented approach.
1.Introduction
Thousands of protein structures have been determined to date, which fundamentally improved our understanding of biochemical and cellular processes. Yet, most often those structures give only static snapshots and lack information on structural dynamics. Infrared (IR) spectroscopy is a label-free method, which reveals dynamic structural information of biomolecules and proteins including their protonation state, hydrogen bonding and orientation. However, the strong absorption of water in the amide I region (1600–1700 cm−1), which is sensitive to protein conformations, makes IR-studies of proteins in aqueous solutions challenging. To achieve reliable signals by transmission IR spectroscopy, high protein concentrations (>3 mg/ml) in combination with very short optical pathlengths (<10 μm) are usually required [32]. The reproducible and stable preparation of those short optical pathlengths is difficult, which makes the subtraction of water from the spectrum of the biomolecule potentially error-prone. Attenuated total reflection Fourier-transform IR (ATR-FTIR) spectroscopy circumvents those issues and represents the method of choice for protein analysis in aqueous solutions. The small penetration depth of the IR beam at the ATR crystal surface into the sample results in a very defined and simultaneously very short optical pathlength (in the order of <1 μm for silicon). Signal-to-noise and sensitivity can be enhanced by the application of multireflection-ATR modules, which increase the effective optical pathlength depending on the number of internal reflections. However, the tendency of proteins to adsorb to plane surfaces can impair the study of proteins by ATR-FTIR spectroscopy. Adsorption of proteins is a complex process involving van der Waals, hydrophobic and electrostatic interactions, and hydrogen bonding. Moreover, this process often leads to structural rearrangements within proteins [16,27], which might interfere with their actual analysis. One of the best known approaches to prevent protein adsorption is the use of polyethylene glycol (PEG) [3,9,11,17]. It is a water-soluble, non-toxic and also non-immunogenic polymer. It was shown that surfaces modified with PEG exhibit protein repellent characteristics [2,5,19,26,29]. An established method to modify silicon surfaces is the use of alkylsilanes, which form bonds between the oxidized silicon surface and the hydrolyzed organosilane molecule [13,30]. Those self-assembled monolayers (SAMs) are highly ordered two-dimensional structures, that form spontaneously on a variety of surfaces [25]. Besides using SAMs to passivate surfaces, they can also be used to immobilize proteins and also other biomolecules specifically at the crystal surface and thereby enhance the local surface concentration [8,14,15,23,24,28]. Thus, good signal-to-noise ratios are achieved while only low sample concentrations are required for measurements.
Fig. 1.
Schematic representation of the ATR-FTIR approach. The silicon (Si) crystal surface is modified with biotin-PEG-silane linkers to prevent unspecific protein adsorption and to enable specific immobilization of biotinylated biomolecules, such as proteins, nucleic acids and ligands via streptavidin.
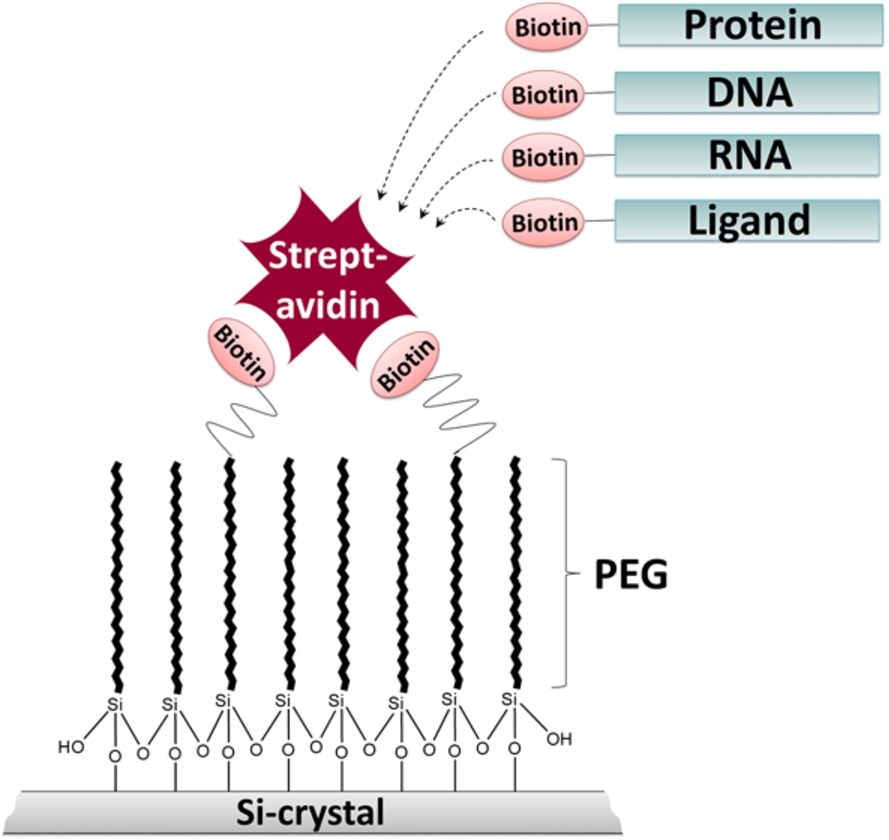
We developed an ATR-FTIR spectroscopic approach which combines a protein-repellent surface and site-specific immobilization of biomolecules (Fig. 1). We demonstrate that the surface modification with PEG-silane linkers efficiently prevents the adsorption of BSA. By using PEG-linkers modified with biotin at their headgroups, streptavidin can be specifically immobilized at the passivated surface. The tetrameric streptavidin itself can serve as a bridge for the specific immobilization of other biotinylated biomolecules. Here, we demonstrate the efficient immobilization of biotinylated DNA via streptavidin at the ATR crystal. Immobilization strongly increased signal intensities due to higher local DNA concentrations at the surface. The streptavidin-biotin interaction is one of the strongest known non-covalent protein-ligand interactions (Kd value of
2.Materials and methods
2.1.ATR-FTIR spectroscopy
ATR-FTIR measurements were performed with a Vertex 70V spectrometer equipped with a BioATR cell II (Bruker). Sample temperature was controlled by an external water bath and set to 20°C. The spectral resolution was 4 cm−1 and each spectrum was an average of 100 scans. The cell was equipped with a multi-reflection silicon crystal. Before each measurement the sample was equilibrated for 20 min.
2.2.Surface passivation of the ATR crystal
To modify the crystal with α-Silane-ω-biotinyl polyethylene glycol (5 kDa, Rapp Polymere), the surface was pretreated two times for 5 min with concentrated H2SO4 followed by 5 min treatment with H2O2: H2SO4 (9:1). Each step was followed by rinsing the surface with water and drying it under a nitrogen stream. Subsequently, the crystal was heated to 50°C. 20 mg/ml PEG-silane in 30 mM sodium acetate pH 5.5 were loaded on the crystal and incubated for 30 min at 50°C. Then, the temperature was set to 20°C. While the crystal was cooling down, the PEG-silane solution was allowed to dry on the surface to achieve condensation of the silanol groups. The surface was washed thoroughly with MilliQ water until the PEG signal was stable. The modified surface was incubated in MilliQ water over night to remove all non-covalently attached PEG-silane molecules. The next day, the MilliQ water was exchanged to the desired buffer and the crystal surface was ready for measurements.
2.3.Adsorption of BSA
The adsorption behavior of BSA was tested by incubating 25 μl of 5 mg/ml BSA solution (Roche) in MilliQ water on the blank or passivated crystal surface for 20 min. Afterwards, the crystal was washed thoroughly several times with MilliQ water to remove unbound protein.
2.4.Immobilization of streptavidin
To immobilize streptavidin on the biotinylated surface, 20 μl of 2.5 μM streptavidin (Sigma) in Tris buffer (50 mM Tris pH 7.4; 150 mM NaCl) were incubated on the modified surface for 20 min. After incubation, non-bound protein was removed by washing several times with Tris buffer.
2.5.Stability of the modified ATR crystal surface
The temperature stability of the modified surface including immobilized streptavidin was analyzed by increasing gradually the temperature of the ATR cell to 45°C, keeping it at this temperature for 10 min and cooling it down to 20°C again. The stability against urea treatment was tested by incubating immobilized streptavidin in 8 M urea for 20 min and washing it afterwards thoroughly with Tris buffer.
2.6.Immobilization of biotinylated DNA
Biotinylated double stranded DNA (Sigma: (Btn)5′-CGAGGAACATGTCCCAACATGTTGCTCGAG-3′; 5′-CTCGAGCAACATGTTGGGACATGTTCCTCG-3′) was immobilized via a streptavidin bridge at the biotinylated surface. Therefore, 50 pmol of streptavidin and 100 pmol of DNA were pre-incubated in a total volume of 15 μl (50 mM Tris pH 7.4; 150 mM NaCl) at room temperature for 10 min and then applied to the biotinylated surface. After 20 min of incubation, the surface was washed several times with Tris buffer to remove unbound streptavidin/DNA. To achieve a maximal level of immobilized DNA, this procedure was repeated once. The spectrum of free DNA was recorded by incubating 20 μl of 60 μM double stranded DNA (Sigma: 5′-CGAGGAACATGTCCCAACATGTTGCTCGAG-3′; 5′-CTCGAG CAACATGTTGGGACATGTTCCTCG-3′) in Tris buffer for 20 min on the blank crystal surface.
3.Results and discussion
3.1.Surface passivation of the ATR crystal
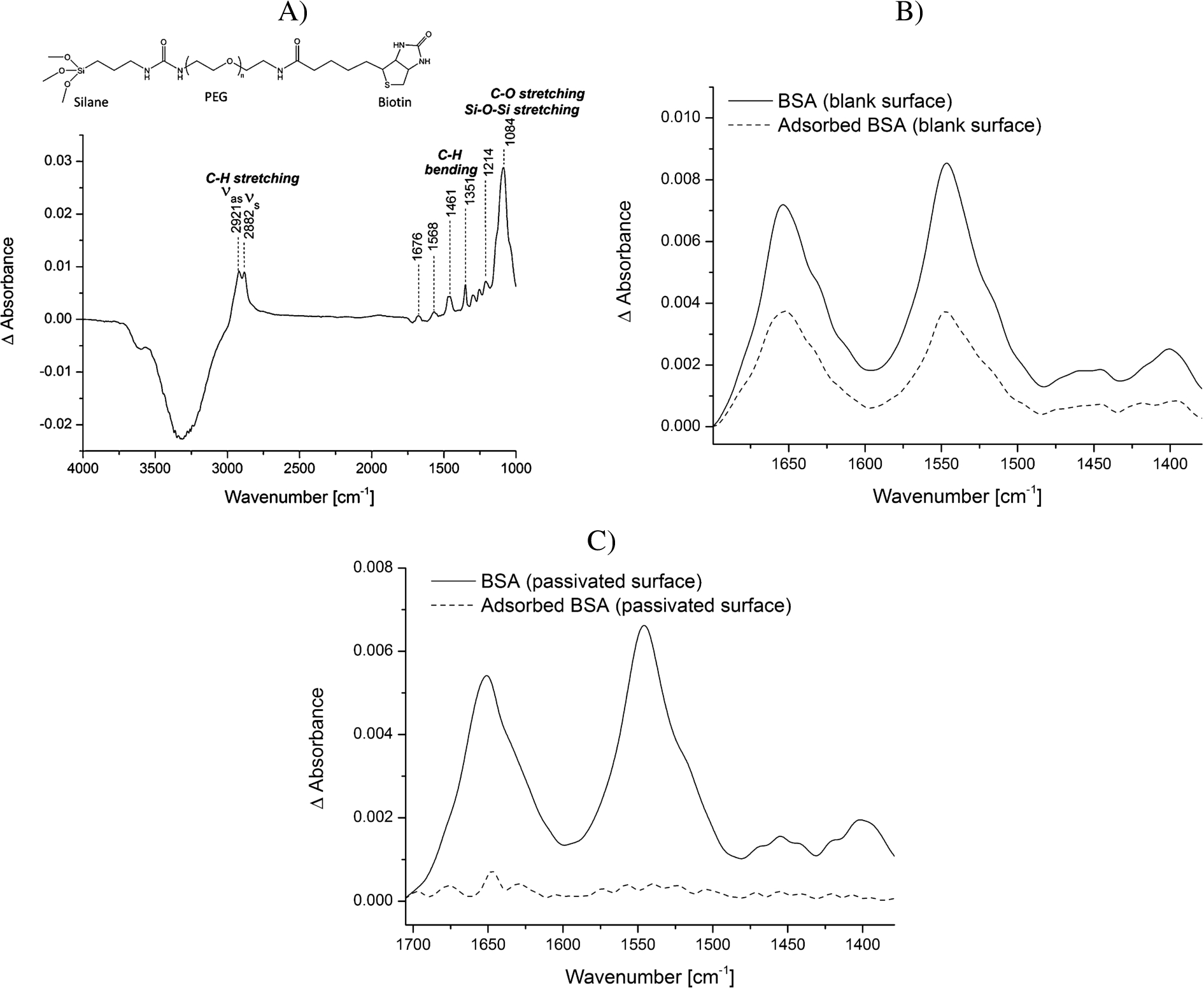
Protein adsorption is a common phenomenon, which is often accompanied with structural rearrangements of proteins. Small conformational changes of proteins, occurring for example upon binding reactions, can only be studied by ATR-FTIR spectroscopy if the adsorption process to the ATR crystal surface is prevented. To passivate the silicon surface of the ATR crystal, the surface was modified using PEG-silane linkers. Many protocols exist for silanization of surfaces. However, many of them use harsh conditions, which might damage the ATR crystal used as optical internal reflection element or which are not applicable as many crystals are permanently installed in the ATR cell. The method we developed was modified from Kumar et al. [21]. They show the efficient immobilization of silanized nucleic acids on glass slides for the generation of DNA microarrays under mild conditions. We demonstrate that similar mild conditions can be applied to modify the silicon ATR crystal with silanized PEG linkers. While Kumar et al. [21] could only indirectly determine the efficiency of immobilization via fluorescence, we could directly follow all surface modification steps by characteristic vibrational modes in the IR spectrum. First, the surface was washed with a mixture of hydrogen peroxide and sulfuric acid to clean the surface and maximize the number of silanol groups at the surface. We tested silanization at room temperature as well as at 50°C and observed that silanization at higher temperature strongly increased the stability of the silane layer. While the IR bands of the PEG-silane layer produced at room temperature gradually decreased over time and were no longer detectable after 12 h, the spectrum of a silane layer produced at 50°C remained stable over this time period (data not shown). Using high temperatures to achieve a stable silane-modified surface is in line with other work [1] and probably due to an increased nucleation process of the silane linkers, which leads to a more homogeneously organized layer. After the incubation step at 50°C, a condensation step followed during which the PEG-silane solution is allowed to dry while the crystal is cooling to room temperature. This achieves a condensation of the silanol groups. The spectrum of the modified crystal surface is shown in Fig. 2A. Water is displaced from the surface by the PEG-silane linkers resulting in negative water bands (e.g. at 3320 cm−1). The spectrum is characterized by the vibrations of the PEG chains including the anti-symmetric and symmetric C–H stretching vibrations (2921 and 2882 cm−1) and the C–H bending vibrations (1461 and 1351 cm−1) [7]. The region from 1000 to 1250 cm−1 is dominated by a combination of the Si–O–Si vibrational modes from the silane groups and the C–O stretching vibration of the PEG chain [1,6]. Moreover, the amide groups within the biotin-PEG-silane molecule (Fig. 2A) absorb at 1676 and 1568 cm−1. We tested the modified surface with regards to unspecific protein adsorption. BSA was used as a model protein as its adsorption behavior to surfaces is a well-known and also intensively studied process [18,20]. We demonstrate that while BSA strongly adsorbed to the plane crystal surface, this could completely be abrogated via the PEG layer (Fig. 2B and C). Notably, the amide I/amide II intensity ratio differs in Fig. 2B before and after rinsing. This variation is likely caused by the concentration-dependent nonproportionality of the observed absorbance and spectral distortions in ATR measurements [10].
Fig. 2.
Analysis of the adsorption of BSA to the blank vs. the PEG-silane passivated ATR crystal surface. A) Structural formula of the biotin-PEG-silane linker and a representative spectrum of the biotin-PEG-silane modified surface in water. The reference spectrum is water. Typical vibrational modes are highlighted. B) BSA (5 mg/ml) was incubated for 20 min on the blank surface. Spectra were recorded before (continuous line) and after (dotted line) washing the surface several times with water. The reference spectrum is water. C) BSA (5 mg/ml) was incubated for 20 min on the PEG-silane modified surface. Spectra were recorded before (continuous line) and after (dotted line) washing the surface several times with water. The reference spectrum is the PEG-modified surface in water.
3.2.Immobilization of streptavidin
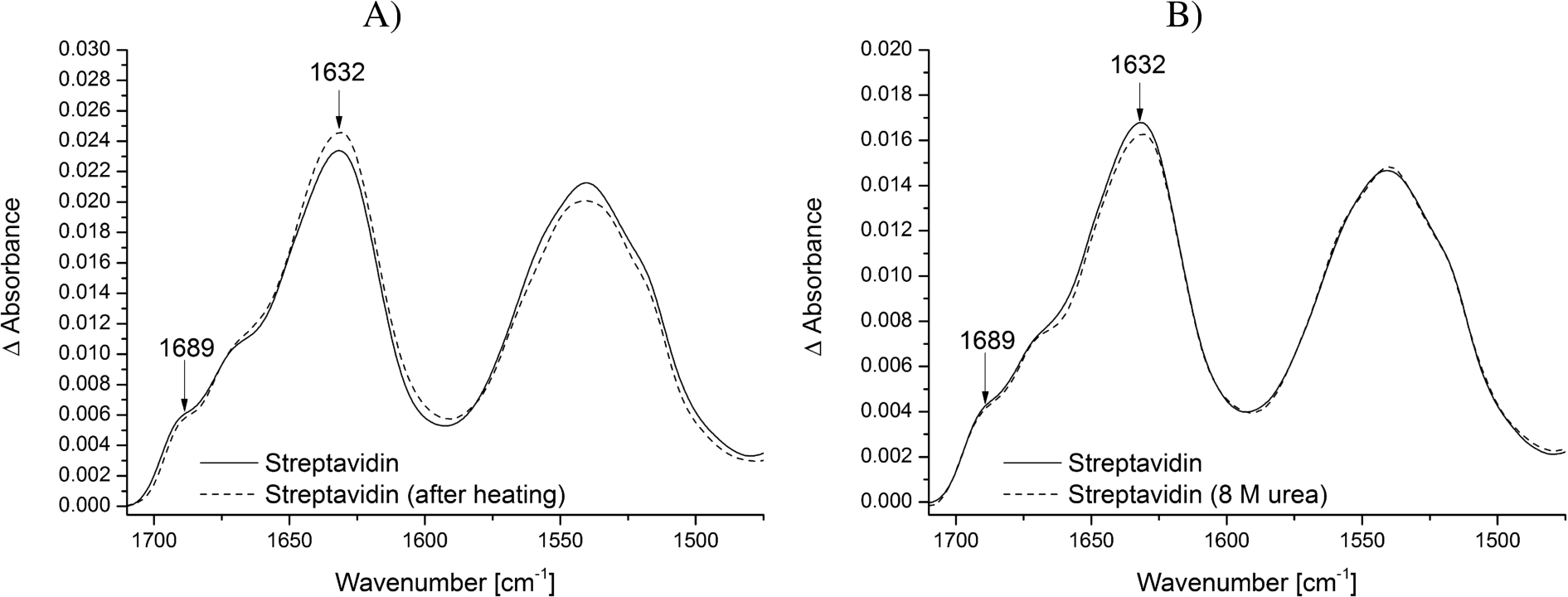
By modifying the surface with biotin-PEG-silane groups the biotin headgroups can be used to specifically immobilize streptavidin at the surface. The tetrameric streptavidin itself can then be employed to immobilize specifically other biotinylated biomolecules such as proteins, DNA or RNA at the surface (Fig. 1). This increases the local surface concentration of the studied biomolecule and thereby improves the signal-to-noise ratio while making it possible to use low concentrations (in the low μM range). The addition of streptavidin resulted in a strong protein signal, which remained constant after several washing steps, proving the efficient immobilization via the biotin headgroups at the crystal surface (Fig. 3A and B). A streptavidin monomer is composed of eight antiparallel β-strands, which fold into a β-barrel. The amide bands at 1689 and 1632 cm−1 are characteristic for this β-sheet rich secondary structure.
Fig. 3.
Stability of the modified ATR crystal surface at different temperatures and biochemical conditions. Streptavidin was immobilized on the biotin-PEG-silane modified surface and tested for its stability. The reference spectrum is the PEG-silane modified surface. The typical vibrational modes of β-sheets of streptavidin are highlighted. A) Spectrum of immobilized streptavidin before (continuous line) and after heating the sample to 45°C (dotted line). B) Spectrum of immobilized streptavidin before (continuous line) and after incubation in 8 M urea (dotted line).
3.3.Stability of the modified ATR crystal surface
The combination of surface passivation and specific immobilization aims to study small conformational changes of proteins and other biomolecules under native conditions. Only if the modified ATR crystal surface including immobilized streptavidin does not change during the measurements, conclusions about structural changes of the studied biomolecule can be drawn. Therefore, we tested the modified ATR crystal surface including immobilized streptavidin for its stability regarding temperature and denaturing buffer additives. Neither the increase of temperature to 45°C nor the addition of 8 M urea affected the structure and intensity of the immobilized streptavidin (Fig. 3A and B). This is in line with other work showing that the streptavidin-biotin interaction is unaffected by extremes of pH, temperature and denaturants [12,22]. Therefore, it can be assumed that the spectrum of the surface modification including streptavidin does not interfere with the spectrum of the biomolecule of interest, even under harsh conditions.
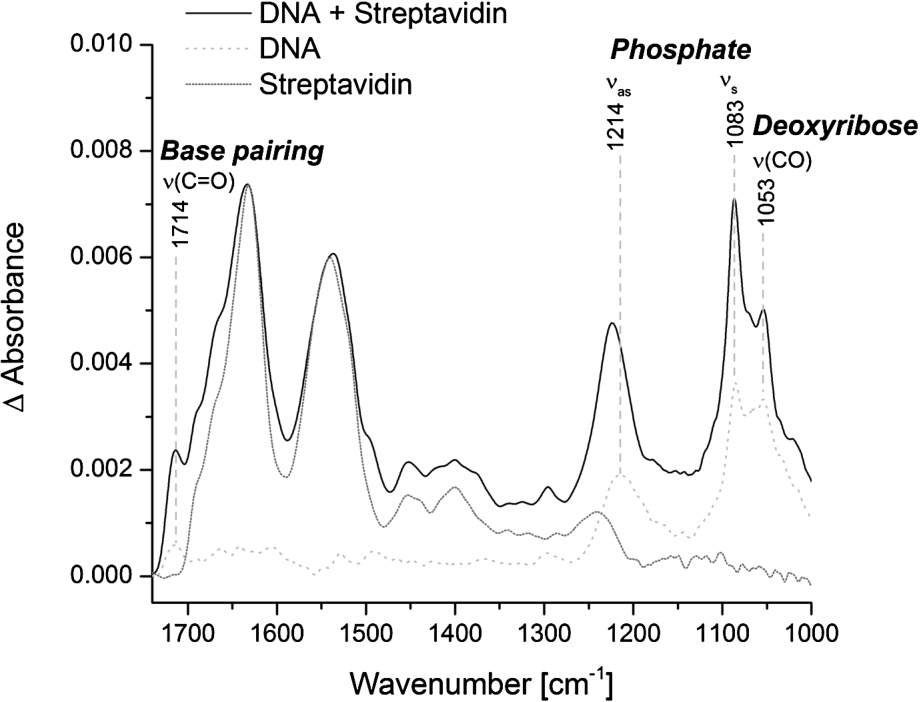
Fig. 4.
Specific immobilization of biotinylated double-stranded DNA at the ATR crystal. Representative spectra of immobilized streptavidin alone (dotted line) and in combination with biotinylated double-stranded DNA (solid line) are shown. The reference spectrum is the PEG-silane modified surface. The spectrum of streptavidin alone is normalized to the intensity of the amide I band of the combined spectrum of DNA and streptavidin. The spectrum of free DNA was measured on the blank crystal (dashed line). Typical vibrational modes of DNA are highlighted.
3.4.Immobilization of biotinylated DNA
The specific immobilization of biomolecules at the ATR crystal increases the local surface concentration and thereby the signal-to-noise ratio. We tested the specific immobilization of biotinylated double-stranded DNA via streptavidin at the PEG modified surface. Surprisingly, the addition of biotinylated DNA to the immobilized streptavidin did not result in a significant DNA signal. Only the pre-incubation of biotinylated DNA with a distinct amount of streptavidin and the subsequent addition to the PEG-biotin modified surface resulted in a distinct and stable DNA signal (Fig. 4). This is probably due to the very strong binding affinity of biotin and streptavidin. As soon as streptavidin alone is added to the PEG-biotin modified surface, all four biotin binding sites of streptavidin are bound to biotin and thereby prevent the additional binding of biotinylated DNA. In contrast, when the biotinylated DNA is pre-incubated with a distinct amount of streptavidin, some biotin-binding sites within the streptavidin tetramer are bound to the biotinylated DNA and some remain free. Those free biotin-binding sites can subsequently bind to the PEG-biotin modified surface and result in the specific immobilization of DNA. The best DNA signal could be achieved by pre-incubating biotinylated DNA and streptavidin tetramers in a 2:1 ratio (Fig. 4). The spectrum of DNA is dominated by the anti-symmetric (1214 cm−1) and symmetric (1083 cm−1) phosphate vibrations. Other DNA characteristic bands are the carbonyl stretching vibration at 1714 cm−1, which is attributed to nucleic bases with strong base pairing and stacking interactions, and the C–O stretching vibration of the backbone at 1053 cm−1 [4,31]. The IR band intensities of immobilized DNA were significantly stronger in comparison to DNA, which was not immobilized, even though the applied concentration of DNA was approximately ten times lower (6.7 μM vs. 60 μM). Therefore, the presented method of immobilization efficiently enhances the local surface concentration resulting in good signal-to-noise ratios with low sample concentrations. The pre-incubation step of the biotinylated biomolecule and streptavidin results in an IR spectrum with contributions of both, streptavidin and the biomolecule of interest. But as we have shown that the immobilized streptavidin resists elevated temperatures and treatment with denaturants (Fig. 3), it can be assumed that it does not undergo structural changes during measurements. Therefore, the presented method of surface immobilization has still a universal applicability to unravel conformational changes of biomolecules in the context of biochemical reactions.
4.Conclusions
The here presented ATR-FTIR approach has many potential biological and biochemical applications and can be used for a variety of biomolecules studied in a range of different conditions. The efficient passivation of the ATR crystal surface with PEG prevents unspecific adsorption of proteins, which is a common problem of surface-sensitive techniques. The direct and specific immobilization of biomolecules via the streptavidin-biotin interaction minimizes the required sample amount and at the same time increases the signal-to-noise ratio. Besides the direct immobilization of biotinylated biomolecules, the method can be adapted to immobilize proteins indirectly via biotin-NTA or biotinylated antibodies. The versatile availability of biotinylated molecules as well as the easy process of biotinylating biomolecules enable the here presented approach to become an universal tool for monitoring a wide spectrum of biochemical processes via ATR-FTIR spectroscopy.
Acknowledgements
This work was supported by the German Research Foundation through the Konstanz Research School Chemical Biology (KoRS-CB), SFB 1214 (A3) and SFB 969 (A2).
References
[1] | N. Aissaoui, L. Bergaoui, J. Landoulsi, J.-F. Lambert et al., Silane layers on silicon surfaces: Mechanism of interaction, stability, and influence on protein adsorption, Langmuir 28: (1) ((2012) ), 656–665. doi:10.1021/la2036778. |
[2] | N.A. Alcantar, E.S. Aydil and J.N. Israelachvili, Polyethylene glycol-coated biocompatible surfaces, J Biomed Mater Res 51: (3) ((2000) ), 343–351. doi:10.1002/1097-4636(20000905)51:3<343::AID-JBM7>3.0.CO;2-D. |
[3] | J.D. Andrade, V. Hlady and S.I. Jeon, Poly(ethylene oxide) and protein resistance, in: Hydrophilic Polymers, American Chemical Society, (1996) , pp. 51–59. doi:10.1021/ba-1996-0248.ch003. |
[4] | M. Banyay, M. Sarkar and A. Gräslund, A library of IR bands of nucleic acids in solution, Biophysical Chemistry 104: (2) ((2003) ), 477–488. doi:10.1016/S0301-4622(03)00035-8. |
[5] | H. Chen, Z. Zhang, Y. Chen, M.A. Brook et al., Protein repellant silicone surfaces by covalent immobilization of poly(ethylene oxide), Biomaterials 26: (15) ((2005) ), 2391–2399. doi:10.1016/j.biomaterials.2004.07.068. |
[6] | B.W. Chieng, N. Ibrahim, W. Yunus and M. Hussein, Effects of graphene nanopletelets on poly(lactic acid)/poly(ethylene glycol) polymer nanocomposites, 6: ((2013) ), 93–104. |
[7] | L.A. Chrisey, G.U. Lee and C.E. O’Ferrall, Covalent attachment of synthetic DNA to self-assembled monolayer films, Nucleic Acids Research 24: (15) ((1996) ), 3031–3039. doi:10.1093/nar/24.15.3031. |
[8] | M.A. Fallah, C. Stanglmair, C. Pacholski and K. Hauser, Devising self-assembled-monolayers for surface-enhanced infrared spectroscopy of pH-driven poly-l-lysine conformational changes, Langmuir 32: (29) ((2016) ), 7356–7364. doi:10.1021/acs.langmuir.6b01742. |
[9] | C.-G. Gölander, J.N. Herron, K. Lim, P. Claesson et al., Properties of immobilized PEG films and the interaction with proteins, in: Poly(Ethylene Glycol) Chemistry: Biotechnical and Biomedical Applications, J.M. Harris, ed., Springer US, Boston, MA, (1992) , pp. 221–245. doi:10.1007/978-1-4899-0703-5_15. |
[10] | M.E. Goldberg and A.F. Chaffotte, Undistorted structural analysis of soluble proteins by attenuated total reflectance infrared spectroscopy, Protein Sci 14: (11) ((2005) ), 2781–2792. doi:10.1110/ps.051678205. |
[11] | W.R. Gombotz, W. Guanghui, T.A. Horbett and A.S. Hoffman, Protein adsorption to poly(ethylene oxide) surfaces, Journal of Biomedical Materials Research 25: (12) ((1991) ), 1547–1562. doi:10.1002/jbm.820251211. |
[12] | M. González, C.E. Argaraña and G.D. Fidelio, Extremely high thermal stability of streptavidin and avidin upon biotin binding, Biomolecular Engineering 16: (1) ((1999) ), 67–72. doi:10.1016/S1050-3862(99)00041-8. |
[13] | J.J. Gooding and S. Ciampi, The molecular level modification of surfaces: From self-assembled monolayers to complex molecular assemblies, Chemical Society Reviews 40: (5) ((2011) ), 2704–2718. doi:10.1039/c0cs00139b. |
[14] | A.C. Gouget-Laemmel, J. Yang, M.A. Lodhi, A. Siriwardena et al., Functionalization of azide-terminated silicon surfaces with glycans using click chemistry: XPS and FTIR study, The Journal of Physical Chemistry C 117: (1) ((2013) ), 368–375. |
[15] | K. Henke, W. Welte and K. Hauser, Direct monitoring of β-sheet formation in the outer membrane protein TtoA assisted by TtOmp85, Biochemistry 55: (31) ((2016) ), 4333–4343. doi:10.1021/acs.biochem.6b00691. |
[16] | V. Hlady and J. Buijs, Protein adsorption on solid surfaces, Current Opinion in Biotechnology 7: (1) ((1996) ), 72–77. doi:10.1016/S0958-1669(96)80098-X. |
[17] | K. Holmberg, K. Bergström and M.-B. Stark, Immobilization of proteins via PEG chains, in: Poly(Ethylene Glycol) Chemistry: Biotechnical and Biomedical Applications, J.M. Harris, ed., Springer US, Boston, MA, (1992) , pp. 303–324. doi:10.1007/978-1-4899-0703-5_19. |
[18] | Y.L. Jeyachandran, E. Mielczarski, B. Rai and J.A. Mielczarski, Quantitative and qualitative evaluation of adsorption/desorption of bovine serum albumin on hydrophilic and hydrophobic surfaces, Langmuir 25: (19) ((2009) ), 11614–11620. doi:10.1021/la901453a. |
[19] | P. Kingshott and H.J. Griesser, Surfaces that resist bioadhesion, Current Opinion in Solid State and Materials Science 4: (4) ((1999) ), 403–412. |
[20] | K. Kubiak-Ossowska, K. Tokarczyk, B. Jachimska and P.A. Mulheran, Bovine serum albumin adsorption at a silica surface explored by simulation and experiment, The Journal of Physical Chemistry B 121: (16) ((2017) ), 3975–3986. doi:10.1021/acs.jpcb.7b01637. |
[21] | A. Kumar, O. Larsson, D. Parodi and Z. Liang, Silanized nucleic acids: A general platform for DNA immobilization, Nucleic Acids Research 28: (14) ((2000) ), e71. doi:10.1093/nar/28.14.e71. |
[22] | G.P. Kurzban, E.A. Bayer, M. Wilchek and P.M. Horowitz, The quaternary structure of streptavidin in urea, J Biol Chem 266: (22) ((1991) ), 14470–14477. |
[23] | N.A. Lapin and Y.J. Chabal, Infrared characterization of biotinylated silicon oxide surfaces, surface stability and specific attachment of streptavidin, The Journal of Physical Chemistry B 113: (25) ((2009) ), 8776–8783. |
[24] | W. Liao, F. Wei, D. Liu, M.X. Qian et al., FTIR-ATR detection of proteins and small molecules through DNA conjugation, Sensors and Actuators B: Chemical 114: (1) ((2006) ), 445–450. doi:10.1016/j.snb.2005.06.021. |
[25] | S. Onclin, B.J. Ravoo and D.N. Reinhoudt, Engineering silicon oxide surfaces using self-assembled monolayers, Angewandte Chemie International Edition 44: (39) ((2005) ), 6282–6304. doi:10.1002/anie.200500633. |
[26] | K.L. Prime and G.M. Whitesides, Adsorption of proteins onto surfaces containing end-attached oligo(ethylene oxide): A model system using self-assembled monolayers, Journal of the American Chemical Society 115: (23) ((1993) ), 10714–10721. doi:10.1021/ja00076a032. |
[27] | M. Rabe, D. Verdes and S. Seeger, Understanding protein adsorption phenomena at solid surfaces, Advances in Colloid and Interface Science 162: (1) ((2011) ), 87–106. doi:10.1016/j.cis.2010.12.007. |
[28] | J. Schartner, J. Güldenhaupt, B. Mei, M. Rögner et al., Universal method for protein immobilization on chemically functionalized germanium investigated by ATR-FTIR difference spectroscopy, Journal of the American Chemical Society 135: (10) ((2013) ), 4079–4087. doi:10.1021/ja400253p. |
[29] | S.J. Sofia, V. Premnath and E.W. Merrill, Poly(ethylene oxide) grafted to silicon surfaces: Grafting density and protein adsorption, Macromolecules 31: (15) ((1998) ), 5059–5070. doi:10.1021/ma971016l. |
[30] | A. Ulman, Part three – Self–assembled monolayers, in: An Introduction to Ultrathin Organic Films, Academic Press, San Diego, (1991) , pp. 237–304. doi:10.1016/B978-0-08-092631-5.50010-5. |
[31] | D.R. Whelan, K.R. Bambery, P. Heraud, M.J. Tobin et al., Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy, Nucleic Acids Research 39: (13) ((2011) ), 5439–5448. doi:10.1093/nar/gkr175. |
[32] | H. Yang, S. Yang, J. Kong, A. Dong et al., Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy, Nat. Protocols 10: (3) ((2015) ), 382–396. doi:10.1038/nprot.2015.024. |



